Occurrence and Molecular Characteristics of Polerovirus BVG Isolates from Poland
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. RNA Isolation
2.3. High-Throughput Sequencing (HTS)
2.4. Reverse Transcriptase Polymerase Chain Reaction (RT-PCR)
2.5. Duplex Immunocapture–Polymerase Chain Reactions (Duplex-IC-PCR)
2.6. DNA Sequencing
2.7. Genetic Variability Assessment and Phylogenetic Analysis
3. Result
3.1. Virus Identification by Illumina Sequencing
3.2. BVG Occurrence Assessment
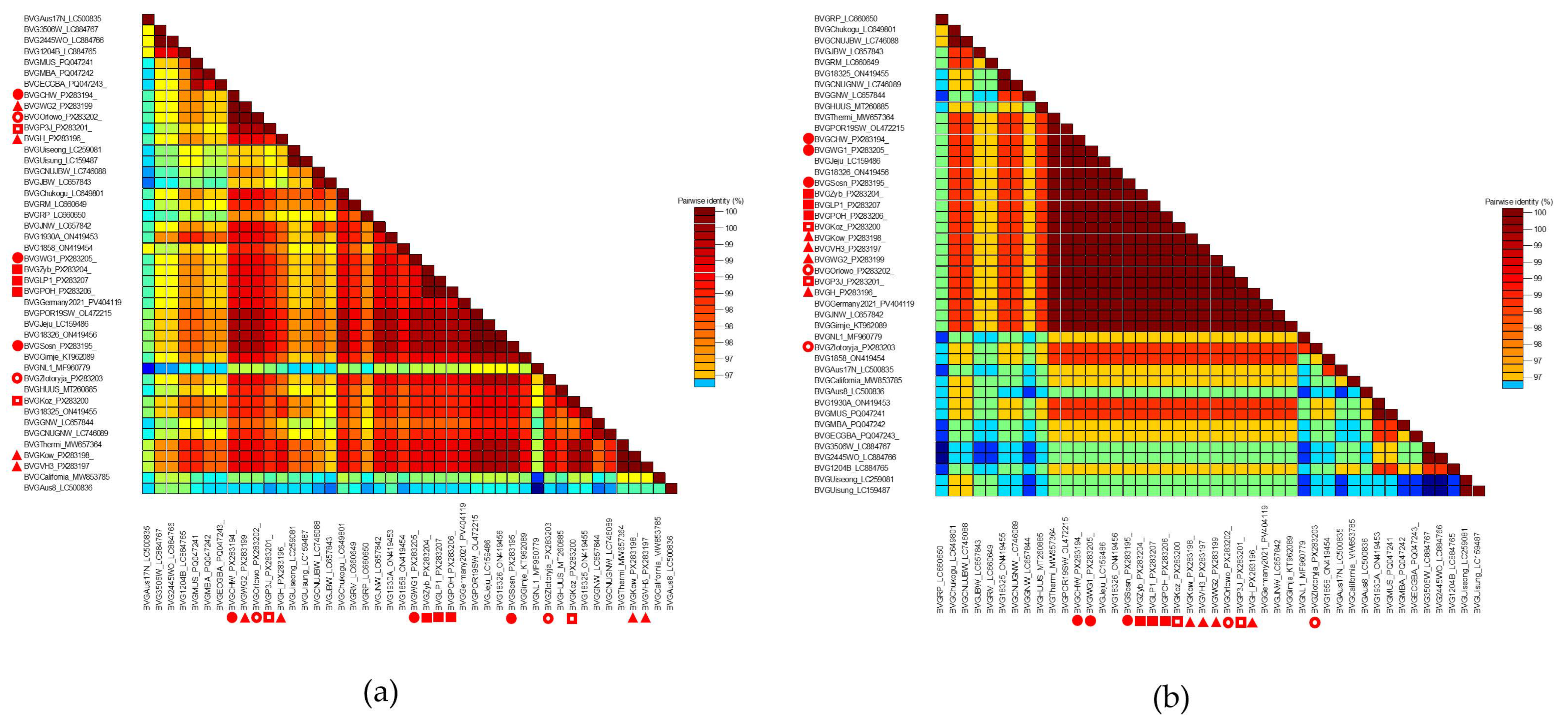
3.3. Genetic Variability of the BVG Population
3.4. Phylogenetic Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, F.; Lim, S.; Yoo, R.H.; Igori, D.; Kim, S.-M.; Kwak, D.Y.; Kim, S.L.; Lee, B.C.; Moon, J.S. The Complete Genomic Sequence of a Tentative New Polerovirus Identified in Barley in South Korea. Arch Virol. 2016, 161, 2047–2050. [Google Scholar] [CrossRef]
- Park, C.Y.; Oh, J.H.; Min, H.-G.; Lee, H.-K.; Lee, S.-H. First Report of Barley Virus G in Proso Millet (Panicum miliaceum) in Korea. Plant Dis. 2017, 101, 393. [Google Scholar] [CrossRef]
- Oh, J.; Park, C.Y.; Min, H.-G.; Lee, H.-K.; Yeom, Y.-A.; Yoon, Y.; Lee, S.-H. First Report of Barley Virus G in Foxtail Millet (Setaria italica) in Korea. Plant Dis. 2017, 101, 1061. [Google Scholar] [CrossRef]
- Kumar, L.M.; Foster, J.A.; McFarland, C.; Malapi-Wight, M. First Report of Barley Virus G in Switchgrass (Panicum virgatum). Plant Dis. 2018, 102, 466. [Google Scholar] [CrossRef]
- Wamaitha, M.J.; Nigam, D.; Maina, S.; Stomeo, F.; Wangai, A.; Njuguna, J.N.; Holton, T.A.; Wanjala, B.W.; Wamalwa, M.; Lucas, T.; et al. Metagenomic Analysis of Viruses Associated with Maize Lethal Necrosis in Kenya. Virol. J. 2018, 15, 90. [Google Scholar] [CrossRef] [PubMed]
- Nancarrow, N.; Aftab, M.; Zheng, L.; Maina, S.; Freeman, A.; Rodoni, B.; Spackman, M.; Trębicki, P. First Report of Barley Virus G in Australia. Plant Dis. 2019, 103, 1799. [Google Scholar] [CrossRef]
- Pasztor, G.; Galbacs, N.Z.; Kossuth, T.; Demian, E.; Nadasy, E.; Takacs, A.P.; Varallyay, E. Millet Could Be Both a Weed and Serve as a Virus Reservoir in Crop Fields. Plants 2020, 9, 954. [Google Scholar] [CrossRef]
- Erickson, A.; Falk, B.W. First Report of Barley Virus G in the United States and California. Plant Dis. 2021, 105, 3312. [Google Scholar] [CrossRef]
- Gavrili, V.; Lotos, L.; Mollov, D.; Grinstead, S.; Tsialtas, I.T.; Katis, N.I.; Maliogka, V.I. First Report of Barley Virus G Infecting Corn in Greece. J. Plant Pathol. 2021, 103, 1331. [Google Scholar] [CrossRef]
- Svanella-Dumas, L.; Vitry, C.; Valade, R.; Robin, N.; Thibord, J.B.; Marais, A.; Candresse, T. First Report of Barley Virus G Infecting Winter Barley (Hordeum vulgare) in France. Plant Dis. 2023, 107, 591. [Google Scholar] [CrossRef]
- Ohki, T. First Report of Barley Virus G Infecting Wheat (Triticum aestivum) in Japan. Plant Dis. 2023, 107, 2569. [Google Scholar] [CrossRef]
- Pasha, A.; Schrader, G.; Ziebell, H. Virus and Viroid Diversity in Hops, Investigating the German Hop Virome. PLoS ONE 2025, 20, e0329289. [Google Scholar] [CrossRef] [PubMed]
- Rivarez, M.P.S.; Pecman, A.; Bačnik, K.; Maksimović, O.; Vučurović, A.; Seljak, G.; Mehle, N.; Gutiérrez-Aguirre, I.; Ravnikar, M.; Kutnjak, D. In-Depth Study of Tomato and Weed Viromes Reveals Undiscovered Plant Virus Diversity in an Agroecosystem. Microbiome 2023, 11, 60. [Google Scholar] [CrossRef]
- Nancarrow, N.; Maina, S.; Zheng, L.; Aftab, M.; Freeman, A.; Kinoti, W.M.; Rodoni, B.; Trębicki, P. Coding-Complete Sequences of Barley Virus G Isolates from Australia, Obtained from a 34-Year-Old and a 1-Year-Old Sample. Microbiol. Resour. Announc. 2019, 8, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Erickson, A.; Falk, B. Barley Virus G (Barley Virus G). 2022. Available online: https://www.cabidigitallibrary.org/doi/abs/10.1079/cabicompendium.64866492 (accessed on 15 May 2025).
- Erickson, A.; Jiang, J.; Kuo, Y.-W.; Falk, B.W. Construction and Use of an Infectious cDNA Clone to Identify Aphid Vectors and Susceptible Monocot Hosts of the Polerovirus Barley Virus G. Virology 2023, 579, 178–185. [Google Scholar] [CrossRef]
- Sõmera, M.; Fargette, D.; Hébrard, E.; Sarmiento, C. ICTV Report Consortium. ICTV Virus Taxonomy Profile: Solemoviridae 2021: This Article Is Part of the ICTV Virus Taxonomy Profiles Collection. J. Gen. Virol. 2021, 102, 001707. [Google Scholar] [CrossRef]
- Lee, H.-J.; Kim, S.-M.; Jeong, R.-D. Analysis of Wheat Virome in Korea Using Illumina and Oxford Nanopore Sequencing Platforms. Plants 2023, 12, 2374. [Google Scholar] [CrossRef]
- Minicka, J.; Taberska, A.; Borodynko-Filas, N.; Kaźmińska, K.; Bartoszewski, G.; Hasiów-Jaroszewska, B. Viruses Infecting Capsicum Crops in Poland and Molecular Characterization of Newly Detected Bell Pepper Alphaendornavirus (BPEV). Crop Prot. 2024, 176, 106478. [Google Scholar] [CrossRef]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for General Users and for Biologist Programmers. In Bioinformatics Methods and Protocols; Misener, S., Krawetz, S.A., Eds.; Humana Press: Totowa, NJ, USA, 1999; pp. 365–386. [Google Scholar] [CrossRef]
- Trzmiel, K. Identification of Barley Yellow Dwarf Viruses in Poland. J. Plant Pathol. 2017, 99, 493–497. [Google Scholar]
- Trzmiel, K.; Hasiów-Jaroszewska, B. Molecular Characteristics of Barley Yellow Dwarf Virus—PAS—The Main Causal Agent of Barley Yellow Dwarf Disease in Poland. Plants 2023, 12, 3488. [Google Scholar] [CrossRef]
- Kundu, J.K. First Report of Barley Yellow Dwarf Virus-PAS in Wheat and Barley Grown in the Czech Republic. Plant Dis. 2008, 92, 1587. [Google Scholar] [CrossRef] [PubMed]
- Kundu, J.K.; Jarošová, J.; Gadiou, S.; Červená, G. Discrimination of Three BYDV Species by One-Step RT-PCR-RFLP and Sequence Based Methods in Cereal Plants from the Czech Republic. Cereal Res. Commun. 2009, 37, 541–550. [Google Scholar] [CrossRef]
- Strażyński, P.; Ruszkowska, M.; Jeżewska, M.; Trzmiel, K. Evaluation of The Autumn Infection of Winter Barley with Barley Yellow Dwarf Viruses Transmitted by Anholocyclic Forms of Bird Cherry-Oat Aphid Rhopalosiphum padi L. in Poland. J. Plant Prot. Res. 2011, 51, 314–321. [Google Scholar] [CrossRef]
- Canning, E.S.G.; Penrose, M.J.; Barker, I.; Coates, D. Improved Detection of Barley Yellow Dwarf Virus in Single Aphids Using RT-PCR. J. Virol. Methods 1996, 56, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Trzmiel, K. Occurrence of Wheat Dwarf Virus and Barley Yellow Dwarf Virus Species in Poland in the Spring of 2019. J. Plant Prot. Res. 2020, 60, 345–350. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. In Nucleic Acids Symposium Series; Oxford University Press: Oxford, UK, 1999; Volume 41, pp. 95–98. [Google Scholar]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A Virus Classification Tool Based on Pairwise Sequence Alignment and Identity Calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and Analysis of Recombination Patterns in Virus Genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Trzmiel, K.; Szydło, W.; Hasiów-Jaroszewska, B. Biological and Molecular Characterisation of the Two Polish Wheat Streak Mosaic Virus Isolates and Their Transmission by Wheat Curl Mites. Plant Prot. Sci. 2021, 57, 171–178. [Google Scholar] [CrossRef]
- Kreuze, J.F.; Perez, A.; Untiveros, M.; Quispe, D.; Fuentes, S.; Barker, I.; Simon, R. Complete Viral Genome Sequence and Discovery of Novel Viruses by Deep Sequencing of Small RNAs: A Generic Method for Diagnosis, Discovery and Sequencing of Viruses. Virology 2009, 388, 1–7. [Google Scholar] [CrossRef]
- Maina, S.; Donovan, N.J.; Plett, K.; Bogema, D.; Rodoni, B.C. High-Throughput Sequencing for Plant Virology Diagnostics and Its Potential in Plant Health Certification. Front. Hortic. 2024, 3, 1388028. [Google Scholar] [CrossRef]
- Maree, H.J.; Fox, A.; Al Rwahnih, M.; Boonham, N.; Candresse, T. Application of HTS for Routine Plant Virus Diagnostics: State of the Art and Challenges. Front. Plant Sci. 2018, 9, 1082. [Google Scholar] [CrossRef]
- Singh, K.; Jarošová, J.; Fousek, J.; Chen, H.; Kundu, J.K. Virome Identification in Wheat in the Czech Republic Using Small RNA Deep Sequencing. J. Integr. Agric. 2020, 19, 1825–1833. [Google Scholar] [CrossRef]
- Sõmera, M.; Massart, S.; Tamisier, L.; Sooväli, P.; Sathees, K.; Kvarnheden, A. A Survey Using High-Throughput Sequencing Suggests That the Diversity of Cereal and Barley Yellow Dwarf Viruses Is Underestimated. Front. Microbiol. 2021, 12, 673218. [Google Scholar] [CrossRef]
- Byrne, S.; Schughart, M.; Ballandras, V.; Carolan, J.C.; Sheppard, L.; McNamara, L. The First Survey Using High-Throughput Sequencing of Cereal and Barley Yellow Dwarf Viruses in Irish Spring and Winter Barley Crops. Ir. J. Agric. Food Res. 2024, 63, 1–16. [Google Scholar] [CrossRef]
- Ranabhat, N.B.; Fellers, J.P.; Bruce, M.A.; Rupp, J.L.S. Brome Mosaic Virus Detected in Kansas Wheat Co-Infected with Other Common Wheat Viruses. Front. Plant Sci. 2023, 14, 1096249. [Google Scholar] [CrossRef]
- Ilbağı, H.; Masonbrink, R.; Miller, W.A. The Nearly Complete Genome Sequence of Clade B, Wheat Streak Mosaic Virus Isolate from Türkiye. J. Plant Dis. Prot. 2024, 131, 1755–1759. [Google Scholar] [CrossRef]
- Leybourne, D.J. How Does Vector Diversity Influence the Transmission Efficiency of Yellow Dwarf Virus? Perspectives from a Review. Plant Pathol. 2024, 73, 1042–1059. [Google Scholar] [CrossRef]
- Delfosse, V.C.; Barrios Barón, M.P.; Distéfano, A.J. What We Know about Poleroviruses: Advances in Understanding the Functions of Polerovirus Proteins. Plant Pathol. 2021, 70, 1047–1061. [Google Scholar] [CrossRef]
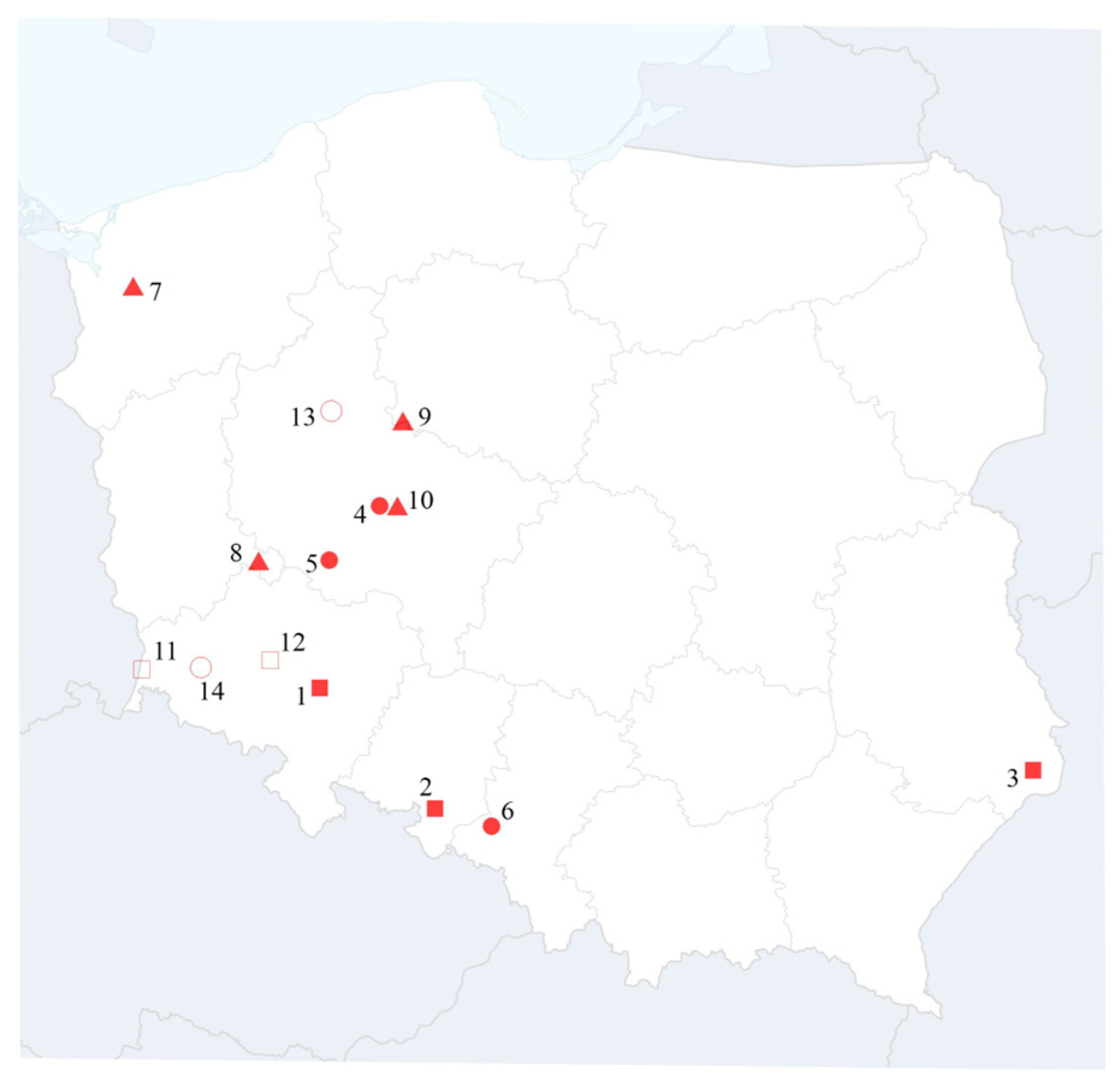


| Name of Isolates | Host | Co-Infection | Geographical Origin | Date of Collection | GenBank Accession Number |
|---|---|---|---|---|---|
| BVG-WG1 | barley | BVG/BYDV-PAS/BYDV-PAV/WDV | Poland: Greater Poland | 2023 | PX283205 |
| BVG-CHW | barley | BVG/BYDV-PAS | Poland: Greater Poland | 2023 | PX283194 |
| BVG-Sosn | barley | BVG/BYDV-PAS | Poland: Silesia | 2023 | PX283195 |
| BVG-Zyb | barley | BVG/BYDV-PAS | Poland: Lower Silesia | 2024 | PX283204 |
| BVG-POH | wheat | BVG/BYDV-PAS | Poland: Opole | 2024 | PX283206 |
| BVG-LP1 | wheat | BVG/WDV | Poland: Lublin | 2024 | PX283207 |
| BVG-H | barley | BVG/BYDV-PAS | Poland: Western Pomerania | 2021 | PX283196 |
| BVG-VH3 | barley | BVG/BYDV-PAS | Poland: Kujavia and Pomerania | 2021 | PX283197 |
| BVG-Kow | barley | BVG/BYDV-PAS | Poland: Lubusz | 2021 | PX283198 |
| BVG-WG2 | barley | BVG/BYDV-PAS | Poland: Greater Poland | 2021 | PX283199 |
| BVG-Koz | barley | BVG/BYDV-PAS | Poland: Lower Silesia | 2019 | PX283200 |
| BVG-P3J | wheat | BVG/BYDV-PAS | Poland: Lower Silesia | 2019 | PX283201 |
| BVG-Orlowo | barley | BVG/BYDV-PAS | Poland: Greater Poland | 2015 | PX283202 |
| BVG-Zlotoryja | barley | BVG/BYDV-PAS | Poland: Lower Silesia | 2015 | PX283203 |
| BVG-S-RM | corn leaf aphid | BVG/BYDV-PAS | Poland: Silesia | 2023 | PX378101 |
| BVG-S-SA | English grain aphid | BVG/BYDV-PAS | Poland: Silesia | 2023 | PX378102 |
| Host Plant | Number of Total Raw Reads | Percent of Reference Genome Covered by Reads | Number of Reads Mapped to Corresponding Reference Sequence from Viral RefSeq | Average Depth of Coverage for Corresponding Viral Species | Name of Identified Virus |
|---|---|---|---|---|---|
| barley | 48,743,038 | 87% | 4890 | 235.00 | BVG |
| barley | 48,743,038 | 95.7% | 1445 | 22.72 | WSMV |
| barley | 48,743,038 | 96.7% | 360,188 | 9030.39 | BYDV-PAV |
| barley | 48,743,038 | 94.3% | 822,505 | 21,152.19 | BYDV-PAS |
| barley | 48,743,038 | 99.9% | 66,402 | 3530.23 | WDV |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trzmiel, K.; Zarzyńska-Nowak, A.; Hasiów-Jaroszewska, B. Occurrence and Molecular Characteristics of Polerovirus BVG Isolates from Poland. Pathogens 2025, 14, 1087. https://doi.org/10.3390/pathogens14111087
Trzmiel K, Zarzyńska-Nowak A, Hasiów-Jaroszewska B. Occurrence and Molecular Characteristics of Polerovirus BVG Isolates from Poland. Pathogens. 2025; 14(11):1087. https://doi.org/10.3390/pathogens14111087
Chicago/Turabian StyleTrzmiel, Katarzyna, Aleksandra Zarzyńska-Nowak, and Beata Hasiów-Jaroszewska. 2025. "Occurrence and Molecular Characteristics of Polerovirus BVG Isolates from Poland" Pathogens 14, no. 11: 1087. https://doi.org/10.3390/pathogens14111087
APA StyleTrzmiel, K., Zarzyńska-Nowak, A., & Hasiów-Jaroszewska, B. (2025). Occurrence and Molecular Characteristics of Polerovirus BVG Isolates from Poland. Pathogens, 14(11), 1087. https://doi.org/10.3390/pathogens14111087








