Gut Microbiotas, Plasma Metabolites, and Autism Spectrum Disorder: A Bidirectional Mendelian Randomization Analysis
Abstract
1. Introduction
- 1.
- Identified causal links between gut microbiota, plasma metabolites, and ASD using Mendelian Randomization.
- 2.
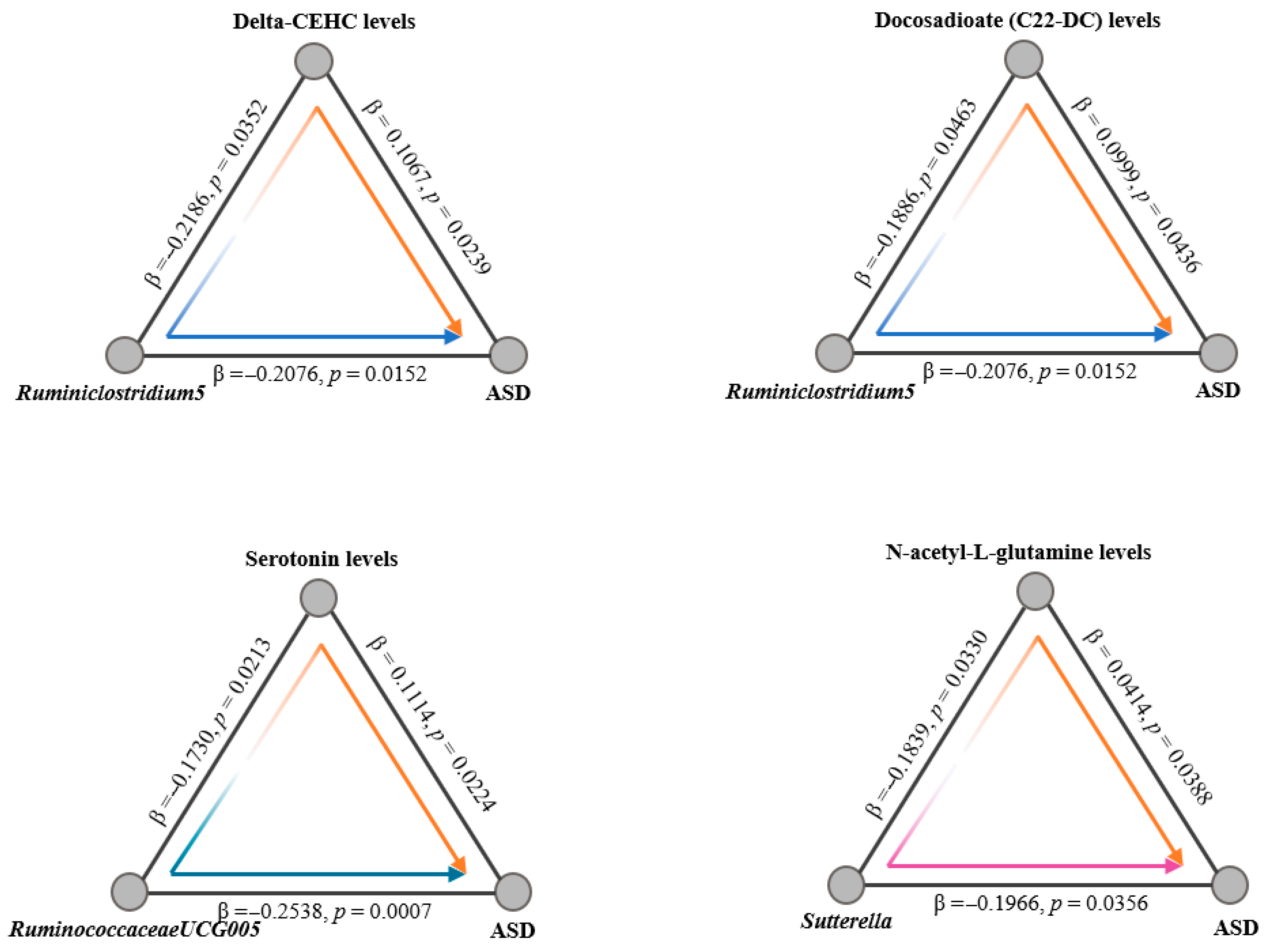
- Revealed key taxa, including Ruminiclostridium5, RuminococcaceaeUCG-005, and Sutterella, influencing ASD through metabolites.
- 3.
- Highlighted metabolites like serotonin, N-acetyl-L-glutamine, Delta-CEHC, and Docosadioate (C22-DC) as mediators in the gut–brain axis.
- 4.
- Proposed diagnostic and therapeutic targets for ASD based on gut microbiota and metabolites.
2. Method
2.1. Study Design Overview
2.2. Data Sources and Instruments
2.2.1. Gut Microbiota
2.2.2. Plasma Metabolome
2.2.3. ASD
2.2.4. Statistical Analysis
2.2.5. Genetic Correlation Analysis
2.2.6. Two-Sample MR
2.2.7. Median MR
3. Result
3.1. Cause Effects of Gut Microbiota on ASD
3.1.1. LDSC Analysis
3.1.2. Two-Sample MR and Sensitivity Analysis
3.1.3. Reverse MR Analysis
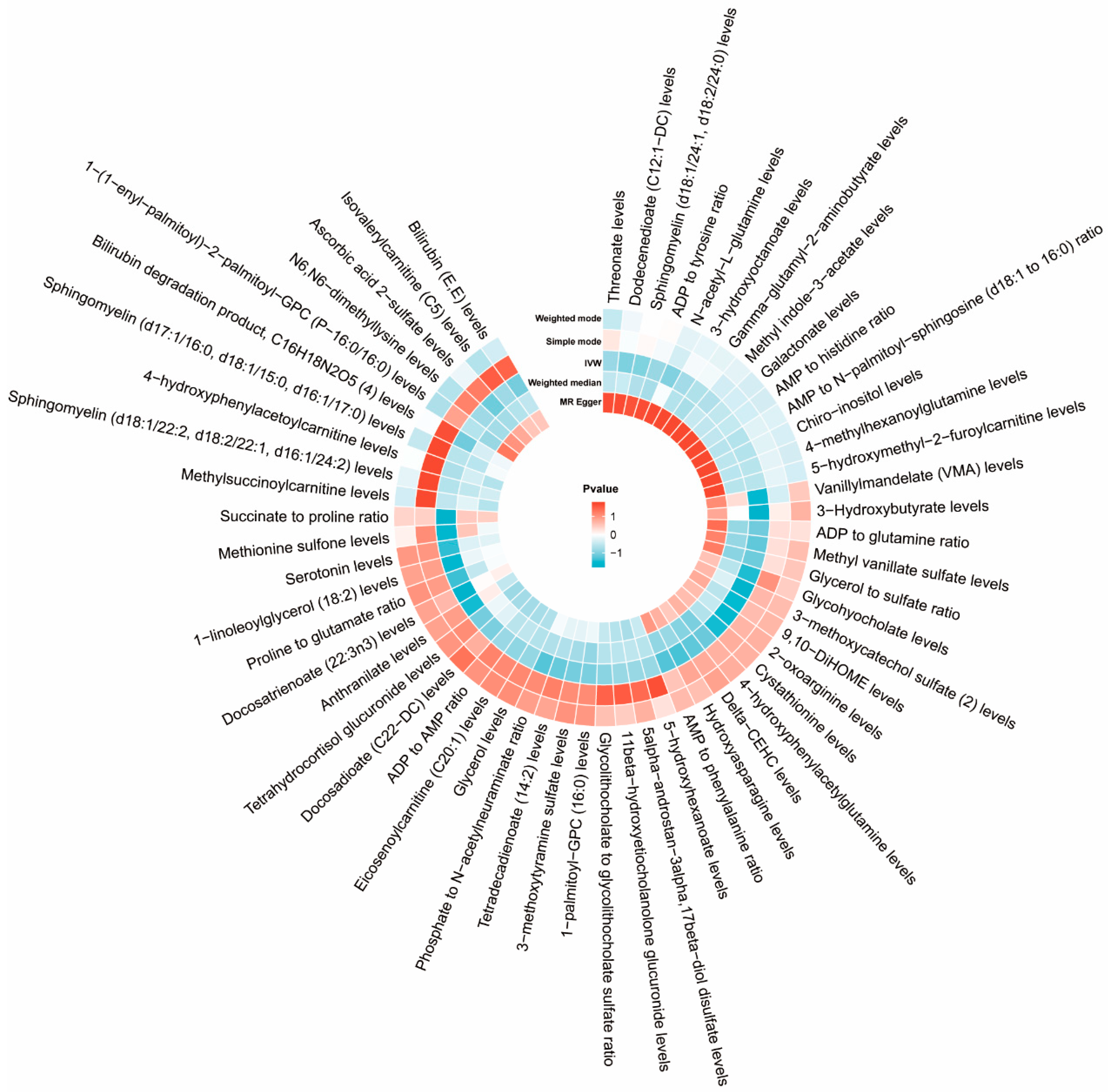
3.2. Cause Effects of Plasma Metabolites on ASD
3.2.1. LDSC Analysis
3.2.2. Two-Sample MR and Sensitivity Analysis
3.2.3. Reverse MR Analysis
3.3. Causal Effects of Cardiometabolic Diseases on Gut-Dependent Metabolites
Median MR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lord, C.; Elsabbagh, M.; Baird, G.; Veenstra-Vanderweele, J. Autism spectrum disorder. Lancet 2018, 392, 508–520. [Google Scholar] [CrossRef]
- Li, Q.; Li, Y.; Liu, B.; Chen, Q.; Xing, X.; Xu, G.; Yang, W. Prevalence of Autism Spectrum Disorder Among Children and Adolescents in the United States From 2019 to 2020. JAMA Pediatr. 2022, 176, 943–945. [Google Scholar] [CrossRef] [PubMed]
- Zwaigenbaum, L.; Bauman, M.L.; Choueiri, R.; Fein, D.; Kasari, C.; Pierce, K.; Stone, W.L.; Yirmiya, N.; Estes, A.; Hansen, R.L.; et al. Early Identification and Interventions for Autism Spectrum Disorder: Executive Summary. Pediatrics 2015, 136 (Suppl. 1), S1–S9. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Deister, C.A.; Gao, X.; Guo, B.; Lynn-Jones, T.; Chen, N.; Wells, M.F.; Liu, R.; Goard, M.J.; Dimidschstein, J.; et al. Dysfunction of cortical GABAergic neurons leads to sensory hyper-reactivity in a Shank3 mouse model of ASD. Nat. Neurosci. 2020, 23, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Penagarikano, O.; Abrahams, B.S.; Herman, E.I.; Winden, K.D.; Gdalyahu, A.; Dong, H.; Sonnenblick, L.I.; Gruver, R.; Almajano, J.; Bragin, A.; et al. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell 2011, 147, 235–246. [Google Scholar] [CrossRef]
- Imbriani, G.; Panico, A.; Grassi, T.; Idolo, A.; Serio, F.; Bagordo, F.; De Filippis, G.; De Giorgi, D.; Antonucci, G.; Piscitelli, P.; et al. Early-Life Exposure to Environmental Air Pollution and Autism Spectrum Disorder: A Review of Available Evidence. Int. J. Environ. Res. Public Health 2021, 18, 1204. [Google Scholar] [CrossRef]
- Love, C.; Sominsky, L.; O’Hely, M.; Berk, M.; Vuillermin, P.; Dawson, S.L. Prenatal environmental risk factors for autism spectrum disorder and their potential mechanisms. BMC Med. 2024, 22, 393. [Google Scholar] [CrossRef]
- Tan, M.; Yang, T.; Zhu, J.; Li, Q.; Lai, X.; Li, Y.; Tang, T.; Chen, J.; Li, T. Maternal folic acid and micronutrient supplementation is associated with vitamin levels and symptoms in children with autism spectrum disorders. Reprod. Toxicol. 2020, 91, 109–115. [Google Scholar] [CrossRef]
- Young, V.B. The role of the microbiome in human health and disease: An introduction for clinicians. BMJ 2017, 356, j831. [Google Scholar] [CrossRef]
- Dan, Z.; Mao, X.; Liu, Q.; Guo, M.; Zhuang, Y.; Liu, Z.; Chen, K.; Chen, J.; Xu, R.; Tang, J.; et al. Altered gut microbial profile is associated with abnormal metabolism activity of Autism Spectrum Disorder. Gut Microbes 2020, 11, 1246–1267. [Google Scholar] [CrossRef]
- He, J.; Gong, X.; Hu, B.; Lin, L.; Lin, X.; Gong, W.; Zhang, B.; Cao, M.; Xu, Y.; Xia, R.; et al. Altered Gut Microbiota and Short-chain Fatty Acids in Chinese Children with Constipated Autism Spectrum Disorder. Sci. Rep. 2023, 13, 19103. [Google Scholar] [CrossRef] [PubMed]
- Retuerto, M.; Al-Shakhshir, H.; Herrada, J.; McCormick, T.S.; Ghannoum, M.A. Analysis of Gut Bacterial and Fungal Microbiota in Children with Autism Spectrum Disorder and Their Non-Autistic Siblings. Nutrients 2024, 16, 3004. [Google Scholar] [CrossRef]
- Fu, Z.; Yang, X.; Jiang, Y.; Mao, X.; Liu, H.; Yang, Y.; Chen, J.; Chen, Z.; Li, H.; Zhang, X.S.; et al. Microbiota profiling reveals alteration of gut microbial neurotransmitters in a mouse model of autism-associated 16p11.2 microduplication. Front. Microbiol. 2024, 15, 1331130. [Google Scholar] [CrossRef]
- Ahmed, H.; Leyrolle, Q.; Koistinen, V.; Karkkainen, O.; Laye, S.; Delzenne, N.; Hanhineva, K. Microbiota-derived metabolites as drivers of gut-brain communication. Gut Microbes 2022, 14, 2102878. [Google Scholar] [CrossRef]
- Hsiao, E.Y.; McBride, S.W.; Hsien, S.; Sharon, G.; Hyde, E.R.; McCue, T.; Codelli, J.A.; Chow, J.; Reisman, S.E.; Petrosino, J.F.; et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 2013, 155, 1451–1463. [Google Scholar] [CrossRef]
- Al-Ayadhi, L.; Zayed, N.; Bhat, R.S.; Moubayed, N.M.S.; Al-Muammar, M.N.; El-Ansary, A. The use of biomarkers associated with leaky gut as a diagnostic tool for early intervention in autism spectrum disorder: A systematic review. Gut Pathog. 2021, 13, 54. [Google Scholar] [CrossRef]
- Tang, X.; Feng, C.; Zhao, Y.; Zhang, H.; Gao, Y.; Cao, X.; Hong, Q.; Lin, J.; Zhuang, H.; Feng, Y.; et al. A study of genetic heterogeneity in autism spectrum disorders based on plasma proteomic and metabolomic analysis: Multiomics study of autism heterogeneity. MedComm 2023, 4, e380. [Google Scholar] [CrossRef]
- Coll-Tane, M.; Gong, N.N.; Belfer, S.J.; van Renssen, L.V.; Kurtz-Nelson, E.C.; Szuperak, M.; Eidhof, I.; van Reijmersdal, B.; Terwindt, I.; Durkin, J.; et al. The CHD8/CHD7/Kismet family links blood-brain barrier glia and serotonin to ASD-associated sleep defects. Sci. Adv. 2021, 7, eabe2626. [Google Scholar] [CrossRef]
- Gevi, F.; Zolla, L.; Gabriele, S.; Persico, A.M. Urinary metabolomics of young Italian autistic children supports abnormal tryptophan and purine metabolism. Mol. Autism 2016, 7, 47. [Google Scholar] [CrossRef] [PubMed]
- Mariani, J.; Coppola, G.; Zhang, P.; Abyzov, A.; Provini, L.; Tomasini, L.; Amenduni, M.; Szekely, A.; Palejev, D.; Wilson, M.; et al. FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell 2015, 162, 375–390. [Google Scholar] [CrossRef] [PubMed]
- Fung, L.K.; Flores, R.E.; Gu, M.; Sun, K.L.; James, D.; Schuck, R.K.; Jo, B.; Park, J.H.; Lee, B.C.; Jung, J.H.; et al. Thalamic and prefrontal GABA concentrations but not GABA(A) receptor densities are altered in high-functioning adults with autism spectrum disorder. Mol. Psychiatry 2021, 26, 1634–1646. [Google Scholar] [CrossRef] [PubMed]
- Golubeva, A.V.; Joyce, S.A.; Moloney, G.; Burokas, A.; Sherwin, E.; Arboleya, S.; Flynn, I.; Khochanskiy, D.; Moya-Perez, A.; Peterson, V.; et al. Microbiota-related Changes in Bile Acid & Tryptophan Metabolism are Associated with Gastrointestinal Dysfunction in a Mouse Model of Autism. EBioMedicine 2017, 24, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Usui, N.; Iwata, K.; Miyachi, T.; Takagai, S.; Wakusawa, K.; Nara, T.; Tsuchiya, K.J.; Matsumoto, K.; Kurita, D.; Kameno, Y.; et al. VLDL-specific increases of fatty acids in autism spectrum disorder correlate with social interaction. EBioMedicine 2020, 58, 102917. [Google Scholar] [CrossRef] [PubMed]
- Bulik-Sullivan, B.; Finucane, H.K.; Anttila, V.; Gusev, A.; Day, F.R.; Loh, P.R.; Consortium, R.; Psychiatric Genomics Consortium; Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3; Duncan, L.; et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015, 47, 1236–1241. [Google Scholar] [CrossRef]
- Birney, E. Mendelian Randomization. Cold Spring Harb. Perspect. Med. 2022, 12, a041302. [Google Scholar] [CrossRef]
- Strati, F.; Cavalieri, D.; Albanese, D.; De Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Renzi, D.; Calabro, A.; et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 2017, 5, 24. [Google Scholar] [CrossRef]
- Maiuolo, J.; Gliozzi, M.; Musolino, V.; Carresi, C.; Scarano, F.; Nucera, S.; Scicchitano, M.; Oppedisano, F.; Bosco, F.; Ruga, S.; et al. The Contribution of Gut Microbiota-Brain Axis in the Development of Brain Disorders. Front. Neurosci. 2021, 15, 616883. [Google Scholar] [CrossRef]
- Tong, Z.; Zhou, X.; Chu, Y.; Zhang, T.; Zhang, J.; Zhao, X.; Wang, Z.; Ding, R.; Meng, Q.; Yu, J.; et al. Implications of oral streptococcal bacteriophages in autism spectrum disorder. npj Biofilms Microbiomes 2022, 8, 91. [Google Scholar] [CrossRef]
- Fan, X.; Zhang, Z.; Gao, W.; Pan, Q.; Luo, K.; He, B.; Pu, Y. An Engineered Butyrate-Derived Polymer Nanoplatform as a Mucosa-Healing Enhancer Potentiates the Therapeutic Effect of Magnolol in Inflammatory Bowel Disease. ACS Nano 2024, 18, 229–244. [Google Scholar] [CrossRef]
- Li, H.; Guo, W.; Li, S.; Sun, B.; Li, N.; Xie, D.; Dong, Z.; Luo, D.; Chen, W.; Fu, W.; et al. Alteration of the gut microbiota profile in children with autism spectrum disorder in China. Front. Microbiol. 2023, 14, 1326870. [Google Scholar] [CrossRef]
- Yang, X.; Li, J.; Zhou, Y.; Zhang, N.; Liu, J. Effect of stigma maydis polysaccharide on the gut microbiota and transcriptome of VPA induced autism model rats. Front. Microbiol. 2022, 13, 1009502. [Google Scholar] [CrossRef]
- Radjabzadeh, D.; Bosch, J.A.; Uitterlinden, A.G.; Zwinderman, A.H.; Ikram, M.A.; van Meurs, J.B.J.; Luik, A.I.; Nieuwdorp, M.; Lok, A.; van Duijn, C.M.; et al. Gut microbiome-wide association study of depressive symptoms. Nat. Commun. 2022, 13, 7128. [Google Scholar] [CrossRef]
- Li, Y.; Xiao, P.; Cao, R.; Le, J.; Xu, Q.; Xiao, F.; Ye, L.; Wang, X.; Wang, Y.; Zhang, T. Effects and microbiota changes following oral lyophilized fecal microbiota transplantation in children with autism spectrum disorder. Front. Pediatr. 2024, 12, 1369823. [Google Scholar] [CrossRef]
- Gerges, P.; Bangarusamy, D.K.; Bitar, T.; Alameddine, A.; Nemer, G.; Hleihel, W. Turicibacter and Catenibacterium as potential biomarkers in autism spectrum disorders. Sci. Rep. 2024, 14, 23184. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, C.; Huang, S.; Wang, X.; Cao, M.; Gu, T.; Ou, X.; Pan, S.; Lin, Z.; Wang, X.; et al. Multi-omics analyses demonstrate the modulating role of gut microbiota on the associations of unbalanced dietary intake with gastrointestinal symptoms in children with autism spectrum disorder. Gut Microbes 2023, 15, 2281350. [Google Scholar] [CrossRef]
- Wu, T.; Wang, H.; Lu, W.; Zhai, Q.; Zhang, Q.; Yuan, W.; Gu, Z.; Zhao, J.; Zhang, H.; Chen, W. Potential of gut microbiome for detection of autism spectrum disorder. Microb. Pathog. 2020, 149, 104568. [Google Scholar] [CrossRef]
- Luna, R.A.; Oezguen, N.; Balderas, M.; Venkatachalam, A.; Runge, J.K.; Versalovic, J.; Veenstra-VanderWeele, J.; Anderson, G.M.; Savidge, T.; Williams, K.C. Distinct Microbiome-Neuroimmune Signatures Correlate with Functional Abdominal Pain in Children With Autism Spectrum Disorder. Cell. Mol. Gastroenterol. Hepatol. 2017, 3, 218–230. [Google Scholar] [CrossRef]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol. Autism 2013, 4, 42. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hu, N.; Cai, Q.; Zhang, F.; Zou, J.; Liu, Y.; Wei, D.; Zhu, Q.; Chen, K.; Zeng, L.; et al. Treatment with the traditional Chinese medicine BuYang HuanWu Tang induces alterations that normalize the microbiome in ASD patients. Biosci. Microbiota Food Health 2020, 39, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Hua, X.; Yang, T.; Guo, M.; Li, Q.; Xiao, L.; Li, L.; Chen, J.; Li, T. Alterations in Gut Vitamin and Amino Acid Metabolism are Associated with Symptoms and Neurodevelopment in Children with Autism Spectrum Disorder. J. Autism Dev. Disord. 2022, 52, 3116–3128. [Google Scholar] [CrossRef]
- Gary, N.C.; Misganaw, B.; Hammamieh, R.; Gautam, A. Exploring metabolomic dynamics in acute stress disorder: Amino acids, lipids, and carbohydrates. Front. Genet. 2024, 15, 1394630. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.N.; Nakamura, M.; Kawashima, H. New Role of the Serotonin as a Biomarker of Gut-Brain Interaction. Life 2024, 14, 1280. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Mendez, R.; Rivas-Arancibia, S. Vitamin C in Health and Disease: Its Role in the Metabolism of Cells and Redox State in the Brain. Front. Physiol. 2015, 6, 397. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, S.; Liu, F.; Dai, N.; Liang, R.; Lv, S.; Bao, L. Gut microbiota and autism spectrum disorders: A bidirectional Mendelian randomization study. Front. Cell. Infect. Microbiol. 2023, 13, 1267721. [Google Scholar] [CrossRef]


| Trait1 | Trait2 | rg | SE | p Value |
|---|---|---|---|---|
| Lachnospiraceae NK4A136 group | ASD | −1.1956 | 0.4208 | 0.0045 |
| Sellimonas | ASD | −0.4673 | 0.1666 | 0.0051 |
| Expoure | Outcome | Beta ± SE | p Value | HR (95% CI) |
|---|---|---|---|---|
| Turicibacter | ASD | 0.1310 ± 0.0630 | 0.0375 | 1.1400 (1.0076–1.2898) |
| Ruminococcaceae UCG005 | ASD | −0.2538 ± 0.0749 | 7 × 10−4 | 0.7758 (0.6699–0.8984) |
| Sutterella | ASD | −0.1966 ± 0.0935 | 0.0356 | 0.8215 (0.6839–0.9868) |
| Ruminiclostridium5 | ASD | −0.2077 ± 0.0856 | 0.0152 | 0.8125 (0.6870–0.9609) |
| Ruminococcus1 | ASD | −0.1846 ± 0.08419 | 0.0284 | 0.8315 (0.7050–0.9807) |
| Dorea | ASD | −0.2098 ± 0.0855 | 0.0142 | 0.8108 (0.6856–0.9587) |
| Expoure | Outcome | Beta ± SE | p Value | HR (95% CI) |
|---|---|---|---|---|
| ASD | Turicibacter | 0.0207± 0.2955 | 0.9443 | 1.0209 (0.5720–1.8219) |
| ASD | Ruminococcaceae UCG005 | 0.0521 ± 0.2215 | 0.8143 | 1.0534 (0.6824–1.6262) |
| ASD | Sutterella | −0.0561 ± 0.1573 | 0.7215 | 0.9455 (0.6946–1.2869) |
| ASD | Ruminiclostridium5 | −0.0332 ± 0.2272 | 0.8839 | 0.9674 (0.6198–1.5100) |
| ASD | Ruminococcus1 | −0.0962 ± 0.1039 | 0.3451 | 0.9082 (0.7410–1.1133) |
| ASD | Dorea | −0.0506 ± 0.1583 | 0.7495 | 0.9507 (0.6970–1.2967) |
| Trait1 | Trait2 | rg | SE | p Value |
|---|---|---|---|---|
| 4-methyl-2-oxopentanoate | ASD | −0.6174 | 0.2172 | 0.0045 |
| Alpha-hydroxyisocaproate | ASD | −0.6501 | 0.2365 | 0.0060 |
| 1-stearoyl-GPI(18:0) | ASD | −1.0537 | 0.4341 | 0.0152 |
| 4-acetylphenolsulfate | ASD | −0.9291 | 0.2787 | 0.0009 |
| Indolepropionate | ASD | −0.3776 | 0.1522 | 0.0131 |
| Pyridoxate | ASD | 0.5543 | 0.2586 | 0.0321 |
| Alpha-hydroxysovalerate | ASD | −0.3251 | 0.1199 | 0.067 |
| Hexanoylglycine | ASD | 1.7316 | 0.8041 | 0.0313 |
| Taurocholenatesulfate | ASD | −0.2675 | 0.1187 | 0.0242 |
| Palmitoylsphingomyelin(d18:1/16:0) | ASD | −0.2580 | 0.1256 | 0.0400 |
| 2s,3R-dihydroxybutyrate | ASD | −0.4529 | 0.1290 | 0.0004 |
| 1-(1-enyl-stearoyl)-GPE(p-18:0) | ASD | −0.2683 | 0.1357 | 0.0480 |
| 2-hydroxydecanoate | ASD | 0.3812 | 0.1873 | 0.0418 |
| 2-aminophenolsulfate | ASD | 0.7891 | 0.3992 | 0.0481 |
| P-cresolglucuronide | ASD | 0.2877 | 0.1087 | 0.0081 |
| Lignoceroylsphingomyelin(d18:1/24:0) | ASD | −0.2622 | 0.1267 | 0.0385 |
| 2-hydroxybutyrate/2-hydroxysobutyrate | ASD | −0.4417 | 0.1758 | 0.0120 |
| Tricosanoylsphingomyelin(d18:1/23:0) | ASD | −0.3056 | 0.1440 | 0.0338 |
| 1-(1-enyl-stearoyl)-2-arachidonoyl-GPE(p-18:0/20:4) | ASD | −0.6361 | 0.2698 | 0.0184 |
| Caffeicacidsulfate | ASD | 0.2972 | 0.1460 | 0.0418 |
| Sphingomyelin(d18:2/24:2) | ASD | −0.2810 | 0.1215 | 0.0207 |
| 2-hydroxy-4-(methylthio)butanoicacid | ASD | 0.2535 | 0.0986 | 0.0101 |
| Pentoseacid | ASD | −0.7062 | 0.3562 | 0.0474 |
| Pantothenate | ASD | 0.2376 | 0.1172 | 0.0427 |
| 2-aminobutyrate | ASD | −0.4337 | 0.1736 | 00125 |
| 4-acetamidobutanoate | ASD | −0.2659 | 0.1127 | 0.0183 |
| Cys-gly,oxidized | ASD | 0.2285 | 0.1076 | 0.0337 |
| Arachidonate(20:4n6) | ASD | −0.4844 | 0.2182 | 0.0264 |
| 3-(4-hydroxyphenyl)lactate | ASD | −0.3912 | 0.1423 | 0.0060 |
| Taurine | ASD | −0.4340 | 0.1997 | 0.0297 |
| Cytidine | ASD | −0.4656 | 0.2223 | 0.0362 |
| Bilirubindegradationproduct,C17H18N2O4(3) | ASD | -0.8692 | 0.3925 | 0.0268 |
| Glycinetopyridoxalratio | ASD | −0.2364 | 0.1127 | 0.0360 |
| Citrullinetodimethylarginine(SDMA+ADMA)ratio | ASD | 0.3116 | 0.1488 | 0.0363 |
| Uridinetocytidineratio | ASD | 0.3035 | 0.1473 | 0.0393 |
| Pyruvateto3-methyl-2-oxobutyrateratio | ASD | 0.2385 | 0.1204 | 0.0477 |
| Cysteinylglycinetotaurineratio | ASD | 0.9990 | 0.3513 | 0.0045 |
| Salicylatetocaprylate(8:0)ratio | ASD | −2.8567 | 0.9801 | 0.0036 |
| Glucosetosucroseratio | ASD | −1.2513 | 0.4655 | 0.0072 |
| Fructosetosucroseratio | ASD | −0.5650 | 0.1929 | 0.0034 |
| Caffeinetotheophyllineratio | ASD | 0.5331 | 0.2424 | 0.0278 |
| Alpha-ketobutyratetopyruvateratio | ASD | −0.5827 | 0.2642 | 0.0274 |
| CytidinetoN-acetylneuraminateratio | ASD | -0.3967 | 0.1625 | 0.0146 |
| CytidinetoN-acetylglucosaminetoN-acetylgalactosan | ASD | −1.7244 | 0.7071 | 0.0147 |
| Arachidonate(20:4n6)toparaxanthineratio | ASD | −0.4253 | 0.1868 | 0.0228 |
| Paraxanthinetolinoleate(18:2n6)ratio | ASD | 0.2857 | 0.1313 | 0.0296 |
| Salicylatetooxalate(ethanedioate)ratio | ASD | −0.9387 | 0.4150 | 0.0237 |
| Alpha-ketobutyrateto3-methyl-2-oxovalerateratio | ASD | −0.5800 | 0.2943 | 0.0487 |
| 3-methyl-2-oxovalerateto4-methyl-2-oxopentanoate | ASD | 0.5238 | 0.1758 | 0.0029 |
| Expoure | Outcome | Beta ± SE | p Value | HR (95% CI) |
|---|---|---|---|---|
| ASD | 9,10-DiHOME | 0.0933 ± 0.0416 | 0.0248 | 1.0978 (1.0119–1.1910) |
| ASD | 1beta-hydroxyetiocholanolone glucuronide | −0.1401 ± 0.0515 | 0.0065 | 0.8692 (0.7858–0.9616) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, J.; Fu, Z.; Gao, Y.; An, C.; Zhang, Z.; Zhong, X.; Tian, L.; Yang, X.; Zhang, J.; Zhang, Q.; et al. Gut Microbiotas, Plasma Metabolites, and Autism Spectrum Disorder: A Bidirectional Mendelian Randomization Analysis. Pathogens 2025, 14, 1137. https://doi.org/10.3390/pathogens14111137
Zhou J, Fu Z, Gao Y, An C, Zhang Z, Zhong X, Tian L, Yang X, Zhang J, Zhang Q, et al. Gut Microbiotas, Plasma Metabolites, and Autism Spectrum Disorder: A Bidirectional Mendelian Randomization Analysis. Pathogens. 2025; 14(11):1137. https://doi.org/10.3390/pathogens14111137
Chicago/Turabian StyleZhou, Jiayi, Zhang Fu, Yunfei Gao, Caiyan An, Zhiqiang Zhang, Xin Zhong, Liusuyan Tian, Xiuyan Yang, Junjing Zhang, Qingyuan Zhang, and et al. 2025. "Gut Microbiotas, Plasma Metabolites, and Autism Spectrum Disorder: A Bidirectional Mendelian Randomization Analysis" Pathogens 14, no. 11: 1137. https://doi.org/10.3390/pathogens14111137
APA StyleZhou, J., Fu, Z., Gao, Y., An, C., Zhang, Z., Zhong, X., Tian, L., Yang, X., Zhang, J., Zhang, Q., Wang, D., & Li, N. (2025). Gut Microbiotas, Plasma Metabolites, and Autism Spectrum Disorder: A Bidirectional Mendelian Randomization Analysis. Pathogens, 14(11), 1137. https://doi.org/10.3390/pathogens14111137







