Abstract
Migratory birds have contributed to the dissemination of multidrug-resistant (MDR) bacteria across the continents. A CTX-M-2-producing Escherichia coli was isolated from a black skimmer (Rynchops niger) in Southeast Brazil. The whole genome was sequenced using the Illumina NextSeq platform and de novo assembled by CLC. Bioinformatic analyses were carried out using tools from the Center for Genomic Epidemiology. The genome size was estimated at 4.9 Mb, with 4790 coding sequences. A wide resistome was detected, with genes encoding resistance to several clinically significant antimicrobials, heavy metals, and biocides. The blaCTX-M-2 gene was inserted in an In229 class 1 integron inside a ∆TnAs3 transposon located in an IncHI2/ST2 plasmid. The strain was assigned to ST5506, CH type fumC19/fimH32, serotype O8:K87, and phylogroup B1. Virulence genes associated with survival in acid conditions, increased serum survival, and adherence were also identified. These data highlight the role of migratory seabirds as reservoirs and carriers of antimicrobial resistance determinants and can help to elucidate the antimicrobial resistance dynamics under a One Health perspective.
1. Introduction
Extended-spectrum β-lactamase (ESBL)-producing Enterobacterales have been considered one of the greatest threats to human health, being listed as critical priority pathogens by the World Health Organization (WHO) [1]. Despite greater emphasis being placed on the human clinical sphere, the One Health approach draws attention to the need for studies involving other ecological spheres, given the interdependence among humans, animals, and the environment [2]. Dissemination of antimicrobial resistance between these different sectors can occur through direct transmission of resistant bacteria or through genetic exchanges, mainly by plasmids and other mobile genetic elements, as integrons and transposons [3]. In this regard, whole-genome sequencing can be a valuable tool for surveillance of antimicrobial resistance, as it allows a deep understanding of the genetic basis of resistance mechanisms, evolution, and dissemination [4].
Since wild animals can also be exposed to human-associated multidrug-resistant (MDR) pathogens, there is a global trend for monitoring the dissemination of medically relevant bacteria in wildlife populations [5]. Recently, migratory birds have been recognized as important reservoirs and vectors for spreading ESBL-producers across the globe [5,6]. When contacting contaminated environments, these animals can incorporate antimicrobial resistant bacteria, as ESBL-producing Escherichia coli, in the gut microbiota; then, it can be spread over distant geographic locations, according to the migratory route [7].
Black skimmers (Rynchops niger) are widespread piscivorous seabirds that inhabit sandy shorelines, being commonly found in the Nearctic and Neotropical regions [8]. Due to their migration behavior, they appear seasonally in large numbers in both the Pacific and Atlantic coasts of South America and are frequently observed in bays, estuaries, beaches, and shallow lagoons, where they prey on populations of small fish [8]. In this study, we aimed to analyze the genomic features of an MDR ESBL-producing E. coli strain recovered from the gut microbiota of a migratory black skimmer.
2. Materials and Methods
In March 2016, as part of a surveillance study on antimicrobial-resistant Gram-negative bacteria among wild animals, a cloacal swab sample was collected from a black skimmer (R. niger), at the admission time to a wildlife rehabilitation center. The bird was rescued by firefighters at a beach on the southeast coast of Brazil (−23.985143, −46.309956) and presented a fracture of the right humerus. Until the sample collection time, no medication had been administered to the bird. The sample was inoculated on MacConkey agar plates supplemented with ceftriaxone (2 μg/mL), colistin (2 μg/mL) and ciprofloxacin (1 μg/mL) (Sigma-Aldrich, St. Louis, MO, USA), for screening of bacteria resistant to cephalosporins, polymyxins, and fluoroquinolones, respectively. Agar plates were incubated for 18–24 h, at 35 °C. The bacterial isolate was identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF). Antimicrobial susceptibility was determined by disk diffusion and E-test methods [9], and the colistin susceptibility test was performed by the broth microdilution method, according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI) [9]. E. coli 25922 was used as the quality control strain.
Genomic DNA extraction was carried out using PureLink™ Quick Gel Extraction Kit (Life Technologies, Carlsbad, CA, USA), and the DNA concentration was measured using a Qubit® 2.0 fluorometer (Life Technologies, Carlsbad, CA, USA). The genomic library was built using a Nextera XT DNA Library Preparation Kit (Illumina Inc., Cambridge, UK), according to the manufacturer’s instructions. The whole genome sequencing was performed on an Illumina NextSeq platform, using paired-end reads (150 bp). Reads with a PHRED quality score below 20 were discarded, and adapters were trimmed using TrimGalore v0.6.5 (https://github.com/FelixKrueger/TrimGalore, accessed on 30 June 2023). De novo genome assembly was performed using CLC Genomics Workbench v. 11 (CLC Bio, Aarhus, Denmark), and the draft genome was annotated using the NCBI Prokaryotic Annotation Pipeline [PGAP (https://www.ncbi.nlm.nih.gov/genome/annotation_prok/, accessed on 17 October 2023)].
Acquired antimicrobial resistance genes (ARGs) were evaluated by ResFinder 4.1 [10], and the genetic context of some ARGs was analyzed by using Geneious Prime® 2022.0.1 (Biomatters, Auckland, New Zealand). Insertion sequences and transposons were identified by ISFinder (https://www-is.biotoul.fr/index.php, accessed on 30 June 2023). The BLASTn tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 30 June 2023) was used for identification of similar genetic environments, and the comparative analysis of these genetic structures was displayed by using Easyfig version 2.1 (http://mjsull.github.io/Easyfig/, accessed on 17 October 2023). Plasmid incompatibility groups and pMLST were predicted with PlasmidFinder 2.1 [11] and pMLST 2.0 [11], respectively. Heavy metal and biocide resistance genes were detected using the BacMet database [12]. MLST, CH type, serotype, and virulence genes were predicted with MLST 2.0 [13], CHTyper 1.0 [14], SerotypeFinder 2.0 [15], and VirulenceFinder 2.0 [16], respectively. E. coli phylogroup was determined through the ClermonTyping online tool [17]. A ≥90% identity threshold was used for all predicted genes. Genomic comparison of ICBTMS1 with other genomes from the same ST—available from EnteroBase (https://enterobase.warwick.ac.uk/species/index/ecoli, accessed on 17 October 2023)—was performed using BRIG 0.95 software [18].
No ethical approval was required for this study. Biological sample collection was authorized by the Authorization System and Information on Biodiversity (SISBIO license number 55804–2).
3. Results and Discussion
A single E. coli strain (ICBTMS1) was recovered from the MacConkey agar plate supplemented with ceftriaxone. No growth was observed on the other agar plates. ICBTMS1 displayed an MDR profile to amoxicillin/clavulanic acid, cefotaxime, ceftazidime, cefepime, ceftriaxone (MIC ≥ 32 μg/mL), aztreonam, gentamicin, trimethoprim/sulfamethoxazole, and tetracycline. Otherwise, it was susceptible to cefoxitin, ciprofloxacin, amikacin, imipenem, ertapenem, meropenem, and colistin. E. coli 25922 presented susceptibility results within acceptable quality control ranges.
A total of 6,161,326 reads, assembled into 154 contigs, was produced with 123x coverage and a G+C content of 48.4%. The genome size was calculated at 4,968,129 bp, comprising 4790 coding sequences (CDS), 2 rRNAs, 44 tRNAs, 9 ncRNAs, 134 pseudogenes, and 2 CRISPR arrays.
Resistome analysis detected the presence of antimicrobial genes conferring resistance to β-lactams (blaCTX-M-2, blaTEM-1C), aminoglycosides [aac(3)-VIa, aadA1, aph(3′)-Ia], sulfamethoxazole (sul1), trimethoprim (dfrA27), and tetracycline (tetA) (Table 1).

Table 1.
Phenotypic and genomic data from Escherichia coli strain ICBTMS1.
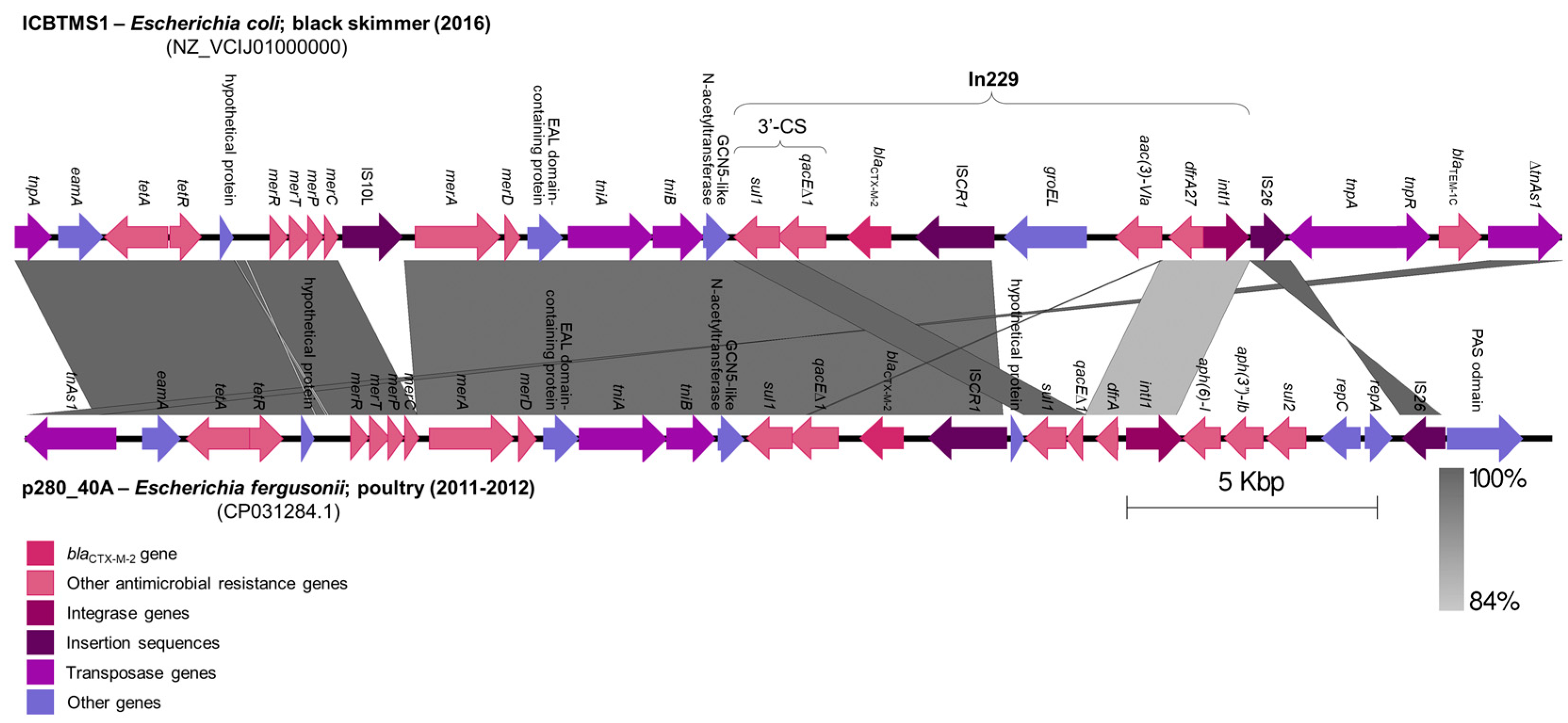
The blaCTX-M-2 gene was inserted in an In229 class 1 integron by an ISCR1, downstream of the 3′-conserved segment (3′-CS), inside a ∆TnAs3 transposon (Figure 1). This genomic structure was located in an IncHI2/ST2 plasmid and showed 73% coverage and 99.71% identity with a 29,941 bp fragment of an IncHI2/ST2 plasmid (CP031284.1) from an Escherichia fergusonii isolated from poultry, in São Paulo State, Brazil, between 2011 and 2012 [19] (Figure 1). Indeed, IncHI2/ST2 plasmids have been previously described in Brazil, in Salmonella spp. isolates from poultry, also carrying the blaCTX-M-2 gene [20,21]. These data infer the occurrence of genetic exchanges among bacterial strains from wild birds and poultry, as already suggested by previous studies [22,23].
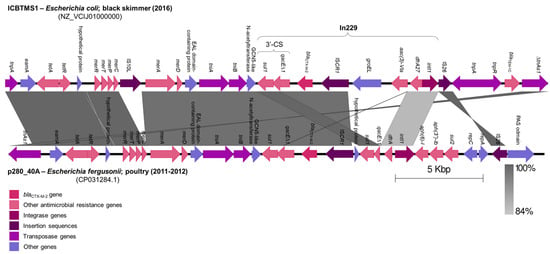
Figure 1.
Comparison of genomic structures from IncHI2/ST2 plasmids harboring the blaCTX-M-2 gene. A fragment of the IncHI2/ST2 plasmid from the ICBTMS1 E. coli strain, isolated from the black skimmer (2016), was compared with a fragment of an IncHI2/ST2 plasmid from an E. fergusonii isolated from poultry (2011–2012), using Easyfig version 2.1 (http://mjsull.github.io/Easyfig/, accessed on 17 October 2023). Regions of homology are marked by gray shading.
ICBTMS1 also carried a variety of genes conferring resistance to heavy metals, as arsenical (arsB), cadmium (ychH and yhcN), chromate (yieF), cobalt (corABD), copper (cueO, cusARS, and cutCEF), iron/manganese (sitABCD), magnesium (mgtA), manganese (mntHPR), mercury (merRTPCAD), molybdate (modABCE), nickel (nikABCDER), silver (silAR), tellurite (tehAB), tellurium (terACDEWZ), zinc/tellurium (pitA), and zinc (zinT, zitB, zntAR, znuAB, zraR, zupT, and zur), in addition to biocide resistance genes (ostA, oxyRkp, qacE∆1, and sugE) (Table 1). Occurrence of these genes in wild animals and the environment is suggestive of heavy metal contamination [24]. Some studies, carried out in the same region where the black skimmer was found, have detected high levels of heavy metals in estuary sediment samples [25,26,27]. In addition, heavy-metal-resistant bacteria have been isolated from wild fish and shrimp in that area [28,29]. The presence of heavy metals in animals and the environment can boost bacterial resistance to both heavy metals and antimicrobials, through co- or cross-selection mechanisms [30].
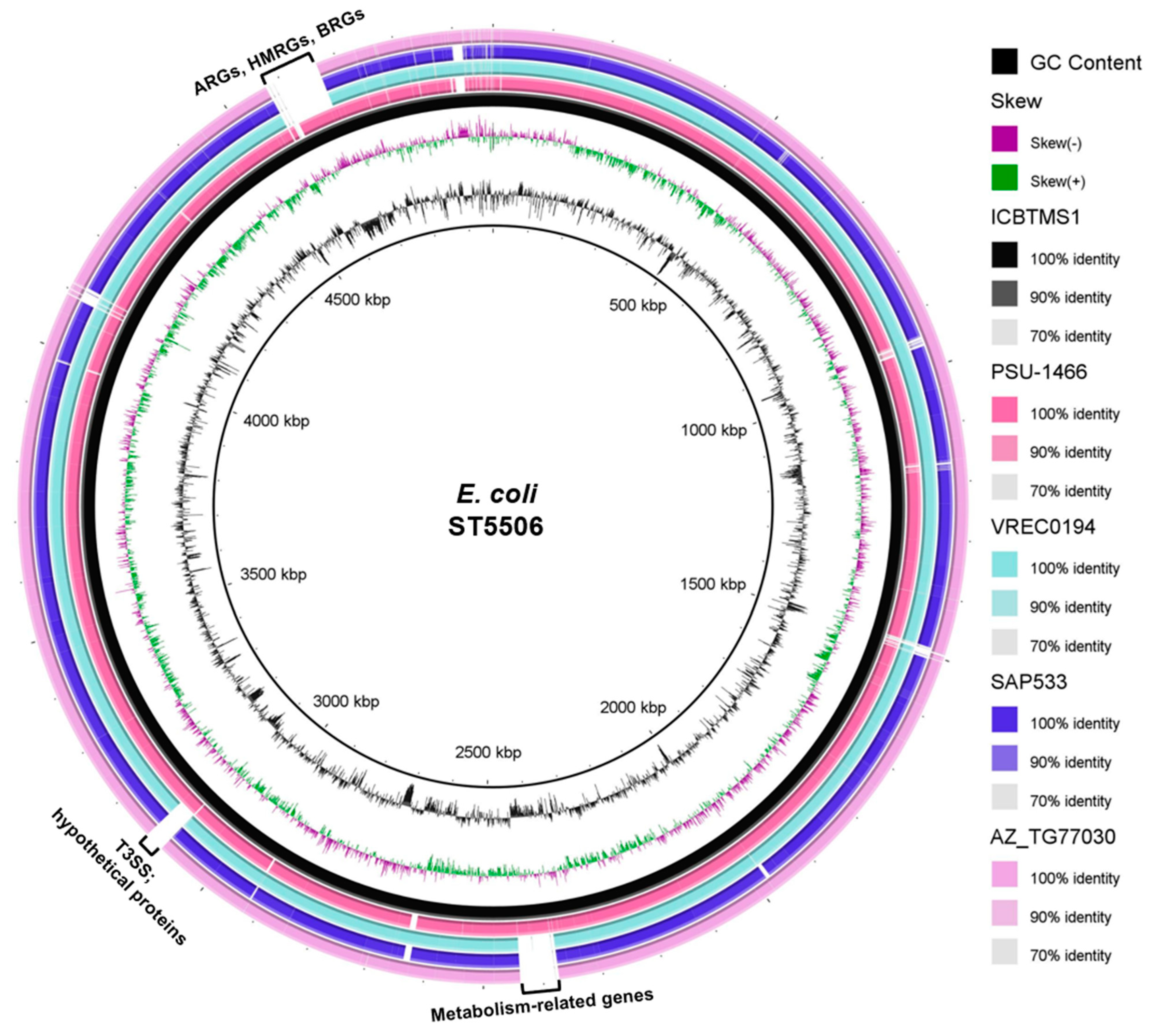
The strain was assigned to the ST5506, which has already been reported in animal and environmental strains (https://enterobase.warwick.ac.uk/species/index/ecoli, accessed on 12 July 2023): the strain PSU-1466 (accession number: AATPIY000000000.1), isolated from turkey, in the USA, in 1990; the strain AZ_TG77030 (accession number: AATISO000000000.1), isolated from chicken meat, in the USA, in 2014; the strain VREC0194 (accession number: DADKSK000000000.1), isolated from water, in England, in 2015; and the strain SAP533 (accession number: SAMEA3697202), isolated in England, in 2015 (unknown source). When compared with the other genomes from ST5506, ICBTMS1 showed differences mainly in the regions of antimicrobial, heavy metal, and biocide resistance genes; the type three secretion system (T3SS); hypothetical proteins; and metabolism-related genes (Figure 2). A higher genetic similarity was found with the strain PSU-1466 (AATPIY000000000.1) (Figure 2). Additionally, the strain belonged to the O8:K87 serotype, known to be an important animal pathogen [31,32]; CH type fumC19/fimH32; and the commensal phylogroup B1 [33]. Virulence genes related to survival in acid conditions (gad), increased serum survival (iss), and adherence (lpfA and papC) were detected, as well.
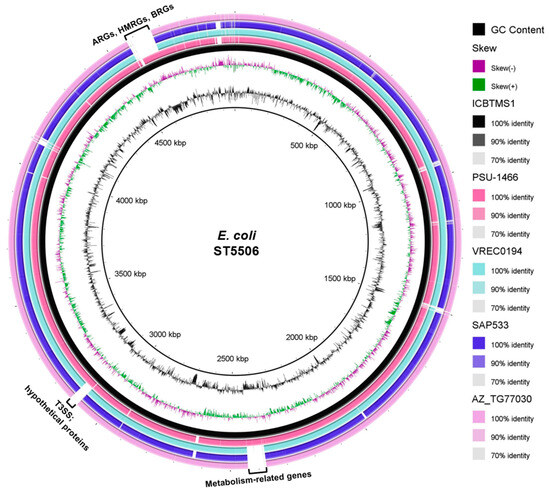
Figure 2.
Genomic comparison of ICBTMS1 against four ST5506 E. coli genomes, made by using the BRIG software (http://brig.sourceforge.net/, accessed on 12 July 2023). The innermost rings depict GC content (black) and GC skew (purple/green), followed by concentric rings of ICBTMS1 (black; accession number: NZ_VCIJ01000000), PSU–1466 (pink; accession number: AATPIY000000000.1), VREC0194 (light blue; accession number: DADKSK000000000.1), SAP533 (purple; accession number: SAMEA3697202), and AZ_TG77030 (light pink; accession number: AATISO000000000.1), colored according to BLASTn identity with 90% and 70% as the upper and lower identity thresholds, respectively. Gapped regions indicate the absence or low similarity among the genomes. ARGs: antimicrobial resistance genes; HMRGs: heavy metal resistance genes; BRGs: biocide resistance genes; T3SS: type III secretion system.
In summary, we report an international E. coli clone, isolated from a migratory seabird, carrying a wide resistome against clinically significant antimicrobials, heavy metals, and biocides, which should be considered a critical epidemiological issue. The broad resistome identified in this WHO critical priority pathogen is likely related to an environment impacted by anthropogenic activities [34]. In addition, these data highlight the importance of monitoring migratory seabirds as reservoirs and carriers of such bacteria and genetic determinants and their role as sentinels of ecosystem health [35]. Last but not least, this study provides valuable genomic information that might be useful in understanding the antimicrobial resistance dynamics under a One Health perspective.
Author Contributions
Conceptualization, Q.M., M.R.F. and F.P.S.; methodology, C.L.N., G.H.P.D., Q.M., M.R.F. and F.P.S.; validation, Q.M. and M.R.F.; formal analysis, Q.M., M.R.F. and B.C.; investigation, Q.M., M.R.F., F.P.S. and B.C.; resources, N.L.; data curation, Q.M., M.R.F. and B.C.; writing—original draft preparation, Q.M.; writing—review and editing, Q.M., F.P.S. and N.L.; visualization, Q.M.; supervision, N.L.; project administration, Q.M.; funding acquisition, N.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Bill and Melinda Gates Foundation, Grand Challenges Explorations Brazil–New approaches to characterize the global burden of antimicrobial resistance (grant number OPP1193112), Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP; grant number 2016/08593-9), and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq; grant number 462042/2014-6).
Institutional Review Board Statement
No ethical approval was required for this study. Biological sample collection was authorized by the Authorization System and Information on Biodiversity (SISBIO license number 55804–2).
Informed Consent Statement
Not applicable.
Data Availability Statement
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under accession number NZ_VCIJ01000000. The version described in this paper is the first version. Additionally, data are also available at the OneBR platform (http://onehealthbr.com, accessed on 1 January 2024), under the ID number ONE134.
Acknowledgments
FAPESP: CNPq and CAPES research grants are gratefully acknowledged. N.L. is a research fellow of CNPq (314336/2021-4). We thank Cefar Diagnóstica Ltda. (São Paulo, Brazil) for kindly supplying antibiotic discs for susceptibility testing.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- World Health Organization (WHO). WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 26 June 2023).
- Aslam, B.; Khurshid, M.; Arshad, M.I.; Muzammil, S.; Rasool, M.; Yasmeen, N.; Shah, T.; Chaudhry, T.H.; Rasool, M.H.; Shahid, A.; et al. Antibiotic Resistance: One Health One World Outlook. Front. Cell. Infect. Microbiol. 2021, 11, 771510. [Google Scholar] [CrossRef]
- Ghaly, T.M.; Gillings, M.R. New perspectives on mobile genetic elements: A paradigm shift for managing the antibiotic resistance crisis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2022, 377, 20200462. [Google Scholar] [CrossRef]
- NIHR Global Health Research Unit on Genomic Surveillance of AMR. Whole-genome sequencing as part of national and international surveillance programmes for antimicrobial resistance: A roadmap. BMJ Glob. Health 2020, 5, e002244. [Google Scholar] [CrossRef]
- Atterby, C.; Ramey, A.M.; Hall, G.G.; Järhult, J.; Börjesson, S.; Bonnedahl, J. Increased prevalence of antibiotic-resistant E. coli in gulls sampled in Southcentral Alaska is associated with urban environments. Infect. Ecol. Epidemiol. 2016, 6, 3233. [Google Scholar]
- Ardiles-Villegas, K.; González-Acuña, D.; Waldenström, J.; Olsen, B.; Hernández, J. Antibiotic resistance patterns in fecal bacteria isolated from Christmas shearwater (Puffinus nativitatis) and masked booby (Sula dactylatra) at remote Easter Island. Avian Dis. 2011, 55, 486–489. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Liang, B.; Jiang, B.W.; Zhu, L.W.; Wang, T.C.; Li, Y.G.; Liu, J.; Guo, X.J.; Ji, X.; Sun, Y. Migratory wild birds carrying multidrug-resistant Escherichia coli as potential transmitters of antimicrobial resistance in China. PLoS ONE 2021, 16, e0261444. [Google Scholar] [CrossRef] [PubMed]
- Davenport, L.C.; Goodenough, K.S.; Haugaasen, T. Birds of Two Oceans? Trans-Andean and Divergent Migration of Black Skimmers (Rynchops niger cinerascens) from the Peruvian Amazon. PLoS ONE 2016, 11, e0144994. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing Twentieth Informational Supplement M100-S30; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Bortolaia, V.; Kaas, R.F.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.R.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Pal, C.; Bengtsson-Palme, J.; Rensing, C.; Kristiansson, E.; Larsson, D.G.J. BacMet: Antibacterial biocide and metal resistance genes database. Nucleic Acids Res. 2014, 42, D737–D743. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef]
- Roer, L.; Johannesen, T.B.; Hansen, F.; Stegger, M.; Tchesnokova, V.; Sokurenko, E.; Garibay, N.; Allesøe, R.; Thomsen, M.C.F.; Lund, O.; et al. CHTyper, a Web Tool for Subtyping of Extraintestinal Pathogenic Escherichia coli Based on the fumC and fimH Alleles. J. Clin. Microbiol. 2018, 56, e00063-18. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Tetzschner, A.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and easy in silico serotyping of Escherichia coli using whole genome sequencing (WGS) data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef] [PubMed]
- Malberg Tetzschner, A.M.; Johnson, J.R.; Johnston, B.D.; Lund, O.; Scheutz, F. In Silico Genotyping of Escherichia coli Isolates for Extraintestinal Virulence Genes by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2020, 58, e01269-20. [Google Scholar] [CrossRef] [PubMed]
- Beghain, J.; Bridier-Nahmias, A.; Le Nagard, H.; Denamur, E.; Clermont, O. ClermonTyping: An easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genom. 2018, 4, e000192. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [PubMed]
- Galetti, R.; Penha Filho, R.A.C.; Ferreira, J.C.; Varani, A.M.; Darini, A.L.C. Antibiotic resistance and heavy metal tolerance plasmids: The antimicrobial bulletproof properties of Escherichia fergusonii isolated from poultry. Infect. Drug Resist. 2019, 12, 1029–1033. [Google Scholar] [CrossRef]
- Galetti, R.; Filho, R.A.C.P.; Ferreira, J.C.; Varani, A.M.; Sazinas, P.; Jelsbak, L.; Darini, A.L.C. The plasmidome of multidrug-resistant emergent Salmonella serovars isolated from poultry. Infect. Genet. Evol. 2021, 89, 104716. [Google Scholar] [CrossRef]
- Núncio, A.S.P.; Webber, B.; Pottker, E.S.; Cardoso, B.; Esposito, F.; Fontana, H.; Lincopan, N.; Girardello, R.; Pilotto, F.; Dos Santos, L.R.; et al. Genomic characterization of multidrug-resistant Salmonella Heidelberg E2 strain isolated from chicken carcass in southern Brazil. Int. J. Food Microbiol. 2022, 379, 109863. [Google Scholar] [CrossRef]
- Borges, C.A.; Beraldo, L.G.; Maluta, R.P.; Cardozo, M.V.; Barboza, K.B.; Guastalli, E.A.; Kariyawasam, S.; DebRoy, C.; Ávila, F.A. Multidrug-resistant pathogenic Escherichia coli isolated from wild birds in a veterinary hospital. Avian Pathol. 2017, 46, 76–83. [Google Scholar] [CrossRef]
- Skarżyńska, M.; Zaja, C.M.; Bomba, A.; Bocian, Ł.; Kozdruń, W.; Polak, M.; Wia Cek, J.; Wasyl, D. Antimicrobial Resistance Glides in the Sky-Free-Living Birds as a Reservoir of Resistant Escherichia coli with Zoonotic Potential. Front. Microbiol. 2021, 12, 656223. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.C., 4th; Kieran, T.J.; Finger, J.W., Jr.; Bayona-Vásquez, N.J.; Oladeinde, A.; Beasley, J.C.; Seaman, J.C.; McArthur, J.V.; Rhodes, O.E., Jr.; Glenn, T.C. Unveiling the Gut Microbiota and Resistome of Wild Cotton Mice, Peromyscus gossypinus, from Heavy Metal- and Radionuclide-Contaminated Sites in the Southeastern United States. Microbiol. Spectr. 2021, 9, e0009721. [Google Scholar] [CrossRef]
- Buruaem, L.M.; Hortellani, M.A.; Sarkis, J.E.; Costa-Lotufo, L.V.; Abessa, D.M. Contamination of port zone sediments by metals from large marine ecosystems of Brazil. Mar. Pollut. Bull. 2012, 64, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.S.M.; Angeli, J.L.F.; Ferreira, P.A.L.; de Mahiques, M.M.; Figueira, R.C.L. A multivariate approach and sediment quality index evaluation applied to Baixada Santista, Southeastern Brazil. Mar. Pollut. Bull. 2019, 143, 72–80. [Google Scholar] [CrossRef]
- Trevizani, T.H.; Domit, C.; Tramonte, K.M.; de Mahiques, M.M.; Figueira, R.C.L. Metals in sediments as indicators of anthropogenic impacts in estuaries of south-southeast Brazil. J. Sediment. Environ. 2021, 6, 417–430. [Google Scholar] [CrossRef]
- Sellera, F.P.; Fernandes, M.R.; Moura, Q.; Carvalho, M.P.N.; Lincopan, N. Extended-spectrum-β-lactamase (CTX-M)-producing Escherichia coli in wild fishes from a polluted area in the Atlantic Coast of South America. Mar. Pollut. Bull. 2018, 135, 183–186. [Google Scholar] [CrossRef]
- Monte, D.F.; Sellera, F.P.; Fernandes, M.R.; Moura, Q.; Landgraf, M.; Lincopan, N. Genome Sequencing of an Escherichia coli Sequence Type 617 Strain Isolated from Beach Ghost Shrimp (Callichirus major) from a Heavily Polluted Ecosystem Reveals a Wider Resistome against Heavy Metals and Antibiotics. Microbiol. Resour. Announc. 2019, 8, e01471-18. [Google Scholar] [CrossRef]
- Nguyen, C.C.; Hugie, C.N.; Kile, M.L.; Navab-Daneshmand, T. Association between heavy metals and antibiotic-resistant human pathogens in environmental reservoirs: A review. Front. Environ. Sci. Eng. 2019, 13, 46. [Google Scholar] [CrossRef]
- Linton, A.H.; Hinton, M.H. Enterobacteriaceae associated with animals in health and disease. Soc. Appl. Bacteriol. Symp. Ser. 1988, 17, 71S–85S. [Google Scholar] [CrossRef] [PubMed]
- Feuerstein, A.; Scuda, N.; Klose, C.; Hoffmann, A.; Melchner, A.; Boll, K.; Rettinger, A.; Fell, S.; Straubinger, R.K.; Riehm, J.M. Antimicrobial Resistance, Serologic and Molecular Characterization of E. coli Isolated from Calves with Severe or Fatal Enteritis in Bavaria, Germany. Antibiotics 2021, 11, 23. [Google Scholar] [CrossRef]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Larsson, D.G.J.; Flach, C.F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Smits, J.E.; Fernie, K.J. Avian wildlife as sentinels of ecosystem health. Comp. Immunol. Microbiol. Infect. Dis. 2013, 36, 333–342. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).