Molecular Dynamics Reveal Key Steps in BAR-Related Membrane Remodeling
Abstract
1. Introduction
2. Materials and Methods
2.1. Systems of MD Simulation
2.2. Time-Resolved Force Distribution Analysis
2.3. Interaction Area, Secondary Structure Analysis, and Helix Kink
3. Results
3.1. Structure Change in the BAR Dimer and Interaction of the BAR Dimer–Membrane
3.2. Brief Analysis of the Bending Mechanism of Membrane Curvature
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xiang, S. Endocytosis and Human Innate Immunity. J. Immunol. Sci. 2018, 2, 65–70. [Google Scholar] [CrossRef]
- Charpentier, J.C.; King, P.D. Mechanisms and Functions of Endocytosis in T Cells. Cell Commun. Signal. 2021, 19, 92. [Google Scholar] [CrossRef]
- Dharmalingam, E.; Haeckel, A.; Pinyol, R.; Schwintzer, L.; Koch, D.; Kessels, M.M.; Qualmann, B. F-BAR Proteins of the Syndapin Family Shape the Plasma Membrane and Are Crucial for Neuromorphogenesis. J. Neurosci. 2009, 29, 13315–13327. [Google Scholar] [CrossRef]
- Kim, B.L.; Bu, W.; Wah, I.G.; Koh, E.; Siew, H.O.; Pawson, T.; Sudhaharan, T.; Ahmed, S. The Cdc42 Effector IRSp53 Generates Filopodia by Coupling Membrane Protrusion with Actin Dynamics. J. Biol. Chem. 2008, 283, 20454–20472. [Google Scholar] [CrossRef]
- Krugmann, S.; Jordens, I.; Gevaert, K.; Driessens, M.; Vandekerckhove, J.; Hall, A. Cdc42 Induces Filopodia by Promoting the Formation of an IRSp53: Mena Complex. Curr. Biol. 2001, 11, 1645–1655. [Google Scholar] [CrossRef]
- Ho, H.Y.H.; Rohatgi, R.; Lebensohn, A.M.; Ma, L.; Li, J.; Gygi, S.P.; Kirschner, M.W. Toca-1 Mediates Cdc42-Dependent Actin Nucleation by Activating the N-WASP-WIP Complex. Cell 2004, 118, 203–216. [Google Scholar] [CrossRef]
- Kamioka, Y.; Fukuhara, S.; Sawa, H.; Nagashima, K.; Masuda, M.; Matsuda, M.; Mochizuki, N. A Novel Dynamin-Associating Molecule, Formin-Binding Protein 17, Induces Tubular Membrane Invaginations and Participates in Endocytosis. J. Biol. Chem. 2004, 279, 40091–40099. [Google Scholar] [CrossRef]
- Salazar, M.A.; Kwiatkowski, A.V.; Pellegrini, L.; Cestra, G.; Butler, M.H.; Rossman, K.L.; Serna, D.M.; Sondek, J.; Gertler, F.B.; De Camilli, P. Tuba, a Novel Protein Containing Bin/Amphiphysin/Rvs and Dbl Homology Domains, Links Dynamin to Regulation of the Actin Cytoskeleton. J. Biol. Chem. 2003, 278, 49031–49043. [Google Scholar] [CrossRef]
- Tian, T.; Zhu, Y.L.; Zhou, Y.Y.; Liang, G.F.; Wang, Y.Y.; Hu, F.H.; Xiao, Z.D. Exosome Uptake through Clathrin-Mediated Endocytosis and Macropinocytosis and Mediating MiR-21 Delivery. J. Biol. Chem. 2014, 289, 22258–22267. [Google Scholar] [CrossRef]
- Faille, D.; El-Assaad, F.; Mitchell, A.J.; Alessi, M.C.; Chimini, G.; Fusai, T.; Grau, G.E.; Combes, V. Endocytosis and Intracellular Processing of Platelet Microparticles by Brain Endothelial Cells. J. Cell. Mol. Med. 2012, 16, 1731–1738. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Tirado-Rives, J. Potential Energy Functions for Atomic-Level Simulations of Water and Organic and Biomolecular Systems. Proc. Natl. Acad. Sci. USA 2005, 102, 6665–6670. [Google Scholar] [CrossRef]
- Zimmerberg, J.; Kozlov, M.M. How Proteins Produce Cellular Membrane Curvature. Nat. Rev. Mol. Cell Biol. 2006, 7, 9–19. [Google Scholar] [CrossRef]
- Sivadon, P.; Bauer, F.; Aigle, M.; Crouzet, M. Actin Cytoskeleton and Budding Pattern Are Altered in the Yeast Rvs161 Mutant: The Rvs161 Protein Shares Common Domains with the Brain Protein Amphiphysin. MGG Mol. Gen. Genet. 1995, 246, 485–495. [Google Scholar] [CrossRef]
- Lichte, B.; Veh, R.W.; Meyer, H.E.; Kilimann, M.W. Amphiphysin, a Novel Protein Associated with Synaptic Vesicles. EMBO J. 1992, 11, 2521–2530. [Google Scholar] [CrossRef]
- Sakamuro, D.; Elliott, K.J.; Wechsler-Reya, R.; Prendergast, G.C. BIN1 Is a Novel MYC-Interacting Protein with Features of a Tumour Suppressor. Nat. Genet. 1996, 14, 69–77. [Google Scholar] [CrossRef]
- Liu, S.; Xiong, X.; Zhao, X.; Yang, X.; Wang, H. F-BAR Family Proteins, Emerging Regulators for Cell Membrane Dynamic Changes—From Structure to Human Diseases. J. Hematol. Oncol. 2015, 8, 47. [Google Scholar] [CrossRef]
- Rao, Y.; Haucke, V. Membrane Shaping by the Bin/Amphiphysin/Rvs (BAR) Domain Protein Superfamily. Cell. Mol. Life Sci. 2011, 68, 3983–3993. [Google Scholar] [CrossRef]
- Carman, P.J.; Dominguez, R. BAR Domain Proteins—A Linkage between Cellular Membranes, Signaling Pathways, and the Actin Cytoskeleton. Biophys. Rev. 2018, 10, 1587–1604. [Google Scholar] [CrossRef]
- Suarez, A.; Ueno, T.; Huebner, R.; McCaffery, J.M.; Inoue, T. Bin/Amphiphysin/Rvs (BAR) Family Members Bend Membranes in Cells. Sci. Rep. 2014, 4, 4693. [Google Scholar] [CrossRef]
- Chan, C.; Wen, H.; Lu, L.; Fan, J. Multiscale Molecular Dynamics Simulations of Membrane Remodeling by Bin/Amphiphysin/Rvs Family Proteins. Chin. Phys. B 2015, 25, 018707. [Google Scholar] [CrossRef]
- Bi, X.; Corpina, R.A.; Goldberg, J. Structure of the Sec23/24-Sar1 Pre-Budding Complex of the COPII Vesicle Coat. Nature 2002, 419, 271–277. [Google Scholar] [CrossRef]
- Cui, H.; Ayton, G.S.; Voth, G.A. Membrane Binding by the Endophilin N-BAR Domain. Biophys. J. 2009, 97, 2746–2753. [Google Scholar] [CrossRef]
- Farsad, K.; Ringstad, N.; Takei, K.; Floyd, S.R.; Rose, K.; De Camilli, P. Generation of High Curvature Membranes Mediated by Direct Endophilin Bilayer Interactions. J. Cell Biol. 2001, 155, 193–200. [Google Scholar] [CrossRef]
- Masuda, M.; Takeda, S.; Sone, M.; Ohki, T.; Mori, H.; Kamioka, Y.; Mochizuki, N. Endophilin BAR Domain Drives Membrane Curvature by Two Newly Identified Structure-Based Mechanisms. EMBO J. 2006, 25, 2889–2897. [Google Scholar] [CrossRef]
- Gallop, J.L.; Jao, C.C.; Kent, H.M.; Butler, P.J.G.; Evans, P.R.; Langen, R.; McMahon, H.T. Mechanism of Endophilin N-BAR Domain-Mediated Membrane Curvature. EMBO J. 2006, 25, 2898–2910. [Google Scholar] [CrossRef]
- Peter, B.J.; Kent, H.M.; Mills, I.G.; Vallis, Y.; Butler, P.J.G.; Evans, P.R.; McMahon, H.T. BAR Domains as Sensors of Membrane Curvature: The Amphiphysin BAR Structure. Science 2004, 303, 495–499. [Google Scholar] [CrossRef] [PubMed]
- Richnau, N.; Fransson, Å.; Farsad, K.; Aspenström, P. RICH-1 Has a BIN/Amphiphysin/Rvsp Domain Responsible for Binding to Membrane Lipids and Tubulation of Liposomes. Biochem. Biophys. Res. Commun. 2004, 320, 1034–1042. [Google Scholar] [CrossRef] [PubMed]
- Weissenhorn, W. Crystal Structure of the Endophilin-A1 BAR Domain. J. Mol. Biol. 2005, 351, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zelhof, A.C. Amphiphysins: Raising the BAR for Synaptic Vesicle Recycling and Membrane Dynamics. Traffic 2002, 3, 452–460. [Google Scholar] [CrossRef]
- Stevens, A.O.; Kazan, I.C.; Ozkan, B.; He, Y. Investigating the Allosteric Response of the PICK1 PDZ Domain to Different Ligands with All-atom Simulations. Protein Sci. 2022, 31, e4474. [Google Scholar] [CrossRef]
- Hendrix, E.; Motta, S.; Gahl, R.F.; He, Y. Insight into the Initial Stages of the Folding Process in Onconase Revealed by UNRES. J. Phys. Chem. B 2022, 126, 7934–7942. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Motta, S.; Stevens, A.O.; Song, S.; Hendrix, E.; Pandini, A.; He, Y. Recognizing the Binding Pattern and Dissociation Pathways of the P300 Taz2-P53 TAD2 Complex. J. Am. Chem. Soc. 2022, 2, 1935–1945. [Google Scholar] [CrossRef] [PubMed]
- Stevens, A.O.; He, Y. Residue-Level Contact Reveals Modular Domain Interactions of PICK1 Are Driven by Both Electrostatic and Hydrophobic Forces. Front. Mol. Biosci. 2021, 7, 485. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Stevens, A.O.; Gil Pineda, L.I.; Song, S.; Ameyaw Baah, C.A.; He, Y. Changes in Structure and Flexibility of P53 TAD2 upon Binding to P300 Taz2. J. Theor. Comput. Chem. 2020, 19, 2040007. [Google Scholar] [CrossRef]
- Gil Pineda, L.I.; Milko, L.N.; He, Y. Performance of CHARMM36m with Modified Water Model in Simulating Intrinsically Disordered Proteins: A Case Study. Biophys. Rep. 2020, 6, 80–87. [Google Scholar] [CrossRef]
- He, Y.; Maisuradze, G.G.; Yin, Y.; Kachlishvili, K.; Rackovsky, S.; Scheraga, H.A. Sequence-, Structure-, and Dynamics-Based Comparisons of Structurally Homologous CheY-like Proteins. Proc. Natl. Acad. Sci. USA 2017, 114, 1578–1583. [Google Scholar] [CrossRef]
- He, Y.; Mozolewska, M.A.; Krupa, P.; Sieradzan, A.K.; Wirecki, T.K.; Liwo, A.; Kachlishvili, K.; Rackovsky, S.; Jagieła, D.; Ślusarz, R.; et al. Lessons from Application of the UNRES Force Field to Predictions of Structures of CASP10 Targets. Proc. Natl. Acad. Sci. USA 2013, 110, 14936–14941. [Google Scholar] [CrossRef]
- He, Y.; Liwo, A.; Weinstein, H.; Scheraga, H.A. PDZ Binding to the BAR Domain of PICK1 Is Elucidated by Coarse-Grained Molecular Dynamics. J. Mol. Biol. 2011, 405, 298–314. [Google Scholar] [CrossRef]
- Yin, Y.; Arkhipov, A.; Schulten, K. Simulations of Membrane Tubulation by Lattices of Amphiphysin N-BAR Domains. Structure 2009, 17, 882–892. [Google Scholar] [CrossRef]
- Yu, H.; Schulten, K. Membrane Sculpting by F-BAR Domains Studied by Molecular Dynamics Simulations. PLoS Comput. Biol. 2013, 9, 1002892. [Google Scholar] [CrossRef]
- Takemura, K.; Hanawa-Suetsugu, K.; Suetsugu, S.; Kitao, A. Salt Bridge Formation between the I-BAR Domain and Lipids Increases Lipid Density and Membrane Curvature. Sci. Rep. 2017, 7, 6808. [Google Scholar] [CrossRef] [PubMed]
- Arkhipov, A.; Yin, Y.; Schulten, K. Membrane-Bending Mechanism of Amphiphysin N-BAR Domains. Biophys. J. 2009, 97, 2727–2735. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Li, T.; Stevens, A.O.; Raad, T.; He, Y. Investigating the Mechanical Properties and Flexibility of N-BAR Domains in PICK1 by Molecular Dynamics Simulations. Curr. Protein Pept. Sci. 2023, 24, 865–877. [Google Scholar] [CrossRef]
- Rose, P.W.; Prlić, A.; Altunkaya, A.; Bi, C.; Bradley, A.R.; Christie, C.H.; Di Costanzo, L.; Duarte, J.M.; Dutta, S.; Feng, Z.; et al. The RCSB Protein Data Bank: Integrative View of Protein, Gene and 3D Structural Information. Nucleic Acids Res. 2017, 45, D271–D281. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An Improved Force Field for Folded and Intrinsically Disordered Proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef]
- Huang, J.; MacKerell, A.D. CHARMM36 All-Atom Additive Protein Force Field: Validation Based on Comparison to NMR Data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.C.; van der Spoel, D.; van Drunen, R. GROMACS: A Message-Passing Parallel Molecular Dynamics Implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindah, E. Gromacs: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Post, J.A.; Langer, G.A.; Op den Kamp, J.A.F.; Verkleij, A.J. Phospholipid Asymmetry in Cardiac Sarcolemma. Analysis of Intact Cells and ‘Gas-Dissected’ Membranes. Biochim. Biophys. Acta-Biomembr. 1988, 943, 256–266. [Google Scholar] [CrossRef] [PubMed]
- Matos, M.J.; Post, J.A.; Roelofsen, B.; Op den Kamp, J.A.F. Composition and Organization of Sarcolemmal Fatty Acids in Cultured Neonatal Rat Cardiomyocytes. Cell Biol. Int. Rep. 1990, 14, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Daum, B.; Auerswald, A.; Gruber, T.; Hause, G.; Balbach, J.; Kühlbrandt, W.; Meister, A. Supramolecular Organization of the Human N-BAR Domain in Shaping the Sarcolemma Membrane. J. Struct. Biol. 2016, 194, 375–382. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Evans, D.J.; Holian, B.L. The Nose-Hoover Thermostat. J. Chem. Phys. 1985, 83, 4069–4074. [Google Scholar] [CrossRef]
- Blood, P.D.; Voth, G.A. Direct Observation of Bin/Amphiphysin/Rvs (BAR) Domain-Induced Membrane Curvature by Means of Molecular Dynamics Simulations. Proc. Natl. Acad. Sci. USA 2006, 103, 15068–15072. [Google Scholar] [CrossRef] [PubMed]
- Blood, P.D.; Swenson, R.D.; Voth, G.A. Factors Influencing Local Membrane Curvature Induction by N-BAR Domains as Revealed by Molecular Dynamics Simulations. Biophys. J. 2008, 95, 1866–1876. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald: An N·Log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Apostolakis, J.; Ferrara, P.; Caflisch, A. Calculation of Conformational Transitions and Barriers in Solvated Systems: Application to the Alanine Dipeptide in Water. J. Chem. Phys. 1999, 110, 2099–2108. [Google Scholar] [CrossRef]
- Costescu, B.I.; Gräter, F. Time-Resolved Force Distribution Analysis. BMC Biophys. 2013, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Stacklies, W.; Seifert, C.; Graeter, F. Implementation of Force Distribution Analysis for Molecular Dynamics Simulations. BMC Bioinform. 2011, 12, 101. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.; Hess, B.; van der Spoel, D.; Lindahl, E. GROMACS User Manual-2023.1. Available online: https://manual.gromacs.org/documentation/2023.1/download.html (accessed on 1 January 2023).
- Eisenhaber, F.; Lijnzaad, P.; Argos, P.; Sander, C.; Scharf, M. The Double Cubic Lattice Method: Efficient Approaches to Numerical Integration of Surface Area and Volume and to Dot Surface Contouring of Molecular Assemblies. J. Comput. Chem. 1995, 16, 273–284. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of Protein Secondary Structure: Pattern Recognition of Hydrogen-Bonded and Geometrical Features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef]
- Wilman, H.R.; Shi, J.; Deane, C.M. Helix Kinks Are Equally Prevalent in Soluble and Membrane Proteins. Proteins Struct. Funct. Bioinform. 2014, 82, 1960–1970. [Google Scholar] [CrossRef]
- Thomson, A.R.; Wood, C.W.; Burton, A.J.; Bartlett, G.J.; Sessions, R.B.; Brady, R.L.; Woolfson, D.N. Computational Design of Water-Soluble α-Helical Barrels. Science 2014, 346, 485–488. [Google Scholar] [CrossRef]
- Langelaan, D.N.; Wieczorek, M.; Blouin, C.; Rainey, J.K. Improved Helix and Kink Characterization in Membrane Proteins Allows Evaluation of Kink Sequence Predictors. J. Chem. Inf. Model. 2010, 50, 2213–2220. [Google Scholar] [CrossRef]
- Team, G. Development GROMACS 2023. 2023. Available online: https://manual.gromacs.org/documentation/ (accessed on 1 January 2023).
- Frost, A.; Perera, R.; Roux, A.; Spasov, K.; Destaing, O.; Egelman, E.H.; De Camilli, P.; Unger, V.M. Structural Basis of Membrane Invagination by F-BAR Domains. Cell 2008, 132, 807–817. [Google Scholar] [CrossRef]
- Mim, C.; Cui, H.; Gawronski-Salerno, J.A.; Frost, A.; Lyman, E.; Voth, G.A.; Unger, V.M. Structural Basis of Membrane Bending by the N-BAR Protein Endophilin. Cell 2012, 149, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Shimada, A.; Niwa, H.; Tsujita, K.; Suetsugu, S.; Nitta, K.; Hanawa-Suetsugu, K.; Akasaka, R.; Nishino, Y.; Toyama, M.; Chen, L.; et al. Curved EFC/F-BAR-Domain Dimers Are Joined End to End into a Filament for Membrane Invagination in Endocytosis. Cell 2007, 129, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Grigoryan, G.; Keating, A.E. Structural Specificity in Coiled-Coil Interactions. Curr. Opin. Struct. Biol. 2008, 18, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Chothia, C. The Nature of the Accessible and Buried Surfaces in Proteins. J. Mol. Biol. 1976, 105, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Fersht, A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding; Macmillan: New York, NY, USA, 1999; ISBN 0716732688. [Google Scholar]
- Kuhlman, B.; Baker, D. Native Protein Sequences Are Close to Optimal for Their Structures. Proc. Natl. Acad. Sci. USA 2000, 97, 10383–10388. [Google Scholar] [CrossRef]
- Henne, W.M.; Kent, H.M.; Ford, M.G.J.; Hegde, B.G.; Daumke, O.; Butler, P.J.G.; Mittal, R.; Langen, R.; Evans, P.R.; McMahon, H.T. Structure and Analysis of FCHo2 F-BAR Domain: A Dimerizing and Membrane Recruitment Module That Effects Membrane Curvature. Structure 2007, 15, 839–852. [Google Scholar] [CrossRef]
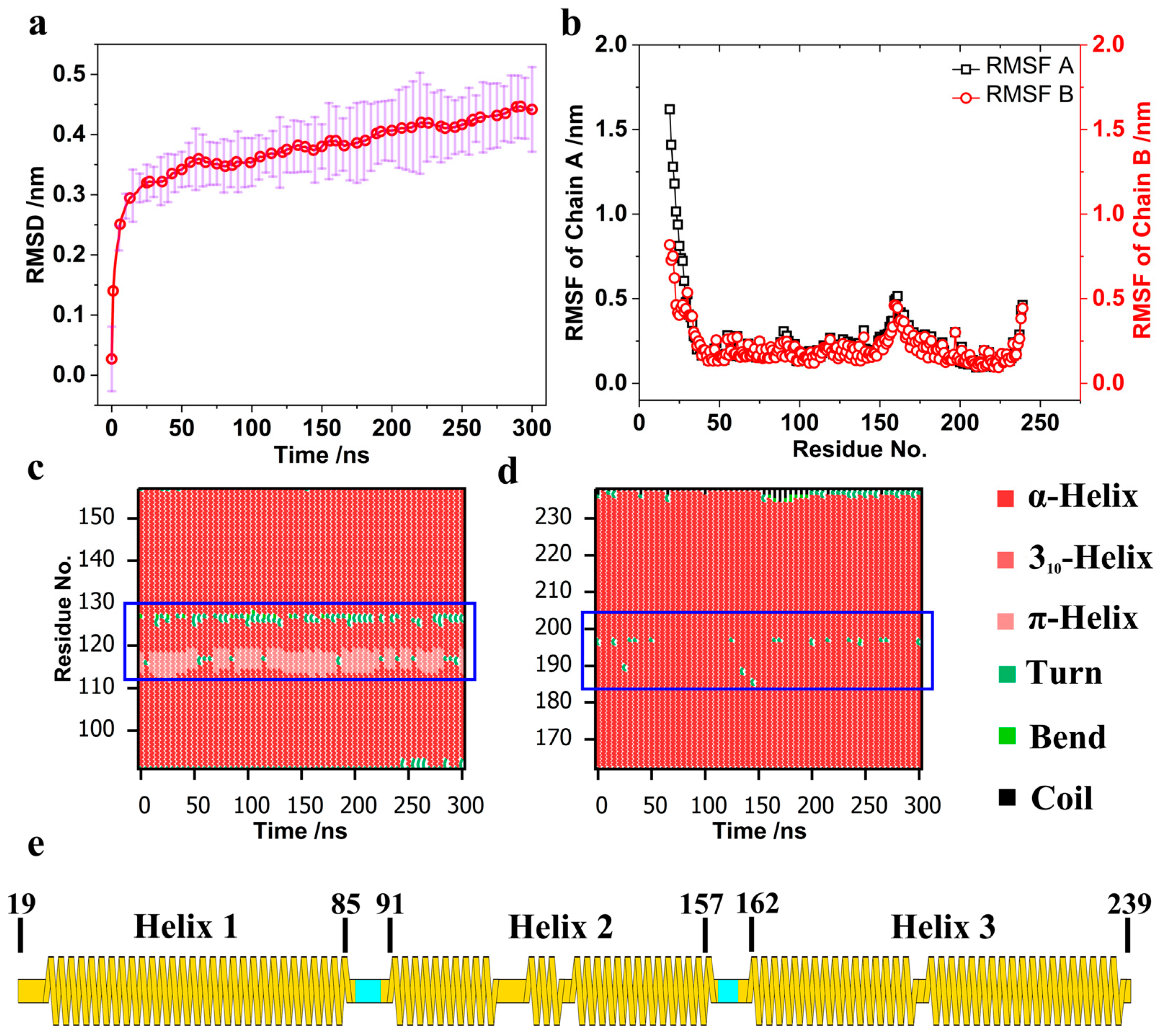
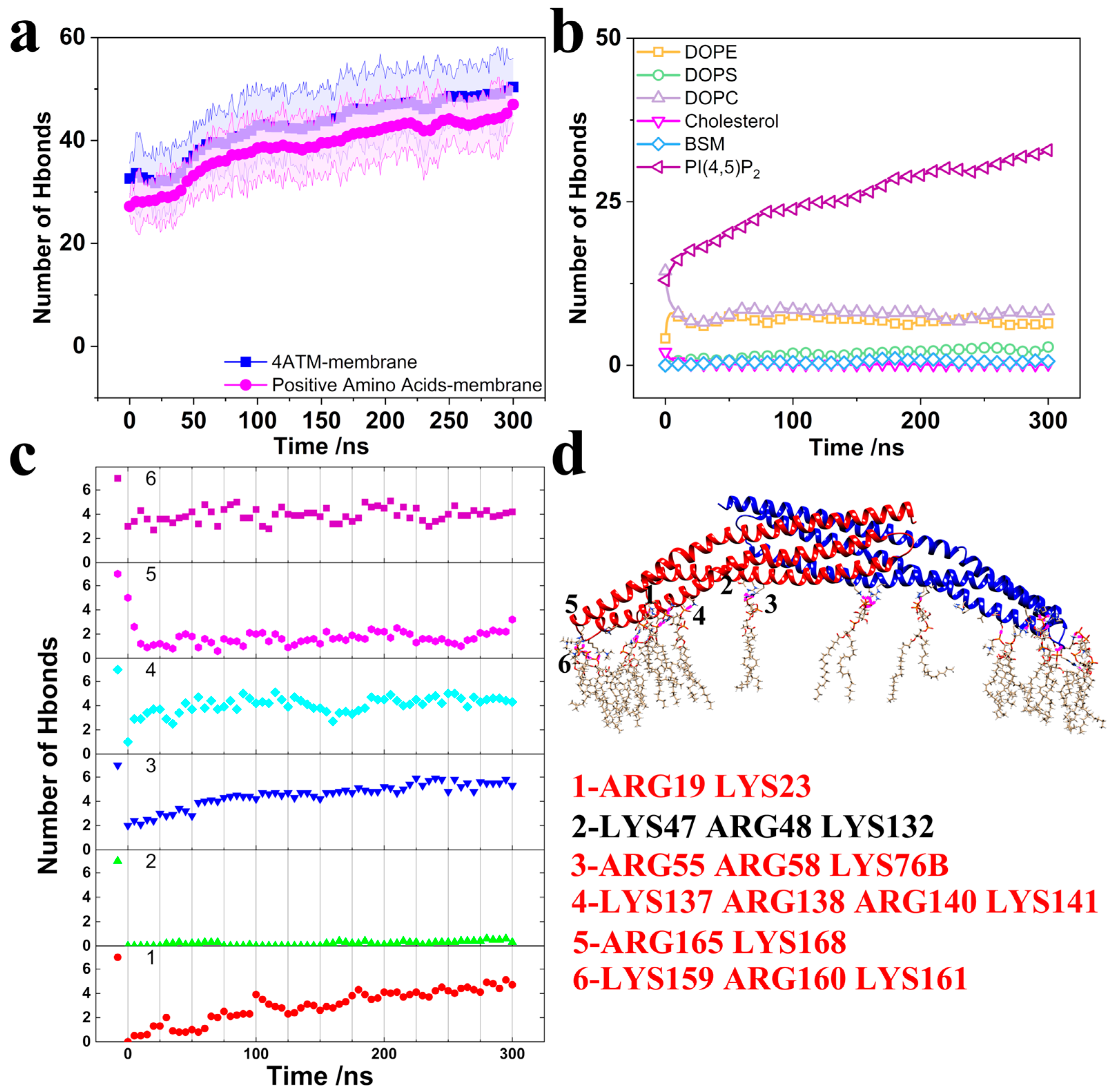


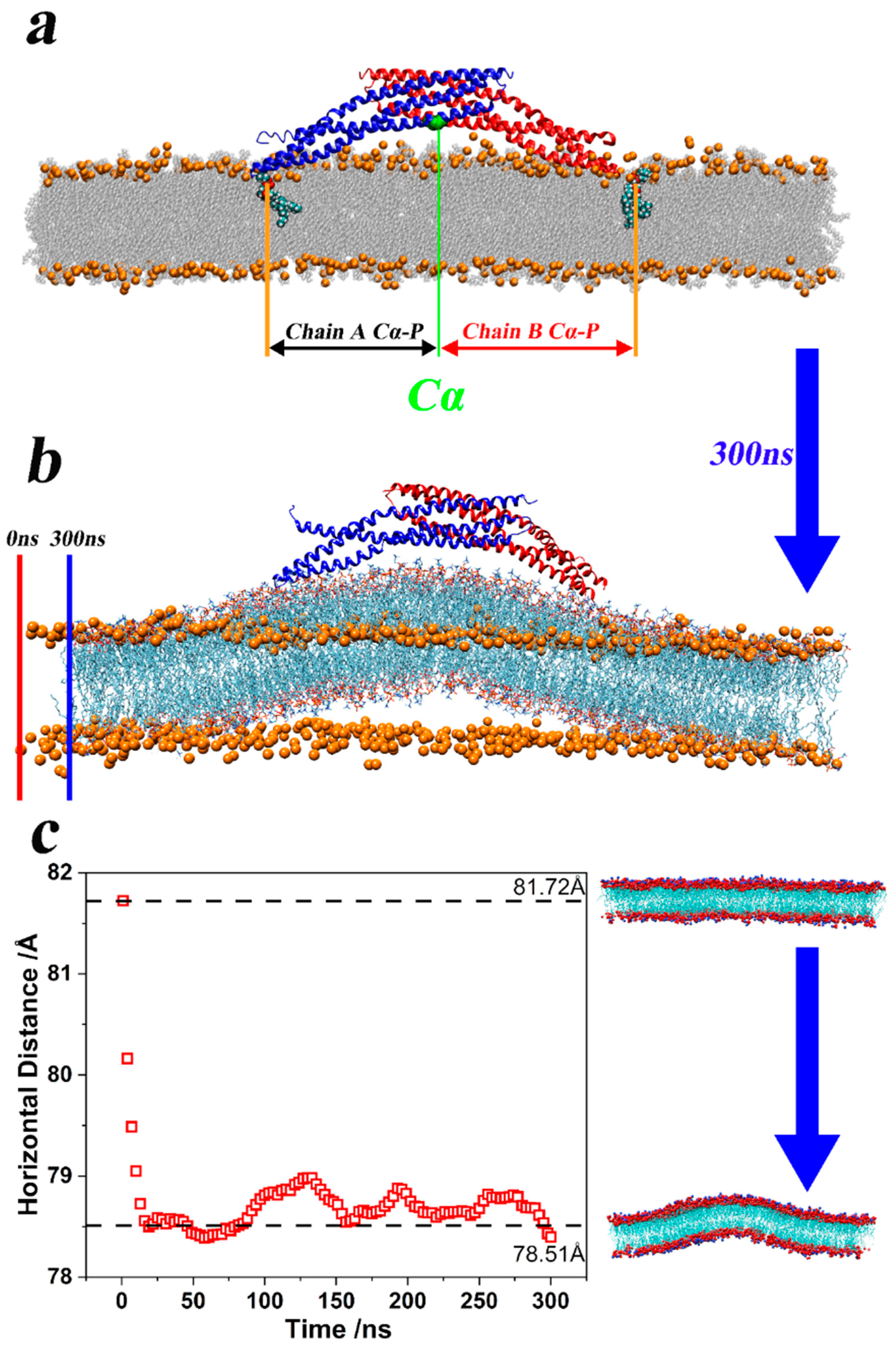
| System | Number | Time/ns | Size |
|---|---|---|---|
| 4ATM + membrane | 20 | 300 | 10.0 × 34.5 × 20.9 nm3 |
| Membrane only | 1 | 300 | 7.6 × 23.0 × 15.2 nm3 |
| Lipid Molecule | Proportion |
|---|---|
| Cholesterol | ~32% |
| DOPC | ~35% |
| DOPE | ~17% |
| DOPS | ~2% |
| PI (4,5) P2 | ~3% |
| Brain SM | ~11% |
| Ranking | Helix 1–Helix 2 | Helix 1–Helix 3 | Helix 2–Helix 3 |
|---|---|---|---|
| 1 | LYS 132 | GLU 52 | ARG 134 |
| 2 | GLU 50 | ARG 198 | ARG 138 |
| 3 | GLU 153 | GLN 49 | TYR 145 |
| 4 | TRP 106 | TRP 195 | PHE 180 |
| 5 | PHE 46 | TYR 202 | PHE 173 |
| 6 | GLN 71 | TYP 63 | LEU 212 |
| 7 | ARG 138 | GLU 213 | PHE 183 |
| 8 | ARG 149 | TYP 42 | GLU 190 |
| 9 | HIS 82 | LEU 191 | PHE 216 |
| 10 | LEU 64 | GLN 188 | LEU 223 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, S.; Li, T.; Stevens, A.O.; Shorty, T.; He, Y. Molecular Dynamics Reveal Key Steps in BAR-Related Membrane Remodeling. Pathogens 2024, 13, 902. https://doi.org/10.3390/pathogens13100902
Song S, Li T, Stevens AO, Shorty T, He Y. Molecular Dynamics Reveal Key Steps in BAR-Related Membrane Remodeling. Pathogens. 2024; 13(10):902. https://doi.org/10.3390/pathogens13100902
Chicago/Turabian StyleSong, Shenghan, Tongtong Li, Amy O. Stevens, Temair Shorty, and Yi He. 2024. "Molecular Dynamics Reveal Key Steps in BAR-Related Membrane Remodeling" Pathogens 13, no. 10: 902. https://doi.org/10.3390/pathogens13100902
APA StyleSong, S., Li, T., Stevens, A. O., Shorty, T., & He, Y. (2024). Molecular Dynamics Reveal Key Steps in BAR-Related Membrane Remodeling. Pathogens, 13(10), 902. https://doi.org/10.3390/pathogens13100902






