Diversity and Functional Predictions of Gut Microbiota in Vietnamese Children Aged 6–24 Months with Persistent Diarrhea of Unknown Etiology
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Participants
2.2. Sample Collection, Processing, and DNA Extraction
2.3. 16S rRNA Gene Sequencing and Bioinformatics Analysis
2.4. Statistical Analysis
3. Results
3.1. Characteristics of Study Participants
3.2. Alpha Diversity of the Gut Microbiota
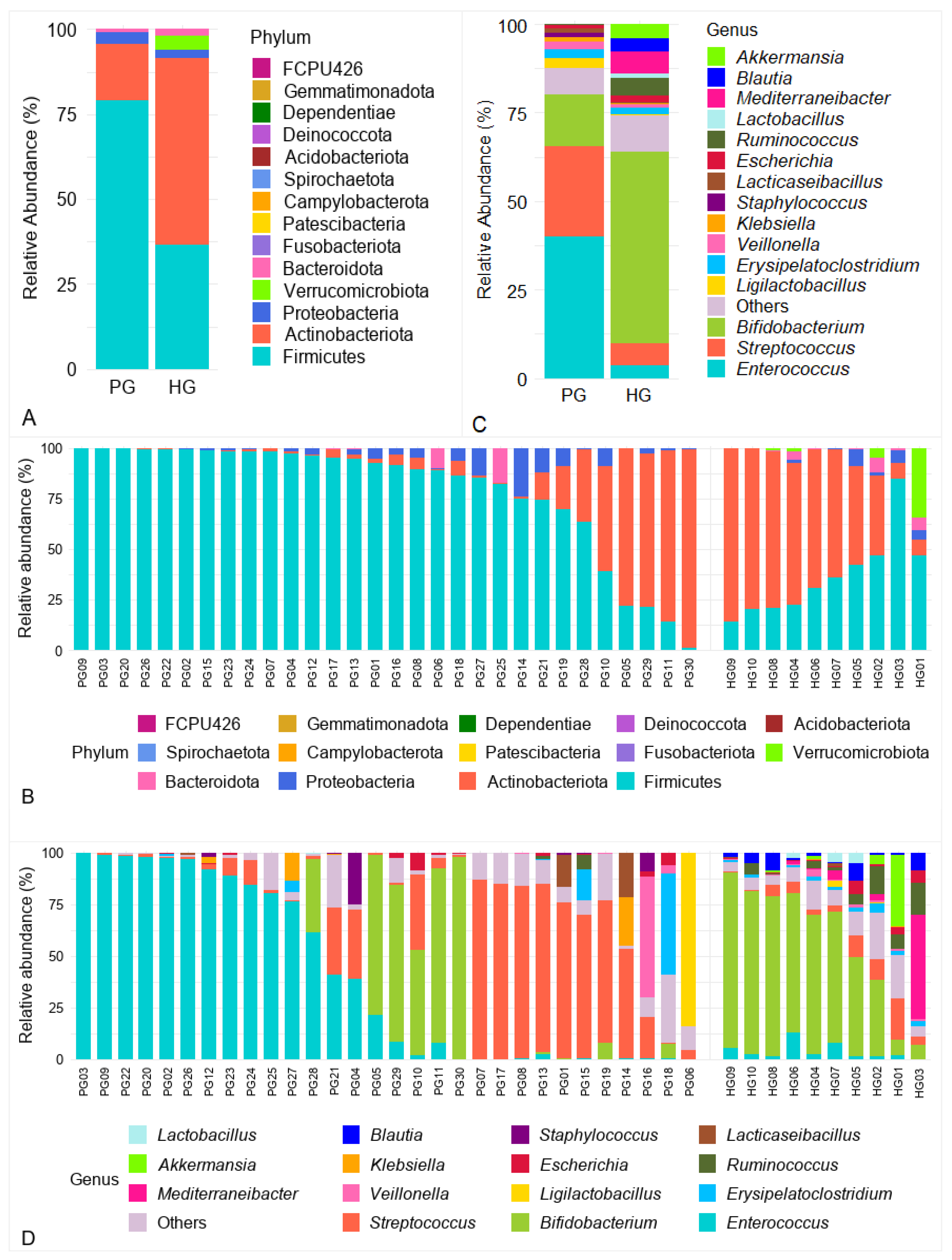
3.3. Gut Microbiota Composition
3.3.1. Analysis of Gut Microbiota at the Phylum Level
3.3.2. Analysis of Gut Microbiota at the Genus Level
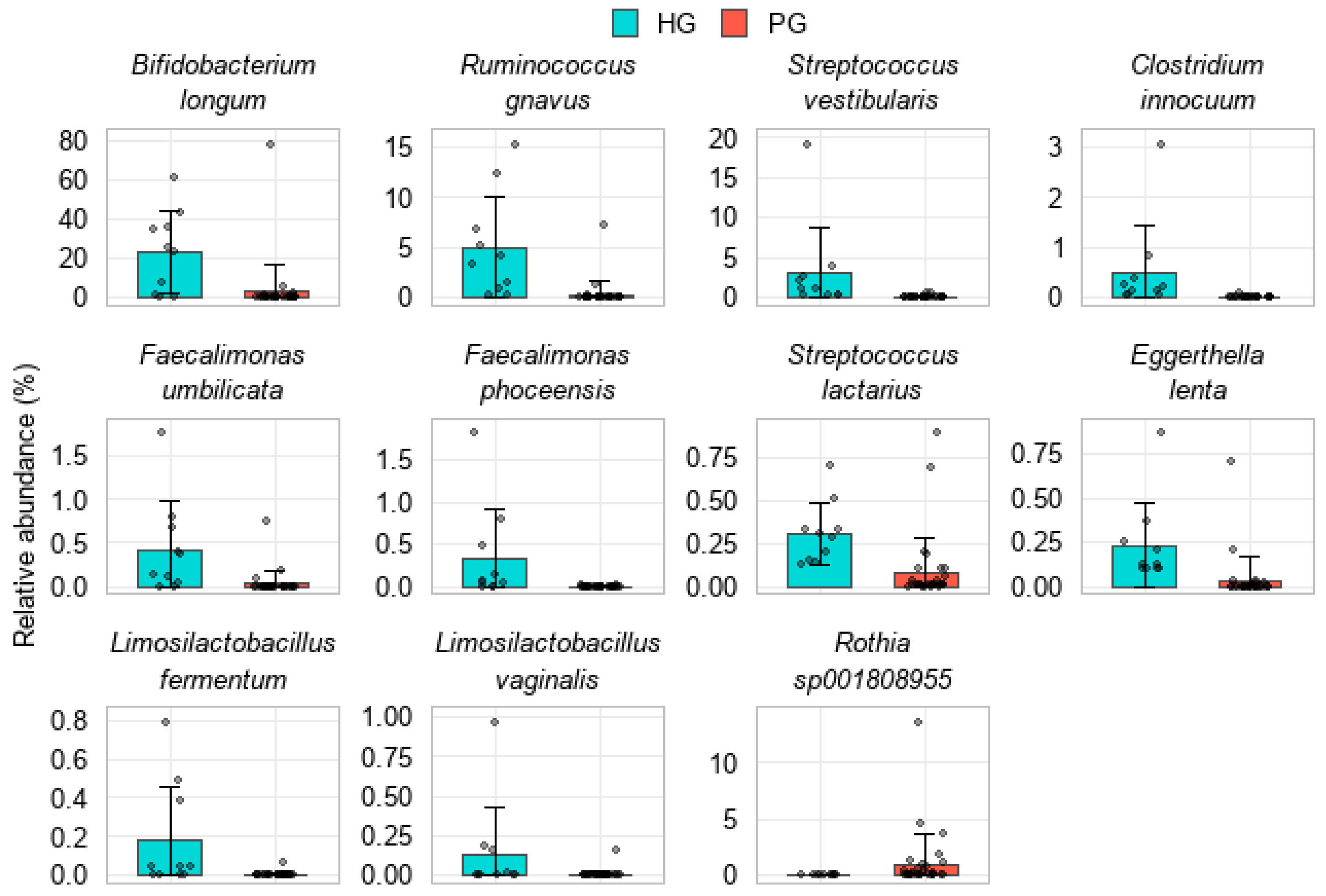
3.3.3. Analysis of Gut Microbiota at the Species Level
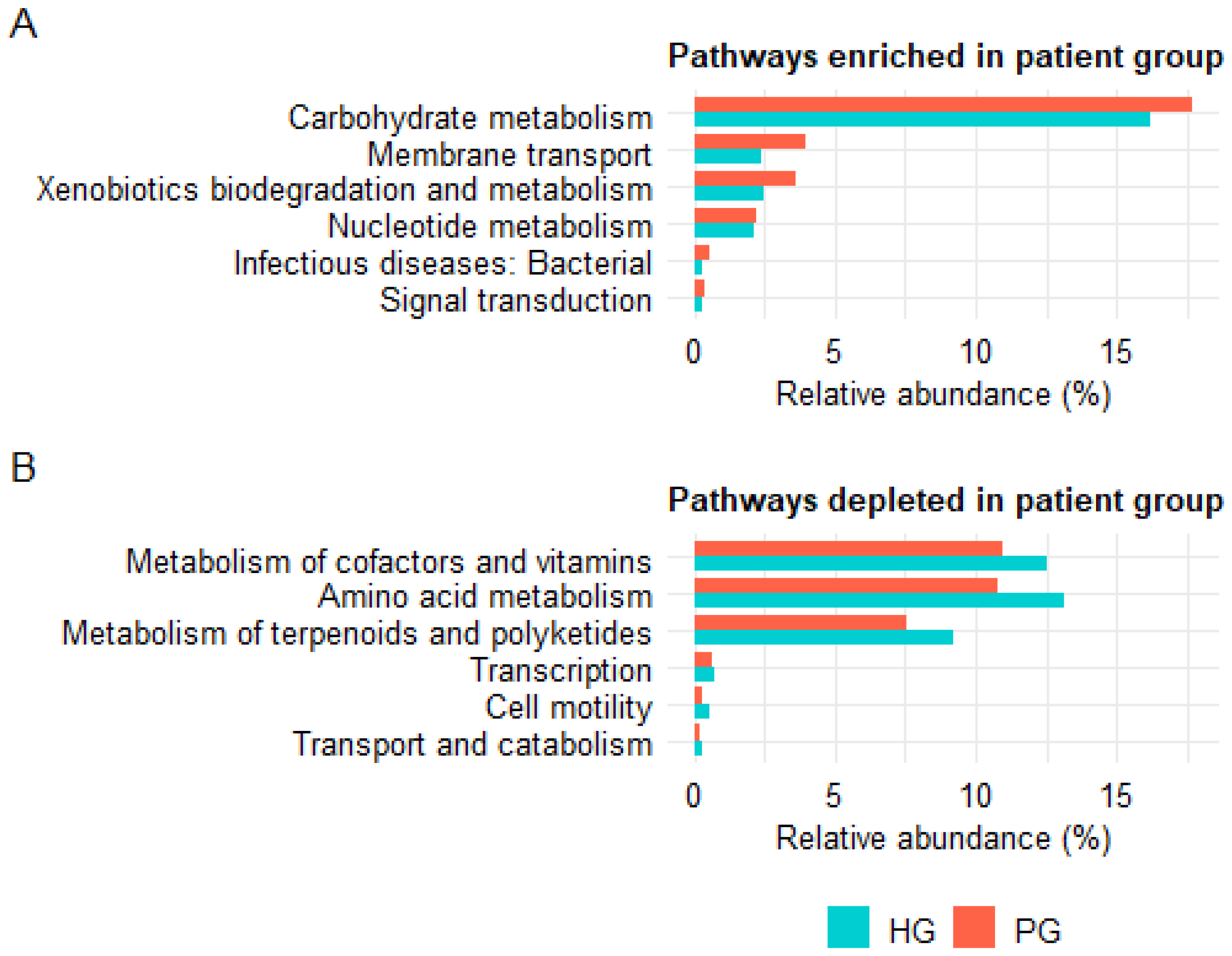
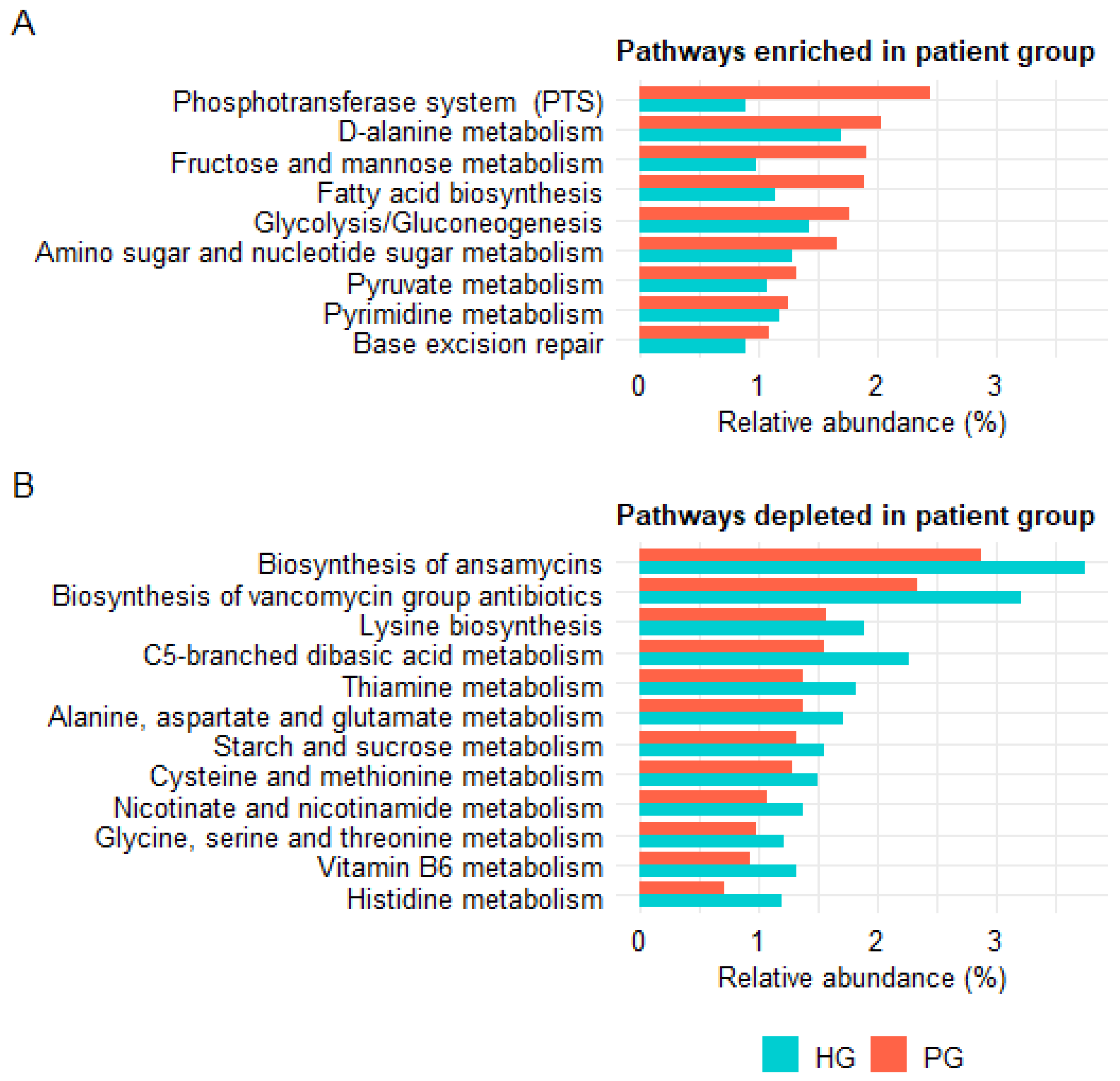
3.3.4. Analysis of Predicted Functional Profiles of the Gut Microbiota
4. Discussion
4.1. Alterations in Alpha Diversity
4.2. Gut Microbiota Dysbiosis and the Firmicutes/Bacteroidota Ratio
4.3. Deficiency of SCFA-Producing and Beneficial Bacteria
4.4. Expansion of Opportunistic and Pathogenic Taxa in the PG
4.5. Functional Predictions from PICRUSt2 and Their Biological Implications
4.6. Study Limitations and Future Research Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PG | Patient Group |
| HG | Healthy Group |
| NGS | Next-generation sequencing |
| WHO | World Health Organization |
| OTUs | Operational taxonomic units |
| RDP | Ribosomal Database Project |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| FDR | False discovery rate |
| IQR | Interquartile range |
| F/B ratio | Firmicutes/Bacteroidota ratio |
| SCFAs | Short-chain fatty acids |
| PTS | Phosphotransferase system |
References
- World Health Organization. Diarrhoeal Disease. 7 March 2024. Available online: https://www.who.int/news-room/fact-sheets/detail/diarrhoeal-disease (accessed on 2 April 2024).
- UNICEF. Diarrhoea Remains a Leading Killer of Young Children, Despite the Availability of a Simple Treatment. November 2024. Available online: https://data.unicef.org/topic/child-health/diarrhoeal-disease/ (accessed on 2 October 2025).
- Iwashita, H.; Tokizawa, A.; Thiem, V.D.; Takemura, T.; Nguyen, T.H.; Doan, H.T.; Pham, A.H.Q.; Tran, N.L.; Yamashiro, T. Risk Factors Associated with Diarrheal Episodes in an Agricultural Community in Nam Dinh Province, Vietnam: A Prospective Cohort Study. Int. J. Environ. Res. Public Health 2022, 19, 2456. [Google Scholar] [CrossRef] [PubMed]
- Infectious Diseases Society of America (IDSA). 2017 Clinical Practice Guidelines for the Diagnosis and Management of Infectious Diarrhea. Available online: https://www.idsociety.org/practice-guideline/infectious-diarrhea/ (accessed on 19 October 2024).
- American College of Gastroenterology (ACG). Diarrheal Diseases—Acute & Chronic. Available online: https://gi.org/topics/diarrhea-acute-and-chronic/ (accessed on 13 August 2025).
- National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). Chronic Diarrhea in Children. Available online: https://www.niddk.nih.gov/health-information/digestive-diseases/chronic-diarrhea-children (accessed on 16 August 2025).
- American College of Gastroenterology (ACG). Diarrhea in Children. Available online: https://gi.org/topics/diarrhea-in-children/ (accessed on 16 August 2025).
- Moore, S.R. Update on Prolonged and Persistent Diarrhea in Children. Curr. Opin. Gastroenterol. 2011, 27, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Durairaj, P.; Raju, S.; Thirumalaikumarasamy, S. Clinical Profile and Risk Factors for Persistent Diarrhoea in Children under Five Years of Age in an Urban Referral Centre. Int. J. Contemp. Pediatr. 2017, 4, 1986–1994. [Google Scholar] [CrossRef][Green Version]
- Ugboko, H.U.; Nwinyi, O.C.; Oranusi, S.U.; Oyewale, J.O. Childhood Diarrhoeal Diseases in Developing Countries. Heliyon 2020, 6, e03690. [Google Scholar] [CrossRef]
- D’Onofrio, A.; Crawford, J.M.; Stewart, E.J.; Witt, K.; Gavrish, E.; Epstein, S.; Clardy, J.; Lewis, K. Siderophores from Neighboring Organisms Promote the Growth of Uncultured Bacteria. Chem. Biol. 2010, 17, 254–264. [Google Scholar] [CrossRef]
- Platts-Mills, J.A.; Operario, D.J.; Houpt, E.R. Molecular Diagnosis of Diarrhea: Current Status and Future Potential. Curr. Infect. Dis. Rep. 2012, 14, 41–46. [Google Scholar] [CrossRef]
- Denno, D.M.; Klein, E.J.; Young, V.B.; Fox, J.G.; Wang, D.; Tarr, P.I. Explaining Unexplained Diarrhea and Associating Risks and Infections. Anim. Health Res. Rev. 2007, 8, 69–80. [Google Scholar] [CrossRef]
- Abba, K.; Sinfield, R.; Hart, C.A.; Garner, P. Pathogens Associated with Persistent Diarrhoea in Children in Low and Middle Income Countries: Systematic Review. BMC Infect. Dis. 2009, 9, 88. [Google Scholar] [CrossRef]
- Kotloff, K.L.; Nataro, J.P.; Blackwelder, W.C.; Nasrin, D.; Farag, T.H.; Panchalingam, S.; Wu, Y.; Sow, S.O.; Sur, D.; Breiman, R.F.; et al. Burden and Aetiology of Diarrhoeal Disease in Infants and Young Children in Developing Countries (the Global Enteric Multicenter Study, GEMS): A Prospective, Case-Control Study. Lancet 2013, 382, 209–222. [Google Scholar] [CrossRef]
- Li, Y.; Xia, S.; Jiang, X.; Feng, C.; Gong, S.; Ma, J.; Fang, Z.; Yin, J.; Yin, Y. Gut Microbiota and Diarrhea: An Updated Review. Front. Cell. Infect. Microbiol. 2021, 11, 625210. [Google Scholar] [CrossRef]
- Tap, J.; Mondot, S.; Levenez, F.; Pelletier, E.; Caron, C.; Furet, J.-P.; Ugarte, E.; Muñoz-Tamayo, R.; Paslier, D.L.E.; Nalin, R.; et al. Towards the Human Intestinal Microbiota Phylogenetic Core. Environ. Microbiol. 2009, 11, 2574–2584. [Google Scholar] [CrossRef]
- Zhu, B.; Wang, X.; Li, L. Human Gut Microbiome: The Second Genome of Human Body. Protein Cell 2010, 1, 718–725. [Google Scholar] [CrossRef]
- Petersen, C.; Round, J.L. Defining Dysbiosis and Its Influence on Host Immunity and Disease. Cell. Microbiol. 2014, 16, 1024–1033. [Google Scholar] [CrossRef]
- Tesfaw, G.; Siraj, D.S.; Abdissa, A.; Jakobsen, R.R.; Johansen, Ø.H.; Zangenberg, M.; Hanevik, K.; Mekonnen, Z.; Langeland, N.; Bjørang, O.; et al. Gut Microbiota Patterns Associated with Duration of Diarrhea in Children under Five Years of Age in Ethiopia. Nat. Commun. 2024, 15, 7532. [Google Scholar] [CrossRef] [PubMed]
- De, R.; Mukhopadhyay, A.K.; Dutta, S. Metagenomic Analysis of Gut Microbiome and Resistome of Diarrheal Fecal Samples from Kolkata, India, Reveals the Core and Variable Microbiota Including Signatures of Microbial Dark Matter. Gut Pathog. 2020, 12, 32. [Google Scholar] [CrossRef]
- Xiao, Q.; Chen, B.; Zhu, Z.; Yang, T.; Tao, E.; Hu, C.; Zheng, W.; Tang, W.; Shu, X.; Jiang, M. Alterations in the Fecal Microbiota Composition in Pediatric Acute Diarrhea: A Cross-Sectional and Comparative Study of Viral and Bacterial Enteritis. Infect. Drug Resist. 2023, 16, 5473. [Google Scholar] [CrossRef]
- Pop, M.; Walker, A.W.; Paulson, J.; Lindsay, B.; Antonio, M.; Hossain, M.A.; Oundo, J.; Tamboura, B.; Mai, V.; Astrovskaya, I.; et al. Diarrhea in Young Children from Low-Income Countries Leads to Large-Scale Alterations in Intestinal Microbiota Composition. Genome Biol. 2014, 15, R76. [Google Scholar] [CrossRef]
- Ward, D.V.; Gevers, D.; Giannoukos, G.; Earl, A.M.; Methé, B.A.; Sodergren, E.; Feldgarden, M.; Ciulla, D.M.; Tabbaa, D.; Arze, C.; et al. Evaluation of 16s rDNA-Based Community Profiling for Human Microbiome Research. PLoS ONE 2012, 7, e39315. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for Prediction of Metagenome Functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Nguyen, T.Q.; Dao, T.K.; Nguyen, H.D.; Phung, T.B.T.; Pham, T.T.N.; Nguyen, T.V.H.; Trinh, T.H.; Le, H.C.; Le, T.T.H.; Do, T.H. Application of PCR-Based Techniques for the Identification of Genetic Fingerprint Diversity of Dominant Bacteria in Fecal Samples of Children with Diarrhea in Vietnam. Infect. Dis. Rep. 2024, 16, 932–951. [Google Scholar] [CrossRef]
- Dao, T.K.; Pham, T.T.N.; Nguyen, H.D.; Dam, Q.T.; Phung, T.B.T.; Nguyen, T.V.H.; Nguyen, T.Q.; Hoang, K.C.; Do, T.H. Metagenomic Analysis of the Gastrointestinal Phageome and Incorporated Dysbiosis in Children with Persistent Diarrhea of Unknown Etiology in Vietnam. Pathogens 2025, 14, 985. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2025; Available online: https://www.r-project.org/ (accessed on 9 August 2025).
- CD Genomics. The Use and Types of Alpha-Diversity Metrics in Microbial NGS. Available online: https://www.cd-genomics.com/microbioseq/the-use-and-types-of-alpha-diversity-metrics-in-microbial-ngs.html (accessed on 19 July 2025).
- CD Genomics. Understanding Alpha Diversity: What It Is and Why It Matters. Available online: https://www.cd-genomics.com/resource-understanding-alpha-diversity-what-it-is-why-it-matters.html (accessed on 16 July 2025).
- EzBioCloud Knowledge Base. Alpha Diversity. Available online: https://kb.ezbiocloud.net/home/science-blogs/analyze/alpha-diversity (accessed on 19 July 2025).
- Cassol, I.; Ibañez, M.; Bustamante, J.P. Key Features and Guidelines for the Application of Microbial Alpha Diversity Metrics. Sci. Rep. 2025, 15, 622. [Google Scholar] [CrossRef]
- The, H.C.; Florez de Sessions, P.; Jie, S.; Pham Thanh, D.; Thompson, C.N.; Nguyen Ngoc Minh, C.; Chu, C.W.; Tran, T.-A.; Thomson, N.R.; Thwaites, G.E.; et al. Assessing Gut Microbiota Perturbations during the Early Phase of Infectious Diarrhea in Vietnamese Children. Gut Microbes 2018, 9, 38–54. [Google Scholar] [CrossRef]
- Spragge, F.; Bakkeren, E.; Jahn, M.T.; Araujo, E.B.N.; Pearson, C.F.; Wang, X.; Pankhurst, L.; Cunrath, O.; Foster, K.R. Microbiome Diversity Protects against Pathogens by Nutrient Blocking. Science 2023, 382, eadj3502. [Google Scholar] [CrossRef]
- Shen, Y.; Fan, N.; Ma, S.; Cheng, X.; Yang, X.; Wang, G. Gut Microbiota Dysbiosis: Pathogenesis, Diseases, Prevention, and Therapy. MedComm 2025, 6, e70168. [Google Scholar] [CrossRef]
- Rupa Health. Firmicutes/Bacteroidetes Ratio. Available online: https://www.rupahealth.com/biomarkers/firmicutes-bacteroidetes-ratio (accessed on 3 June 2025).
- Stojanov, S.; Berlec, A.; Štrukelj, B. The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel Disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef]
- Deering, K.E.; Devine, A.; O’Sullivan, T.A.; Lo, J.; Boyce, M.C.; Christophersen, C.T. Characterizing the Composition of the Pediatric Gut Microbiome: A Systematic Review. Nutrients 2020, 12, 16. [Google Scholar] [CrossRef]
- Lobionda, S.; Sittipo, P.; Kwon, H.Y.; Lee, Y.K. The Role of Gut Microbiota in Intestinal Inflammation with Respect to Diet and Extrinsic Stressors. Microorganisms 2019, 7, 271. [Google Scholar] [CrossRef]
- Fusco, W.; Lorenzo, M.B.; Cintoni, M.; Porcari, S.; Rinninella, E.; Kaitsas, F.; Lener, E.; Mele, M.C.; Gasbarrini, A.; Collado, M.C.; et al. Short-Chain Fatty-Acid-Producing Bacteria: Key Components of the Human Gut Microbiota. Nutrients 2023, 15, 2211. [Google Scholar] [CrossRef]
- Singh, V.; Lee, G.; Son, H.; Koh, H.; Kim, E.S.; Unno, T.; Shin, J.-H. Butyrate Producers, “The Sentinel of Gut”: Their Intestinal Significance with and beyond Butyrate, and Prospective Use as Microbial Therapeutics. Front. Microbiol. 2023, 13, 1103836. [Google Scholar] [CrossRef]
- Nogal, A.; Valdes, A.M.; Menni, C. The Role of Short-Chain Fatty Acids in the Interplay between Gut Microbiota and Diet in Cardio-Metabolic Health. Gut Microbes 2021, 13, 1897212. [Google Scholar] [CrossRef]
- Rastogi, S.; Singh, A. Gut Microbiome and Human Health: Exploring How the Probiotic Genus Lactobacillus Modulate Immune Responses. Front. Pharmacol. 2022, 13, 1042189. [Google Scholar] [CrossRef]
- Elshaghabee, F.M.F.; Rokana, N.; Gulhane, R.D.; Sharma, C.; Panwar, H. Bacillus As Potential Probiotics: Status, Concerns, and Future Perspectives. Front. Microbiol. 2017, 8, 1490. [Google Scholar] [CrossRef]
- Ilinskaya, O.N.; Ulyanova, V.V.; Yarullina, D.R.; Gataullin, I.G. Secretome of Intestinal Bacilli: A Natural Guard against Pathologies. Front. Microbiol. 2017, 8, 1666. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.H.; Jimenez, D.M.R.; Meisel, M. Limosilactobacillus reuteri—A Probiotic Gut Commensal with Contextual Impact on Immunity. Gut Microbes 2025, 17, 2451088. [Google Scholar] [CrossRef]
- Arzola-Martínez, L.; Ravi, K.; Huffnagle, G.B.; Lukacs, N.W.; Fonseca, W. Lactobacillus johnsonii and Host Communication: Insight into Modulatory Mechanisms during Health and Disease. Front. Microbiomes. 2024, 2, 1345330. [Google Scholar] [CrossRef]
- Bengoa, A.A.; Dardis, C.; Garrote, G.L.; Abraham, A.G. Health-Promoting Properties of Lacticaseibacillus paracasei: A Focus on Kefir Isolates and Exopolysaccharide-Producing Strains. Foods 2021, 10, 2239. [Google Scholar] [CrossRef]
- Hao, Y.; Jiang, L.; Han, D.; Si, D.; Sun, Z.; Wu, Z.; Dai, Z. Limosilactobacillus mucosae and Lactobacillus amylovorus Protect Against Experimental Colitis via Upregulation of Colonic 5-Hydroxytryptamine Receptor 4 and Transforming Growth Factor-Β2. J. Nutr. 2023, 153, 2512–2522. [Google Scholar] [CrossRef]
- Lu, S. The Role of Lactic Acid Bacteria in Gut Microbiota and Mucosal Immune System. South Fla. J. Dev. 2021, 2, 3818–3825. [Google Scholar] [CrossRef]
- O’Callaghan, A.; van Sinderen, D. Bifidobacteria and Their Role as Members of the Human Gut Microbiota. Front. Microbiol. 2016, 7, 925. [Google Scholar] [CrossRef]
- Abdulqadir, R.; Engers, J.; Al-Sadi, R. Role of Bifidobacterium in Modulating the Intestinal Epithelial Tight Junction Barrier: Current Knowledge and Perspectives. Curr. Dev. Nutr. 2023, 7, 102026. [Google Scholar] [CrossRef]
- Shin, J.H.; Tillotson, G.; MacKenzie, T.N.; Warren, C.A.; Wexler, H.M.; Goldstein, E.J.C. Bacteroides and Related Species: The Keystone Taxa of the Human Gut Microbiota. Anaerobe 2024, 85, 102819. [Google Scholar] [CrossRef]
- Turroni, F.; Duranti, S.; Milani, C.; Lugli, G.A.; van Sinderen, D.; Ventura, M. Bifidobacterium Bifidum: A Key Member of the Early Human Gut Microbiota. Microorganisms 2019, 7, 544. [Google Scholar] [CrossRef]
- Chung The, H.; Nguyen Ngoc Minh, C.; Tran Thi Hong, C.; Nguyen Thi Nguyen, T.; Pike, L.J.; Zellmer, C.; Pham Duc, T.; Tran, T.-A.; Ha Thanh, T.; Van, M.P.; et al. Exploring the Genomic Diversity and Antimicrobial Susceptibility of Bifidobacterium pseudocatenulatum in a Vietnamese Population. Microbiol. Spectr. 2021, 9, e00526-21. [Google Scholar] [CrossRef] [PubMed]
- Engevik, M.A.; Luk, B.; Chang-Graham, A.L.; Hall, A.; Herrmann, B.; Ruan, W.; Endres, B.T.; Shi, Z.; Garey, K.W.; Hyser, J.M.; et al. Bifidobacterium dentium Fortifies the Intestinal Mucus Layer via Autophagy and Calcium Signaling Pathways. mBio 2019, 10, e01087-19. [Google Scholar] [CrossRef] [PubMed]
- Monira, S.; Shabnam, S.A.; Alam, N.H.; Endtz, H.P.; Cravioto, A.; Alam, M. 16S rRNA Gene-Targeted TTGE in Determining Diversity of Gut Microbiota during Acute Diarrhoea and Convalescence. J. Health Popul. Nutr. 2012, 30, 250–256. [Google Scholar] [CrossRef]
- Gao, F.; Cheng, C.; Li, R.; Chen, Z.; Tang, K.; Du, G. The Role of Akkermansia muciniphila in Maintaining Health: A Bibliometric Study. Front. Med. 2025, 12, 1484656. [Google Scholar] [CrossRef]
- Paone, P.; Cani, P.D. Mucus Barrier, Mucins and Gut Microbiota: The Expected Slimy Partners? Gut 2020, 69, 2232–2243. [Google Scholar] [CrossRef]
- Kim, S.; Shin, Y.-C.; Kim, T.-Y.; Kim, Y.; Lee, Y.-S.; Lee, S.-H.; Kim, M.-N.; O, E.; Kim, K.S.; Kweon, M.-N. Mucin Degrader Akkermansia Muciniphila Accelerates Intestinal Stem Cell-Mediated Epithelial Development. Gut Microbes 2021, 13, 1892441. [Google Scholar] [CrossRef]
- Rokhsefat, S.; Lin, A.; Comelli, E.M. Mucin-Microbiota Interaction During Postnatal Maturation of the Intestinal Ecosystem: Clinical Implications. Dig. Dis. Sci. 2016, 61, 1473–1486. [Google Scholar] [CrossRef]
- Guo, P.; Zhang, K.; Ma, X.; He, P. Clostridium Species as Probiotics: Potentials and Challenges. J. Anim. Sci. Biotechnol. 2020, 11, 24. [Google Scholar] [CrossRef]
- Moreira de Gouveia, M.I.; Bernalier-Donadille, A.; Jubelin, G. Enterobacteriaceae in the Human Gut: Dynamics and Ecological Roles in Health and Disease. Biology 2024, 13, 142. [Google Scholar] [CrossRef]
- Quaye, E.K.; Adjei, R.L.; Isawumi, A.; Allen, D.J.; Caporaso, J.G.; Quaye, O. Altered Faecal Microbiota Composition and Structure of Ghanaian Children with Acute Gastroenteritis. Int. J. Mol. Sci. 2023, 24, 3607. [Google Scholar] [CrossRef]
- Fan, Q.; Yi, M.; Liu, H.; Wang, Y.; Li, X.; Yuan, J.; Wang, L.; Hou, B.; Li, M. The Impact of Age and Pathogens Type on the Gut Microbiota in Infants with Diarrhea in Dalian, China. Can. J. Infect. Dis. Med. Microbiol. 2020, 2020, 8837156. [Google Scholar] [CrossRef] [PubMed]
- Donskey, C.J.; Chowdhry, T.K.; Hecker, M.T.; Hoyen, C.K.; Hanrahan, J.A.; Hujer, A.M.; Hutton-Thomas, R.A.; Whalen, C.C.; Bonomo, R.A.; Rice, L.B. Effect of Antibiotic Therapy on the Density of Vancomycin-Resistant Enterococci in the Stool of Colonized Patients. N. Engl. J. Med. 2000, 343, 1925–1932. [Google Scholar] [CrossRef]
- Taur, Y. Intestinal Microbiome Changes and Stem Cell Transplantation: Lessons Learned. Virulence 2016, 7, 930–938. [Google Scholar] [CrossRef] [PubMed]
- Ubeda, C.; Taur, Y.; Jenq, R.R.; Equinda, M.J.; Son, T.; Samstein, M.; Viale, A.; Socci, N.D.; van den Brink, M.R.M.; Kamboj, M.; et al. Vancomycin-Resistant Enterococcus Domination of Intestinal Microbiota Is Enabled by Antibiotic Treatment in Mice and Precedes Bloodstream Invasion in Humans. J. Clin. Investig. 2010, 120, 4332–4341. [Google Scholar] [CrossRef]
- Dubin, K.; Pamer, E.G. Enterococci and Their Interactions with the Intestinal Microbiome. Microbiol. Spectr. 2017, 5, 10-1128. [Google Scholar] [CrossRef]
- Van Tyne, D.; Gilmore, M.S. A Delicate Balance: Maintaining Mutualism to Prevent Disease. Cell Host Microbe 2014, 16, 425–427. [Google Scholar] [CrossRef][Green Version]
- Kastl, A.J.; Terry, N.A.; Wu, G.D.; Albenberg, L.G. The Structure and Function of the Human Small Intestinal Microbiota: Current Understanding and Future Directions. Cell. Mol. Gastroenterol. Hepatol. 2019, 9, 33–45. [Google Scholar] [CrossRef]
- Wen, H.; Yin, X.; Yuan, Z.; Wang, X.; Su, S. Comparative Analysis of Gut Microbial Communities in Children under 5 Years Old with Diarrhea. J. Microbiol. Biotechnol. 2018, 28, 652–662. [Google Scholar] [CrossRef]
- Winter, S.E.; Lopez, C.A.; Bäumler, A.J. The Dynamics of Gut-Associated Microbial Communities during Inflammation. EMBO Rep. 2013, 14, 319–327. [Google Scholar] [CrossRef]
- David, L.A.; Weil, A.; Ryan, E.T.; Calderwood, S.B.; Harris, J.B.; Chowdhury, F.; Begum, Y.; Qadri, F.; LaRocque, R.C.; Turnbaugh, P.J. Gut Microbial Succession Follows Acute Secretory Diarrhea in Humans. mBio 2015, 6, e00381-15. [Google Scholar] [CrossRef]
- Chung The, H.; Le, S.-N.H. Dynamic of the Human Gut Microbiome under Infectious Diarrhea. Curr. Opin. Microbiol. 2022, 66, 79–85. [Google Scholar] [CrossRef] [PubMed]
- West, S.R.; Suddaby, A.B.; Lewin, G.R.; Ibberson, C.B. Rothia. Trends Microbiol. 2024, 32, 720–721. [Google Scholar] [CrossRef] [PubMed]
- Torres-Morales, J.; Mark Welch, J.L.; Dewhirst, F.E.; Borisy, G.G. Site-Specialization of Human Oral Gemella Species. J. Oral Microbiol. 2023, 15, 2225261. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Ren, Y.; Mu, Y.; Zhang, L.; Chen, B.; Li, S.; Fang, Q.; Zhang, Z.; Zhang, K.; Li, S.; et al. Microbial Imbalance in Chinese Children with Diarrhea or Constipation. Sci. Rep. 2024, 14, 13516. [Google Scholar] [CrossRef]
- Granger, M.-F.; Kelly, M.; Fortier, L.-C.; Fournier, E.; Côté-Gravel, J.; Malouin, F.; Valiquette, L.; Lévesque, S. Chronic Diarrhea Caused by a Klebsiella oxytoca Toxin Producer Strain Following Antibiotic-Associated Hemorrhagic Colitis: Successful Treatment by Fecal Microbiota Transplant. Clin. Infect. Dis. 2023, 77, 1700–1703. [Google Scholar] [CrossRef]
- Cheng, V.C.C.; Yam, W.-C.; Tsang, L.-L.; Yau, M.C.Y.; Siu, G.K.H.; Wong, S.C.Y.; Chan, J.F.W.; To, K.K.W.; Tse, H.; Hung, I.F.N.; et al. Epidemiology of Klebsiella oxytoca-Associated Diarrhea Detected by Simmons Citrate Agar Supplemented with Inositol, Tryptophan, and Bile Salts. J. Clin. Microbiol. 2012, 50, 1571–1579. [Google Scholar] [CrossRef]
- Dione, N.; Mlaga, K.D.; Liang, S.; Jospin, G.; Marfori, Z.; Alvarado, N.; Scarsella, E.; Uttarwar, R.; Ganz, H.H. Comparative Genomic and Phenotypic Description of Escherichia ruysiae: A Newly Identified Member of the Gut Microbiome of the Domestic Dog. Front. Microbiol. 2025, 16, 1558802. [Google Scholar] [CrossRef]
- Campos-Madueno, E.I.; Aldeia, C.; Sendi, P.; Endimiani, A. Escherichia ruysiae May Serve as a Reservoir of Antibiotic Resistance Genes across Multiple Settings and Regions. Microbiol. Spectr. 2023, 11, e01753-23. [Google Scholar] [CrossRef]
- Lifshitz, F.; Coello-Ramirez, P.; Gutierrez-Topete, G.; Cornado-Cornet, M.C. Carbohydrate Intolerance in Infants with Diarrhea. J. Pediatr. 1971, 79, 760–767. [Google Scholar] [CrossRef] [PubMed]
- Hammer, H.F.; Hammer, J. Diarrhea Caused By Carbohydrate Malabsorption. Gastroenterol. Clin. N. Am. 2012, 41, 611–627. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, M.; Hartke, A.; Huycke, M. The Physiology and Metabolism of Enterococci. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. Available online: https://www.ncbi.nlm.nih.gov/books/NBK190432/ (accessed on 4 October 2025).
- Wang, Y.; Wu, J.; Lv, M.; Shao, Z.; Hungwe, M.; Wang, J.; Bai, X.; Xie, J.; Wang, Y.; Geng, W. Metabolism Characteristics of Lactic Acid Bacteria and the Expanding Applications in Food Industry. Front. Bioeng. Biotechnol. 2021, 9, 612285. [Google Scholar] [CrossRef]
- Culp, E.J.; Goodman, A.L. Cross-Feeding in the Gut Microbiome: Ecology and Mechanisms. Cell Host Microbe 2023, 31, 485–499. [Google Scholar] [CrossRef]
- Lange, O.; Proczko-Stepaniak, M.; Mika, A. Short-Chain Fatty Acids—A Product of the Microbiome and Its Participation in Two-Way Communication on the Microbiome-Host Mammal Line. Curr. Obes. Rep. 2023, 12, 108–126. [Google Scholar] [CrossRef]
- Oliphant, K.; Allen-Vercoe, E. Macronutrient Metabolism by the Human Gut Microbiome: Major Fermentation by-Products and Their Impact on Host Health. Microbiome 2019, 7, 91. [Google Scholar] [CrossRef]
- Iddrisu, I.; Monteagudo-Mera, A.; Poveda, C.; Pyle, S.; Shahzad, M.; Andrews, S.; Walton, G.E. Malnutrition and Gut Microbiota in Children. Nutrients 2021, 13, 2727. [Google Scholar] [CrossRef]
- Wan, Z.; Zheng, J.; Zhu, Z.; Sang, L.; Zhu, J.; Luo, S.; Zhao, Y.; Wang, R.; Zhang, Y.; Hao, K.; et al. Intermediate Role of Gut Microbiota in Vitamin B Nutrition and Its Influences on Human Health. Front. Nutr. 2022, 9, 1031502. [Google Scholar] [CrossRef]
- Wibowo, S.; Pramadhani, A. Vitamin B, Role of Gut Microbiota and Gut Health. In Vitamin B and Vitamin E—Pleiotropic and Nutritional Benefits; IntechOpen: London, UK, 2024; ISBN 978-1-83768-379-6. [Google Scholar] [CrossRef]
- Radjabzadeh, D.; Boer, C.G.; Beth, S.A.; van der Wal, P.; Kiefte-De Jong, J.C.; Jansen, M.A.E.; Konstantinov, S.R.; Peppelenbosch, M.P.; Hays, J.P.; Jaddoe, V.W.V.; et al. Diversity, Compositional and Functional Differences between Gut Microbiota of Children and Adults. Sci. Rep. 2020, 10, 1040. [Google Scholar] [CrossRef]
- Dinleyici, E.C.; Martínez-Martínez, D.; Kara, A.; Karbuz, A.; Dalgic, N.; Metin, O.; Yazar, A.S.; Guven, S.; Kurugol, Z.; Turel, O.; et al. Time Series Analysis of the Microbiota of Children Suffering From Acute Infectious Diarrhea and Their Recovery After Treatment. Front. Microbiol. 2018, 9, 1230. [Google Scholar] [CrossRef]
- World Health Organization. Antimicrobial Resistance in Viet Nam. Available online: https://www.who.int/vietnam/health-topics/antimicrobial-resistance (accessed on 2 November 2025).
- Torumkuney, D.; Kundu, S.; Vu, G.V.; Nguyen, H.A.; Pham, H.V.; Kamble, P.; Truong Ha Lan, N.; Keles, N. Country Data on AMR in Vietnam in the Context of Community-Acquired Respiratory Tract Infections: Links between Antibiotic Susceptibility, Local and International Antibiotic Prescribing Guidelines, Access to Medicines and Clinical Outcome. J. Antimicrob. Chemother. 2022, 77, i26–i34. [Google Scholar] [CrossRef]


| Characteristics | HG (n = 30) | PG (n = 30) | p-Value | |
|---|---|---|---|---|
| Age, months (Mean ± SD) | 11.4 ± 3.8 | 7.2 ± 2.0 | <0.001 a | |
| Age group, n (%) | 6–12 months | 19 (63.3%) | 29 (96.7%) | 0.002 b |
| 13–24 months | 11 (36.7%) | 1 (3.3%) | ||
| Sex, n (%) | Male | 16 (53.3%) | 18 (60.0%) | 0.602 c |
| Female | 14 (46.7%) | 12 (40.0%) | ||
| Residence, n (%) | Rural | 12 (40.0%) | 19 (63.3%) | 0.070 c |
| Urban | 18 (60.0%) | 11 (36.7%) | ||
| Mode of delivery, n (%) | Vaginal | 16 (53.3%) | 11 (36.7%) | 0.195 c |
| Cesarean | 14 (46.7%) | 19 (63.3%) | ||
| Feeding type (0–4 months) | Exclusive breastfeeding | 11 (36.7%) | 11 (36.7%) | 1.000 b |
| Mixed feeding | 15 (50.0%) | 16 (53.3%) | ||
| Exclusive formula feeding | 4 (13.3%) | 3 (10.0%) | ||
| Timing of complementary feeding | Not yet introduced | 1 (3.3%) | 7 (23.3%) | 0.073 b |
| Introduced at 4–6 months | 14 (46.7%) | 13 (43.3%) | ||
| Introduced after 6 months | 15 (50.0%) | 10 (33.4%) | ||
| Drinking water source | Tap water | 22 (73.3%) | 21 (70.0%) | 0.775 c |
| Other (well, rain, river) | 8 (26.7%) | 9 (30.0%) | ||
| Number of children | ≤2 children | 22 (73.3%) | 24 (80.0%) | 0.543 c |
| ≥3 children | 8 (26.7%) | 6 (20.0%) | ||
| Childcare | At home | 29 (96.7%) | 25 (83.3%) | 0.196 b |
| Daycare | 1 (3.3%) | 5 (16.7%) | ||
| WAZ | WAZ < −2 SD | 0 (0.0%) | 1 (3.3%) | 1.000 b |
| −2 SD ≤ WAZ ≤ 2 SD | 29 (96.7%) | 29 (96.7%) | ||
| WAZ > 2 SD | 1 (3.3%) | 0 (0.0%) | ||
| HAZ | HAZ < −2 SD | 2 (6.7%) | 4 (13.3%) | 0.748 b |
| −2 SD ≤ HAZ ≤ 2 SD | 26 (86.6%) | 25 (83.4%) | ||
| HAZ > 2 SD | 2 (6.7%) | 1 (3.3%) | ||
| Antibiotic use in the past 30 days, n (%) | 0 (0%) | 26 (86.7%) | - | |
| Species | Mean PG | SD PG | FDR | Patient Positive n (%) |
|---|---|---|---|---|
| Enterococcus lactis | 34.94 | 43.81 | <0.0001 | 30 (100) |
| Streptococcus thermophilus | 5.28 | 9.64 | <0.0001 | 30 (100) |
| Streptococcus parasanguinis | 3.70 | 7.16 | <0.0001 | 30 (100) |
| Staphylococcus aureus | 1.31 | 4.79 | <0.0001 | 25 (83.3) |
| Lacticaseibacillus rhamnosus | 1.28 | 4.76 | 0.002 | 19 (63.3) |
| Klebsiella oxytoca | 1.04 | 4.38 | 0.004 | 18 (60.0) |
| Escherichia ruysiae | 0.87 | 1.94 | <0.0001 | 29 (96.7) |
| Streptococcus mitis | 0.15 | 0.25 | <0.0001 | 27 (90.0) |
| Species | Mean HG | SD HG | FDR | Healthy Positive n (%) |
|---|---|---|---|---|
| Bifidobacterium pseudocatenulatum | 12.59 | 21.64 | <0.0001 | 7 (70) |
| Mediterraneibacter faecis | 6.06 | 15.81 | <0.0001 | 7 (70) |
| Akkermansia muciniphila | 4.18 | 10.74 | <0.0001 | 6 (60) |
| Bifidobacterium dentium | 3.68 | 11.20 | <0.0001 | 8 (80) |
| Enterococcus avium | 3.28 | 3.73 | <0.0001 | 10 (100) |
| Blautia luti | 1.97 | 2.23 | <0.0001 | 8 (80) |
| Lactobacillus johnsonii | 0.98 | 1.61 | <0.0001 | 6 (60) |
| Blautia argi | 0.84 | 0.96 | <0.0001 | 9 (90) |
| Eubacterium callanderi | 0.74 | 0.95 | <0.0001 | 10 (100) |
| Bifidobacterium bifidum | 0.72 | 0.95 | 0.002 | 5 (50) |
| Acetilactobacillus jinshanensis | 0.69 | 0.27 | <0.0001 | 10 (100) |
| Collinsella aerofaciens | 0.66 | 1.08 | <0.001 | 6 (60) |
| Limosilactobacillus mucosae | 0.48 | 0.88 | 0.002 | 5 (50) |
| Fusicatenibacter saccharivorans | 0.42 | 0.86 | 0.002 | 5 (50) |
| Anaerostipes hadrus | 0.33 | 0.57 | 0.002 | 5 (50) |
| Lacticaseibacillus paracasei | 0.30 | 0.58 | 0.002 | 5 (50) |
| Anaeroglobus micronuciformis | 0.26 | 0.69 | <0.001 | 7 (70) |
| Klebsiella pneumoniae 723684 | 0.15 | 0.21 | <0.001 | 9 (90) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pham, T.T.N.; Dao, T.K.; Nguyen, T.V.H.; Phung, T.B.T.; Nguyen, H.D.; Nguyen, T.Q.; Le, T.T.H.; Do, T.H. Diversity and Functional Predictions of Gut Microbiota in Vietnamese Children Aged 6–24 Months with Persistent Diarrhea of Unknown Etiology. Pathogens 2025, 14, 1136. https://doi.org/10.3390/pathogens14111136
Pham TTN, Dao TK, Nguyen TVH, Phung TBT, Nguyen HD, Nguyen TQ, Le TTH, Do TH. Diversity and Functional Predictions of Gut Microbiota in Vietnamese Children Aged 6–24 Months with Persistent Diarrhea of Unknown Etiology. Pathogens. 2025; 14(11):1136. https://doi.org/10.3390/pathogens14111136
Chicago/Turabian StylePham, Thi Thanh Nga, Trong Khoa Dao, Thi Viet Ha Nguyen, Thi Bich Thuy Phung, Hong Duong Nguyen, Thi Quy Nguyen, Thi Thu Hong Le, and Thi Huyen Do. 2025. "Diversity and Functional Predictions of Gut Microbiota in Vietnamese Children Aged 6–24 Months with Persistent Diarrhea of Unknown Etiology" Pathogens 14, no. 11: 1136. https://doi.org/10.3390/pathogens14111136
APA StylePham, T. T. N., Dao, T. K., Nguyen, T. V. H., Phung, T. B. T., Nguyen, H. D., Nguyen, T. Q., Le, T. T. H., & Do, T. H. (2025). Diversity and Functional Predictions of Gut Microbiota in Vietnamese Children Aged 6–24 Months with Persistent Diarrhea of Unknown Etiology. Pathogens, 14(11), 1136. https://doi.org/10.3390/pathogens14111136






