Application of Machine Learning Algorithms in Urinary Tract Infections Diagnosis Based on Non-Microbiological Parameters
Abstract
1. Introduction
2. Materials and Methods
2.1. Population, Sample and Source of Information
2.2. Variables and Statistical Analysis
- Descriptive Analysis: Qualitative variables were summarized using frequencies and percentages, while quantitative variables were described using means and standard deviations (for symmetric distributions) or medians and interquartile ranges (for asymmetric distributions).
- Estimation of the Percentage of Positive Urine Cultures (positive urine cultures divided by the total number of urine cultures). This was accompanied by a 95% confidence interval.
- Bivariate Analysis: Associations between quantitative variables and urine culture results were tested using Student’s t-test (for equal variances) or Welch’s t-test (for unequal variances). Associations with qualitative variables were assessed using Pearson’s chi-square test. Statistical significance was set at p < 0.05.
- Machine Learning Algorithm Application:
- Variable Recoding: All categorical variables were previously converted to dichotomous (positive-negative), except for months and sample type that were recoded as dummy variables.
- Dataset Splitting: The dataset was divided into a training set (80%) for model development and a test set (20%) for validation, ensuring sufficient data for both stages to enhance model generalizability [12].
- Missing Data Imputation: Missing values were imputed in the training set using the k-Nearest Neighbors (kNN) method, which identifies the k closest neighbors based on dataset variables and imputes missing values using the mean. Subsequently, imputation was performed on the test set using the imputation model learned on the training set.
- Variable Normalization: To ensure all variables contributed equally to the model and to mitigate differences in measurement scales, quantitative values were rescaled between 0 and 1.
- Pearson Correlation Analysis: Highly correlated variables (r > 0.7) were removed to reduce redundancy.
- Variable Importance Assessment: The most relevant variables for the training set were selected using the Random Forest method with cross validation with 10 folds.
- Machine Learning Algorithm Implementation: The following widely used algorithms were applied (the cross-validation method with 10 iterations of resampling was used for training). All the algorithms were initialized with the same seed to avoid reproducibility:
- ○
- Logistic Regression—A classification model predicting binary outcomes using a logistic function.
- ○
- k-Nearest Neighbors (kNN)—Classifies a data point based on the majority class of its k nearest neighbors using a distance metric.
- ○
- Decision Trees—Splits data into successive decisions based on feature values, forming a tree structure.
- ○
- Support Vector Machine (SVM)—Identifies the hyperplane that maximizes class separation.
- ○
- Random Forest—An ensemble method averaging predictions from multiple decision trees.
- ○
- Gradient Boosting Machine (GBM)—A sequential model where each iteration corrects the errors of the previous one.
- ○
- Artificial Neural Network (NNet)—A multi-layered model that processes data hierarchically through interconnected nodes (neurons).
- Model Prediction and Validation: Performance was evaluated using a confusion matrix (true positives, true negatives, false positives, and false negatives) in the test set.
- Accuracy Calculation: Defined as the proportion of correct predictions over the total number of predictions.
- Sensitivity, Specificity, and ROC Curve Analysis: Performance comparisons among models were conducted using receiver operating characteristic (ROC) curves, with sensitivity and specificity reported alongside 95% confidence intervals.
2.3. Ethical Considerations
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| UTI | Urinary Tract Infection |
| AI | Artificial Intelligent |
| EPINE | Nosocomial Infection Prevalence Study (Spain) |
| CFU | Colony Forming Units |
| RBC | Red Blood Cell Series |
| WBC | White Blood Cell Series |
| PS | Platelet Series |
| RF | Random Forest |
| SVM | Support Vector Machine |
| GBM | Gradient Boost Machine |
| NNet | Neural Network |
| LR | Logistic Regression |
| Tree | Decision Tree |
| kNN | k Nearest-Neighbourg |
| CI95% | 95% Confidence Interval |
| ROC | Receiver Operating Characteristic curve |
| AUC | Area Under ROC curve |
References
- Medina Polo, J.; Arribi Vilela, A.; Candel González, F.J.; Salinas Casado, J. Actualización de la Infección Urinaria en Urología. Available online: https://www.aeu.es/UserFiles/files/ManualInfeccionesUrinarias.pdf (accessed on 8 October 2024).
- Yang, X.; Chen, H.; Zheng, Y.; Qu, S.; Wang, H.; Yi, F. Disease burden and long-term trends of urinary tract infections: A worldwide report. Front. Public Health 2022, 10, 888205. [Google Scholar] [CrossRef]
- Sociedad Española de Medicina Preventiva, Salud Pública y Gestión Sanitaria. Prevalencia de Infecciones (Relacionadas con la Asistencia Sanitaria y Comunitarias) y uso de Antimicrobianos en Hospitales de Agudos; Sociedad Española de Medicina Preventiva, Salud Pública y Gestión Sanitaria: Madrid, Spain, 2024. [Google Scholar]
- Sociedad Española de Medicina Preventiva, Salud Pública y Gestión Sanitaria. Informe Preliminar Epine Año 2024; H. Universitario Virgen de las Nieves: Granada, Spain, 2024. [Google Scholar]
- Dai, Y.; Sullivan, B.; Montout, A.; Dillon, A.; Waller, C.; Acs, P.; Denholm, R.; Williams, P.; Hay, A.D.; Santos-Rodriguez, R.; et al. Explainable AI for Classifying UTI Risk Groups Using a Real-World Linked EHR and Pathology Lab Dataset. arXiv 2025, arXiv:2411.17645. [Google Scholar] [CrossRef]
- Park, J.I.; Bliss, D.Z.; Chi, C.-L.; Delaney, C.W.; Westra, B.L. Knowledge Discovery with Machine Learning for Hospital-Acquired Catheter-Associated Urinary Tract Infections. Comput. Inform. Nurs. 2020, 38, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, R.B.; Velásquez, J.D. Inteligencia artificial al servicio de la salud del futuro. Rev. Med. Clin. Condes 2022, 34, 84–91. [Google Scholar] [CrossRef]
- Naik, N.; Talyshinskii, A.; Shetty, D.K.; Hameed, B.M.Z.; Zhankina, R.; Somani, B.K. Smart Diagnosis of Urinary Tract Infections: Is Artificial Intelligence the Fast-Lane Solution? Curr. Urol. Rep. 2024, 25, 37–47. [Google Scholar] [CrossRef]
- Goździkiewicz, N.; Zwolińska, D.; Polak-Jonkisz, D. The Use of Artificial Intelligence Algorithms in the Diagnosis of Urinary Tract Infections-A Literature Review. J. Clin. Med. 2022, 11, 2734. [Google Scholar] [CrossRef]
- Rojo, M.D.; Bautista, M.F.; Gutiérrez-Fernández, J. Procedimiento Normalizado de Trabajo. Cultivo Cuantitativo de Orina para Estudio de Microorganismos Aerobios/Facultativos de Crecimiento Rápido (Vol. 8). PNT-OR-01. Available online, 2015. Available online: https://doi.org/10.6084/m9.figshare.1149879 (accessed on 4 March 2025).
- Rodríguez Del Águila, M.M.; Sorlózano-Puerto, A.; Fernández-Sierra, M.A.; Navarro Marí, J.M.; Gutiérrez Fernández, J. Características sociodemográficas y factores de riesgo asociados a las bacteriurias significativas en un área de salud del sudeste español [Sociodemographic characteristics and risk factors associated to significative bacteriuria in a Spanish health area]. Rev. Española Quimioter. 2022, 35, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Gholamy, A.; Kreinovich, V.; Kosheleva, O. Why 70/30 or 80/20 Relation Between Training and Testing Sets: A Pedagogical Explanation. Departmental Technical Reports (CS). 2018. Available online: https://scholarworks.utep.edu/cs_techrep/1209 (accessed on 17 March 2025).
- Enshaeifar, S.; Zoha, A.; Skillman, S.; Markides, A.; Acton, S.T.; Elsaleh, T.; Kenny, M.; Rostill, H.; Nilforooshan, R.; Barnaghi, P. Machine learning methods for detecting urinary tract infection and analysing daily living activities in people with dementia. PLoS ONE 2019, 14, e0209909. [Google Scholar] [CrossRef] [PubMed]
- Ozkan, I.A.; Koklu, M.; Sert, I.U. Diagnosis of urinary tract infection based on artificial intelligence methods. Comput. Methods Programs Biomed. 2018, 166, 51–59. [Google Scholar] [CrossRef]
- Aydin, D.B.; Er, O. A new proposal for early stage diagnosis of urinary tract infection using computers aid systems. Sak. Univ. J. Comput. Inf. Sci. 2018, 1, 1–9. [Google Scholar]
- Gadalla, A.A.H.; Friberg, I.M.; Kift-Morgan, A.; Zhang, J.; Eberl, M.; Topley, N.; Weeks, I.; Cuff, S.; Wootton, M.; Gal, M.; et al. Identification of clinical and urine biomarkers for uncomplicated urinary tract infection using machine learning algorithms. Sci. Rep. 2019, 9, 19694. [Google Scholar] [CrossRef]
- Taylor, R.A.; Moore, C.L.; Cheung, K.-H.; Brandt, C. Predicting urinary tract infections in the emergency department with machine learning. PLoS ONE 2018, 13, e0194085. [Google Scholar] [CrossRef] [PubMed]
- Burton, R.J.; Albur, M.; Eberl, M.; Cuff, S.M. Using artificial intelligence to reduce diagnostic workload without compromising detection of urinary tract infections. BMC Med. Inform. Decis. Mak. 2019, 19, 171. [Google Scholar] [CrossRef]
- Tartar, A.S.; Balin, S.O. Geriatric urinary tract infections: The value of laboratory parameters in estimating the need for bacteremia and Intensive Care Unit. Pak. J. Med. Sci. 2019, 35, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Sukri, M. Correlation between Hematologic Parameters and Asymptomatic Bacteriuria in the Full-Term Pregnant Patients in Dr. Kariadi Hospital, Semarang, Indonesia. Diponegoro Int. Med. J. 2022, 3, 7–13. [Google Scholar] [CrossRef]
- Nyman, J. Machine Learning Approaches for Detection of Urinary Tract Infections. OpenAIRE—Explore. Available online: https://explore.openaire.eu/search/publication?articleId=od_______264::cd9221b3567c3df8acb3b5d09a4a89b1 (accessed on 9 June 2023).
- Dedeene, L.; Van Elslande, J.; Dewitte, J.; Martens, G.; De Laere, E.; De Jaeger, P.; De Smet, D. An artificial intelligence-driven support tool for prediction of urine culture test results. Clin. Chim. Acta 2024, 562, 119854. [Google Scholar] [CrossRef]
- Del Ben, F.; Da Col, G.; Cobârzan, D.; Turetta, M.; Rubin, D.; Buttazzi, P.; Antico, A. A fully interpretable machine learning model for increasing the effectiveness of urine screening. Am. J. Clin. Pathol. 2023, 160, 620–632. [Google Scholar] [CrossRef]
- Choi, M.H.; Kim, D.; Bae, H.G.; Kim, A.-R.; Lee, M.; Lee, K.; Lee, K.-R.; Jeong, S.H. Predictive performance of urinalysis for urine culture results according to causative microorganisms: An integrated analysis with artificial intelligence. J. Clin. Microbiol. 2024, 62, e0117524. [Google Scholar] [CrossRef]
- Choi, M.H.; Kim, D.; Park, Y.; Jeong, S.H. Development and validation of artificial intelligence models to predict urinary tract infections and secondary bloodstream infections in adult patients. J. Infect. Public Health 2024, 17, 10–17. [Google Scholar] [CrossRef]
- Domingos, P. A few useful things to know about machine learning. Commun. ACM 2012, 55, 78–87. [Google Scholar] [CrossRef]
- Moreland, R.B.; Brubaker, L.; Wolfe, A.J. Polymicrobial urine cultures: Reconciling contamination with the urobiome while recognizing the pathogens. Front. Cell. Infect. Microbiol. 2025, 15, 1562687. [Google Scholar] [CrossRef] [PubMed]

| Independent Variables | White Blood Cell Series (WBC) |
|---|---|
| Age (years) *† | White blood cell count *† |
| Sex (male, female) * | Neutrophil count |
| Sample type (spontaneous miction, permanent catheter, temporary urethral catheter) * | Neutrophil percentage |
| Origin (hospital, community) *† | Lymphocyte count *† |
| Number of isolations (only in positive urine cultures) *† | Lymphocyte percentage *† |
| Month of urine culture collection (January to December) * | Monocyte count *† |
| Variables from urine dipstick test | Monocyte percentage * |
| Glucose (mg/dL) * | Basophil count * |
| Bilirubin * | Basophil percentage *† |
| Ketone bodies (mg/dL) * | Eosinophil count *† |
| Density * | Eosinophil percentage |
| Ph * | Immature granulocyte count |
| Hematuria (negative, +, ++, +++, 4+ and 5+) *† | Immature granulocyte percentage * |
| Proteins (mg/dL) *† | Platelet Series (PS) |
| Urobilinogen (mg/dL) * | Platelet count * |
| Nitrites (negative, positive) *† | Mean platelet volume *† |
| Leukocytes (negative, +, ++, +++, 4+ and 5+) *† | Other Blood Parameters |
| Red Blood Cell Series (RBC) | Creatinine * |
| Red blood cell count * | Hemolytic index * |
| Mean corpuscular volume * | Icteric index * |
| Red cell distribution width index *† | Lipemic index * |
| Hematocrit | |
| Hemoglobin | |
| Mean corpuscular hemoglobin | |
| Mean corpuscular hemoglobin concentration * |
| Variable | n | Total Sample | Positive Urine Culture (38.6%) | Negative Urine Culture (61.4%) | p-Value |
|---|---|---|---|---|---|
| Independent variables | |||||
| Age | 4283 | 56.4 ± 23.1 | 59.0 ± 24.0 | 54.7 ± 22.4 | <0.001 |
| Sex (woman) | 4283 | 2204 (51.5) | 951 (57.5) | 1253 (47.7) | <0.001 |
| Spontaneous miction | 4283 | 3589 (83.8) | 1271 (76.8) | 2318 (88.2) | <0.001 |
| Temporary urethral catheter | 484 (11.3) | 246 (14.9) | 238 (9.1) | ||
| Permanent catheter | 210 (4.9) | 138 (8.3) | 72 (2.7) | ||
| Origin (hospital) | 4283 | 347 (8.1) | 112 (6.8) | 235 (8.9) | <0.05 |
| January | 4283 | 412 (9.6) | 138 (8.3) | 274 (10.4) | 0.068 |
| February | 408 (9.5) | 162 (9.8) | 246 (9.4) | ||
| March | 370 (8.6) | 140 (8.5) | 230 (8.8) | ||
| April | 379 (8.8) | 139 (8.4) | 240 (9.1) | ||
| May | 390 (9.1) | 145 (8.8) | 245 (9.3) | ||
| June | 388 (9.1) | 143 (8.6) | 245 (9.3) | ||
| July | 319 (7.4) | 130 (7.9) | 189 (7.2) | ||
| August | 293 (6.8) | 126 (7.6) | 167 (6.4) | ||
| September | 321 (7.5) | 149 (9.0) | 172 (6.5) | ||
| October | 359 (8.4) | 134 (8.1) | 225 (8.6) | ||
| November | 361 (8.4) | 134 (8.1) | 227 (8.6) | ||
| December | 283 (6.4) | 115 (6.9) | 168 (6.4) | ||
| Urine dipstick test | |||||
| Glucose | 4272 | 0.275 | |||
| 0 mg/dL | 3863 (90.4) | 1474 (89.3) | 2389 (91.1) | ||
| 1–50 mg/dL | 139 (3.3) | 61 (3.7) | 78 (3.0) | ||
| 51–100 mg/dL | 85 (2.0) | 35 (2.1) | 50 (1.9) | ||
| >100 mg/dL | 185 (4.3) | 80 (4.9) | 105 (4.0) | ||
| Bilirrubin | 4270 | 0.2 ± 0.6 | 0.24 ± 0.56 | 0.23 ± 0.58 | 0.643 |
| Ketone bodies | 4269 | 0.267 | |||
| 0 mg/dL | 3415 (80.0) | 1297 (78.7) | 2118 (80.8) | ||
| 1–20 mg/dL | 643 (15.1) | 274 (16.6) | 369 (14.1) | ||
| 21–40 mg/dL | 19 (0.4) | 7 (0.4) | 12 (0.5) | ||
| 41–60 mg/dL | 109 (2.6) | 41 (2.5) | 68 (2.6) | ||
| >60 mg/dL | 83 (1.9) | 30 (1.8) | 53 (2.0) | ||
| Density | 3962 | 1.018 ± 0.007 | 1.017 ± 0.007 | 1.018 ± 0.008 | <0.001 |
| pH | 4270 | 6.01 ± 0.95 | 6.06 ± 0.97 | 5.98 ± 0.94 | <0.05 |
| Hematuria * | 4249 | <0.001 | |||
| Negative | 1267 (29.8) | 268 (16.3) | 999 (38.3) | ||
| Positive + | 1111 (26.1) | 412 (25.1) | 699 (26.8) | ||
| Positive 2+ | 385 (9.1) | 164 (10.0) | 221 (8.5) | ||
| Positive 3+ | 469 (11.0) | 260 (15.8) | 209 (8.0) | ||
| Positive 4+ and 5+ | 1017 (23.9) | 540 (32.8) | 477 (18.3) | ||
| Proteins | 4270 | <0.001 | |||
| 0 mg/dL | 1887 (44.2) | 498 (30.2) | 1389 (53.0) | ||
| 1–50 mg/dL | 1254 (29.4) | 514 (31.2) | 740 (28.2) | ||
| 51–100 mg/dL | 617 (14.4) | 348 (21.1) | 269 (10.3) | ||
| >100 mg/dL | 512 (12.0) | 289 (17.5) | 223 (8.5) | ||
| Urobilinogen | 4270 | 0.42 ± 1.34 | 0.36 ± 1.20 | 0.46 ± 1.42 | <0.05 |
| Nitrites (positive) | 4270 | 722 (16.9) | 576 (34.9) | 146 (5.6) | <0.001 |
| Leukocytes * | 4270 | <0.001 | |||
| Negative | 1555 (36.4) | 202 (12.2) | 1353 (51.6) | ||
| Positive + | 64 (1.5) | 22 (1.3) | 42 (1.6) | ||
| Positive 2+ | 681 (15.9) | 168 (10.2) | 513 (19.6) | ||
| Positive 3+ | 595 (13.9) | 266 (16.1) | 329 (12.6) | ||
| Positive 4+ and 5+ | 1375 (32.2) | 991 (60.1) | 384 (14.7) | ||
| Red blood Cell Series (RBC) | |||||
| Red blood cell count | 4.44 ± 0.75 | 4.35 ± 0.73 | 4.49 ± 0.74 | <0.001 | |
| Mean corpuscular volumen | 4218 | 88.43 ± 7.24 | 88.36 ± 7.69 | 88.47 ± 6.94 | 0.636 |
| Red cell distribution width index | 4215 | 14.19 ± 2.24 | 14.37 ± 2.31 | 14.07 ± 2.18 | <0.001 |
| Hematocrit | 4218 | 39.08 ± 6.19 | 38.31 ± 6.13 | 39.57 ± 6.19 | <0.001 |
| Hemoglobin | 4218 | 13.00 ± 2.20 | 12.69 ± 2.18 | 13.19 ± 2.20 | <0.001 |
| Mean corpuscular hemoglobin | 4218 | 29.39 ± 2.77 | 29.26 ± 2.89 | 29.48 ± 2.69 | <0.05 |
| Mean corpuscular hemoglobin concentration | 4218 | 33.20 ± 1.83 | 33.04 ± 2.02 | 33.30 ± 1.70 | <0.001 |
| White Blood Cell Series (WBC) | |||||
| White blood cell count | 4218 | 10.30 ± 6.18 | 11.6 ± 6.96 | 9.47 ± 5.48 | <0.001 |
| Neutrophil count | 4218 | 7.55 ± 5.01 | 8.73 ± 5.29 | 6.81 ± 4.68 | <0.001 |
| Neutrophil percentage | 4218 | 70.23 ± 15.14 | 73.04 ± 14.70 | 68.45 ± 15.14 | <0.001 |
| Lymphocyte count | 4218 | 1.77 ± 2.29 | 1.81 ± 3.25 | 1.75 ± 1.38 | 0.465 |
| Lymphocyte percentage | 4218 | 19.72 ± 12.59 | 17.28 ± 12.11 | 21.26 ± 12.65 | <0.001 |
| Monocyte count | 4218 | 0.80 ± 1.05 | 0.89 ± 0.79 | 0.75 ± 1.18 | <0.001 |
| Monocyte percentage | 4218 | 8.06 ± 4.23 | 7.85 ± 4.08 | 8.20 ± 4.32 | <0.05 |
| Basophil count | 4218 | 0.04 ± 0.04 | 0.04 ± 0.06 | 0.04 ± 0.02 | <0.01 |
| Basophil percentage | 4218 | 0.42 ± 0.30 | 0.40 ± 0.31 | 0.44 ± 0.29 | <0.001 |
| Eosinophil count | 4218 | 0.11 ± 0.15 | 0.10 ± 0.13 | 0.12 ± 0.16 | <0.001 |
| Eosinophil percentage | 4218 | 1.36 ± 1.70 | 1.14 ± 1.48 | 1.51 ± 1.81 | <0.001 |
| Immature granulocyte count | 4050 | 0.11 ± 0.66 | 0.15 ± 0.99 | 0.08 ± 0.30 | <0.01 |
| Immature granulocyte percentage | 4058 | 0.80 ± 1.78 | 0.91 ± 2.17 | 0.74 ± 1.47 | <0.01 |
| Platalet Series (PS) | |||||
| Platelet count | 4218 | 241.37 ± 102.50 | 250.45 ± 109.90 | 235.63 ± 97.12 | <0.001 |
| Mean platelet volumen | 4139 | 10.69 ± 1.21 | 10.67 ± 1.21 | 10.69 ± 1.22 | 0.620 |
| Other blood parameters | |||||
| Creatinine | 4148 | 1.18 ± 1.05 | 1.24 ± 1.03 | 1.15 ± 1.07 | <0.05 |
| Hemolytic index | 4209 | 5.0 [2.0–13.0] | 5.0 [2.0–15.0] | 5.0 [2.0–13.0] | 0.125 |
| Icteric index | 4209 | 0.8 [0.6–1.1] | 0.8 [0.5–1.1] | 0.8 [0.6–1.2] | 0.219 |
| Lipemic index | 4209 | 2.0 [0.0–6.0] | 2.0 [0.0–6.0] | 2.0 [0.0–6.0] | 0.316 |
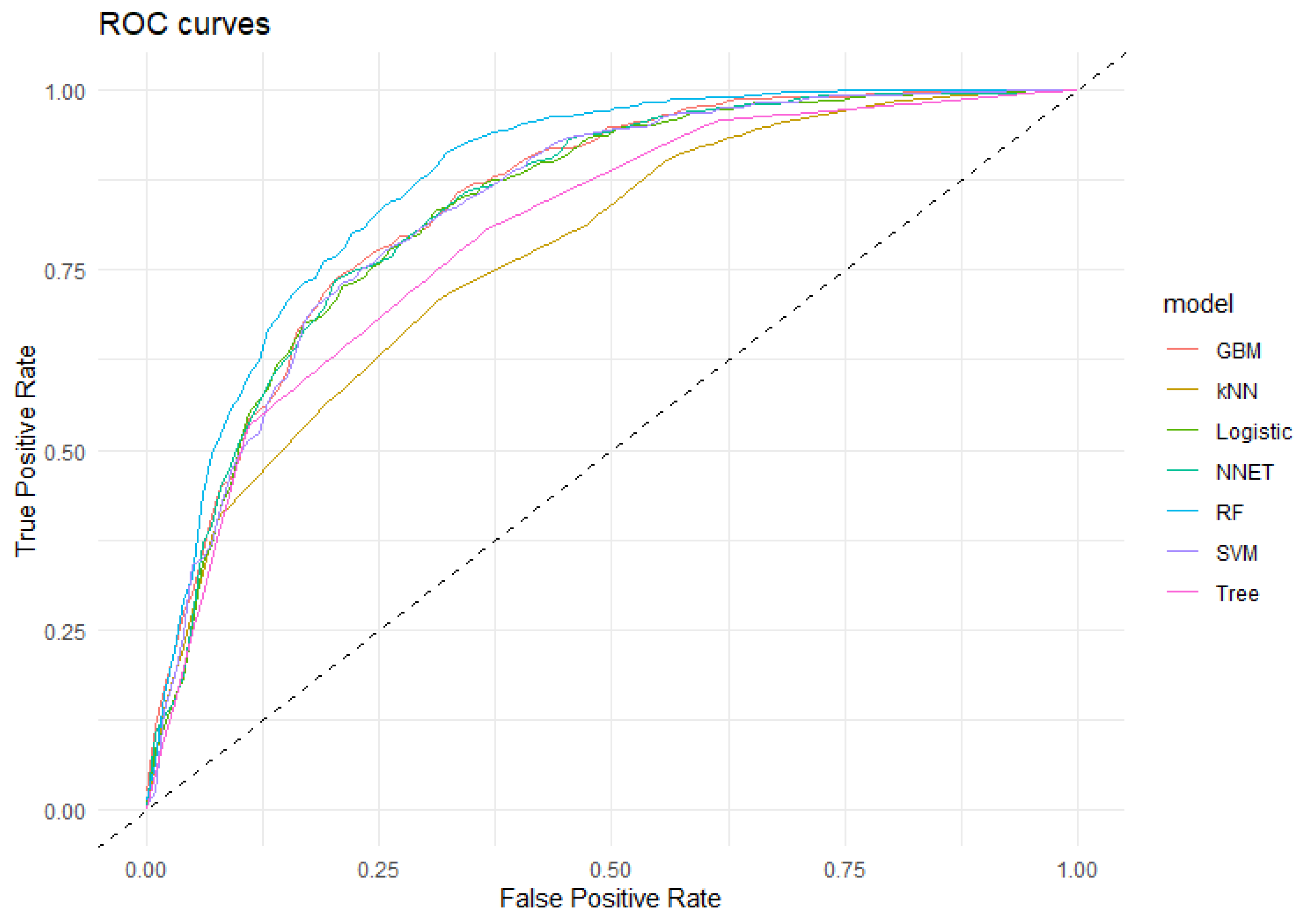
| Algorithm | Sensitivity | Specificity | Accuracy (CI95%) | Kappa | AUC (CI95%) |
|---|---|---|---|---|---|
| RF | 65.56 | 92.76 | 82.24 (79.51–84.75) | 60.90 | 87.10 (84.55–89.66) |
| GBM | 62.24 | 88.19 | 78.15 (75.23–80.88) | 52.26 | 84.08 (81.31–86.85) |
| SVM | 59.21 | 89.52 | 77.80 (74.87–80.55) | 51.02 | 83.45 (80.60–86.31) |
| NNet | 60.12 | 88.76 | 77.69 (74.75–80.44) | 50.96 | 83.60 (80.76–86.44) |
| LR | 62.24 | 87.43 | 77.69 (74.75–80.44) | 51.36 | 83.24 (80.36–86.11) |
| Tree | 63.44 | 80.57 | 73.95 (70.87–76.86) | 44.49 | 79.74 (76.79–82.69) |
| kNN | 53.47 | 80.76 | 70.21 (67.02–73.26) | 35.28 | 77.19 (74.04–80.35) |
| Algorithm | Sensitivity | Specificity | Accuracy (CI95%) | Kappa | AUC (CI95%) |
|---|---|---|---|---|---|
| Tree | 90.50 | 50.46 | 77.27 (72.37–81.68) | 44.36 | 79.01 (74.23–83.79) |
| NNet | 86.43 | 55.96 | 76.36 (71.40–80.84) | 44.24 | 82.10 (77.42–86.78) |
| GBM | 90.95 | 46.79 | 76.36 (71.40–80.84) | 41.40 | 81.44 (76.62–86.27) |
| SVM | 92.31 | 44.04 | 76.36 (71.40–80.84) | 40.49 | 78.45 (73.23–83.67) |
| kNN | 86.43 | 55.05 | 76.06 (71.08–80.56) | 43.39 | 76.77 (71.50–82.05) |
| LR | 91.86 | 42.20 | 75.45 (70.44–80.00) | 38.04 | 81.38 (76.61–86.15) |
| RF | 87.33 | 49.54 | 74.85 (69.81–79.44) | 39.34 | 78.89 (73.79–84.00) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez del Águila, M.M.; Sorlózano-Puerto, A.; Bernier-Rodríguez, C.; Navarro-Marí, J.M.; Gutiérrez-Fernández, J. Application of Machine Learning Algorithms in Urinary Tract Infections Diagnosis Based on Non-Microbiological Parameters. Pathogens 2025, 14, 1034. https://doi.org/10.3390/pathogens14101034
Rodríguez del Águila MM, Sorlózano-Puerto A, Bernier-Rodríguez C, Navarro-Marí JM, Gutiérrez-Fernández J. Application of Machine Learning Algorithms in Urinary Tract Infections Diagnosis Based on Non-Microbiological Parameters. Pathogens. 2025; 14(10):1034. https://doi.org/10.3390/pathogens14101034
Chicago/Turabian StyleRodríguez del Águila, M. Mar, Antonio Sorlózano-Puerto, Cecilia Bernier-Rodríguez, José María Navarro-Marí, and José Gutiérrez-Fernández. 2025. "Application of Machine Learning Algorithms in Urinary Tract Infections Diagnosis Based on Non-Microbiological Parameters" Pathogens 14, no. 10: 1034. https://doi.org/10.3390/pathogens14101034
APA StyleRodríguez del Águila, M. M., Sorlózano-Puerto, A., Bernier-Rodríguez, C., Navarro-Marí, J. M., & Gutiérrez-Fernández, J. (2025). Application of Machine Learning Algorithms in Urinary Tract Infections Diagnosis Based on Non-Microbiological Parameters. Pathogens, 14(10), 1034. https://doi.org/10.3390/pathogens14101034







