Expression of Proteins, Glycoproteins, and Transcripts in the Guts of Fasting, Fed, and Trypanosoma cruzi-Infected Triatomines: A Systematic Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Systematic Search
2.2. Inclusion Criteria
2.3. Information Capture
2.4. Parameters for Transcript, Protein, and Glycoprotein Analysis
3. Results
3.1. General Analysis of Selected Articles
3.2. Characteristics of Included Publications
3.3. Reports of Transcripts, Proteins, and Glycoproteins
3.4. Triatomine Species Used in the Studies
3.5. Feeding Conditions and T. cruzi Infection
3.6. Triatomine Gut Regions
3.7. Analysis of Reported Transcripts
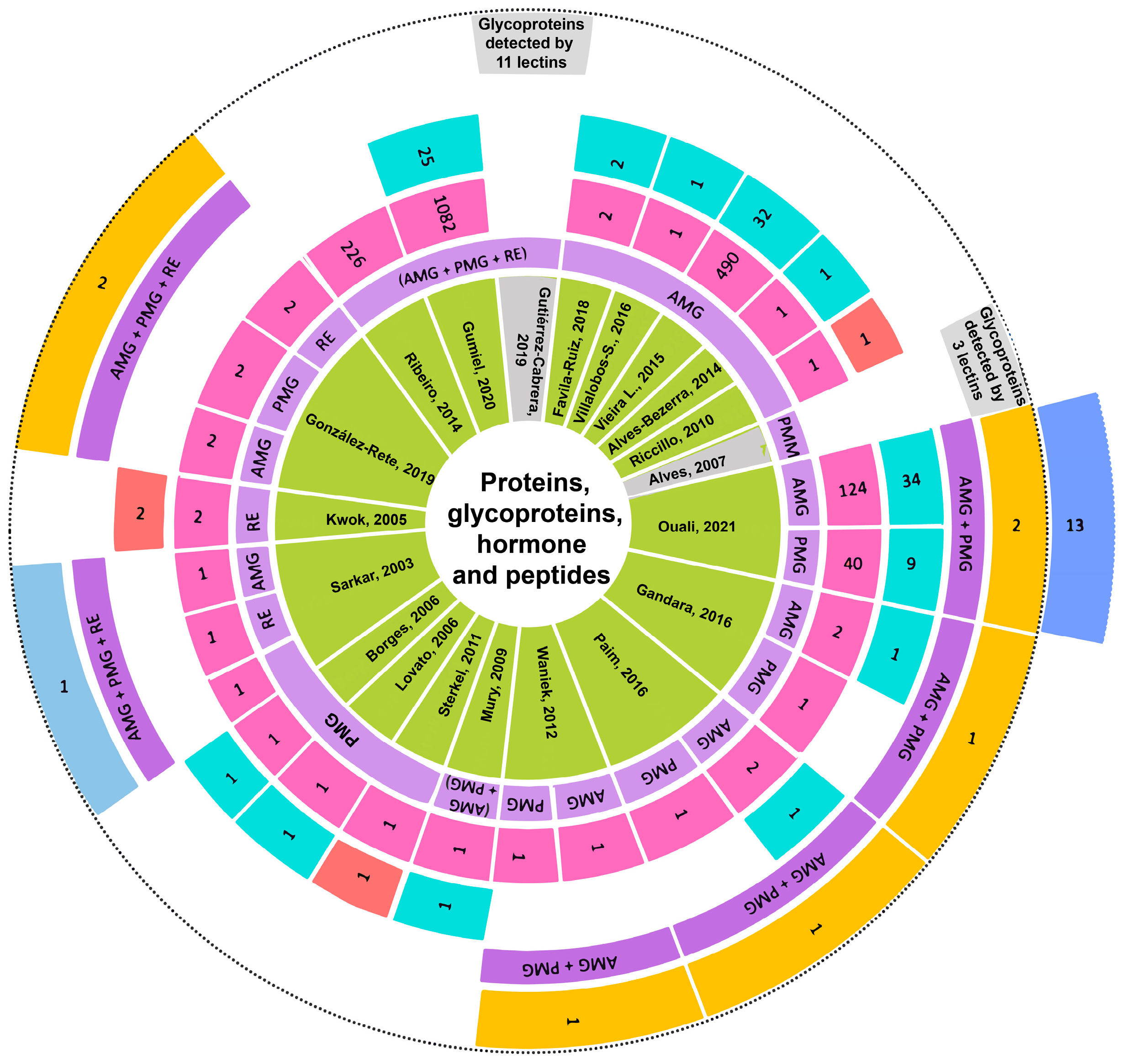
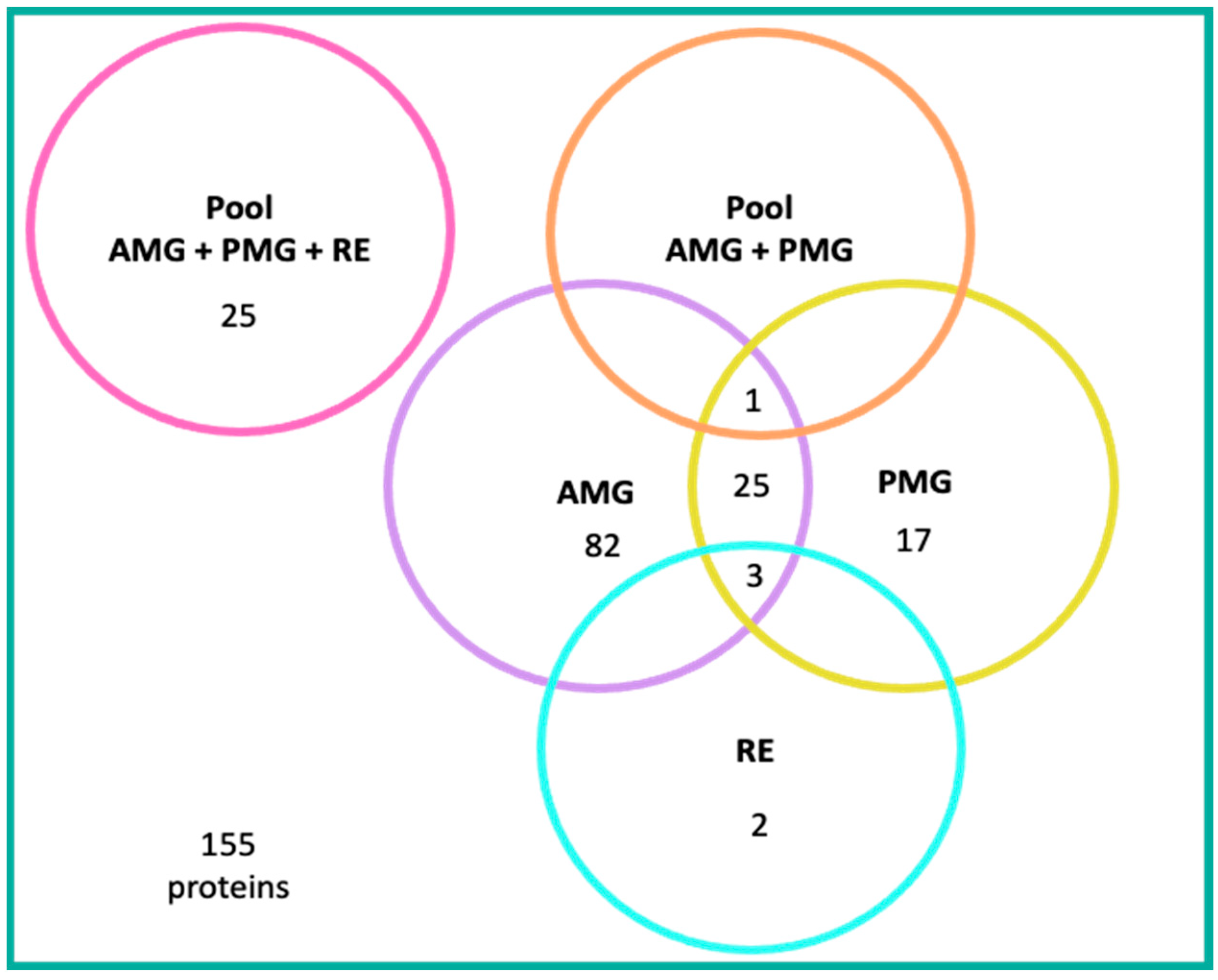
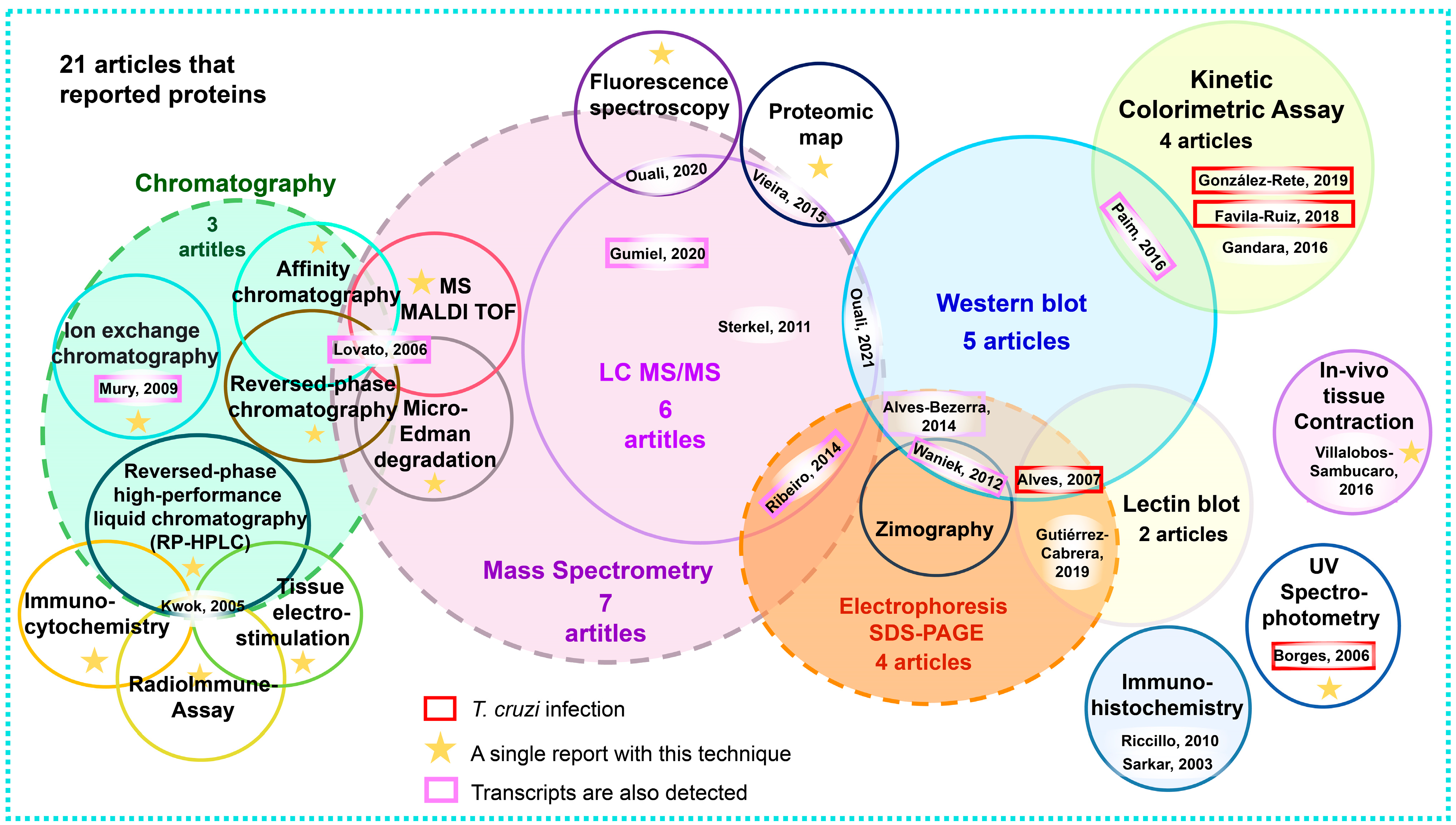
3.8. Analysis of Reported Proteins
4. Discussion
4.1. Transcripts Are More Frequently Reported Than Proteins
4.2. Triatomine Species
4.3. Discussion on Transcripts
4.3.1. Transcripts by Gut Sections
4.3.2. Feeding Stimulates Transcript Expression
4.3.3. T. cruzi Modulates Transcript Expression Differentially Depending on the Strain
4.4. Discussion on Proteins
4.4.1. Proteins Detected in the AMG
4.4.2. Proteins Detected in the PMG
4.4.3. Proteins Expressed in Both AMG and PMG
4.4.4. Proteins Detected in RE
4.4.5. Proteins Detected in Pooled Samples
4.4.6. Food Stimulates Protein Expression
4.4.7. Proteins with Unchanged Expression Levels
4.4.8. T. cruzi Modulates Protein Expression Differentially, Depending on the Strain and Gut Section
4.4.9. Other Proteins and Glycoproteins
4.4.10. Recognition between Glycoproteins in Triatomine Gut and T. cruzi
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- PAHO Chagas Disease–PAHO/WHO|Pan American Health Organization. Available online: https://www.paho.org/en/topics/chagas-disease (accessed on 16 March 2023).
- WHO Chagas Disease (American Trypanosomiasis). Available online: https://www.who.int/health-topics/chagas-disease (accessed on 16 March 2023).
- Salazar-Schettino, P.M.; Bucio Torres, M.I.; Cabrera-Bravo, M.; De Alba-Alvarado, M.; Castillo-Saldaña, D.R.; Zenteno-Galindo, E.; Rojo-Medina, J.; Fernández-Santos, N.; Perera-Salazar, M.G. Enfermedad de Chagas en México. Rev. De La Fac. De Med. De La UNAM 2016, 59, 6–16. [Google Scholar]
- Kollien, A.; Schaub, G. The Development of Trypanosoma cruzi in Triatominae. Parasitol. Today 2000, 16, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Moremen, K.W.; Tiemeyer, M.; Nairn, A.V. Vertebrate Protein Glycosylation: Diversity, Synthesis and Function. Nat. Rev. Mol. Cell Biol. 2012, 13, 448–462. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Devenport, M.; Jacobs-Lorena, M. The Peritrophic Matrix of Hematophagous Insects. Arch. Insect Biochem. Physiol. 2001, 47, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Ouali, R.; Vieira, L.R.; Salmon, D.; Bousbata, S. Early Post-Prandial Regulation of Protein Expression in the Midgut of Chagas Disease Vector Rhodnius prolixus Highlights New Potential Targets for Vector Control Strategy. Microorganisms 2021, 9, 804. [Google Scholar] [CrossRef]
- Garcia, E.S.; Genta, F.A.; de Azambuja, P.; Schaub, G.A. Interactions between Intestinal Compounds of Triatomines and Trypanosoma cruzi. Trends Parasitol. 2010, 26, 499–505. [Google Scholar] [CrossRef]
- Bonay, P.; Molina, R.; Fresno, M. Binding Specificity of Mannose-Specific Carbohydrate-Binding Protein from the Cell Surface of Trypanosoma cruzi. Glycobiology 2001, 11, 719–729. [Google Scholar] [CrossRef][Green Version]
- De Lederkremer, R.M.; Agusti, R. Chapter 7 Glycobiology of Trypanosoma cruzi. In Advances in Carbohydrate Chemistry and Biochemistry; Elsevier: Amsterdam, Netherlands, 2009; Volume 62, pp. 311–366. ISBN 978-0-12-374743-3. [Google Scholar]
- Gonzalez, M.S.; Souza, M.S.; Garcia, E.S.; Nogueira, N.F.S.; Mello, C.B.; Cánepa, G.E.; Bertotti, S.; Durante, I.M.; Azambuja, P.; Buscaglia, C.A. Trypanosoma cruzi TcSMUG L-Surface Mucins Promote Development and Infectivity in the Triatomine Vector Rhodnius prolixus. Public Libr. Sci. Negl. Trop. Dis. 2013, 7, e2552. [Google Scholar] [CrossRef]
- Alves, C.R.; Albuquerque-Cunha, J.M.; Mello, C.B.; Garcia, E.S.; Nogueira, N.F.; Bourguingnon, S.C.; de Souza, W.; Azambuja, P.; Gonzalez, M.S. Trypanosoma cruzi: Attachment to Perimicrovillar Membrane Glycoproteins of Rhodnius prolixus. Exp. Parasitol. 2007, 116, 44–52. [Google Scholar] [CrossRef]
- Mesquita, R.D.; Vionette-Amaral, R.J.; Lowenberger, C.; Rivera-Pomar, R.; Monteiro, F.A.; Minx, P.; Spieth, J.; Carvalho, A.B.; Panzera, F.; Lawson, D.; et al. Genome of Rhodnius prolixus, an Insect Vector of Chagas Disease, Reveals Unique Adaptations to Hematophagy and Parasite Infection. Proc. Natl. Acad. Sci. USA 2015, 112, 14936–14941. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. Br. Med. J. 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Cabrera, A.E.; Zandberg, W.F.; Zenteno, E.; Rodríguez, M.H.; Espinoza, B.; Lowenberger, C. Glycosylation on Proteins of the Intestine and Perimicrovillar Membrane of Triatoma (Meccus) pallidipennis, under Different Feeding Conditions. Insect Sci. 2019, 26, 796–808. [Google Scholar] [CrossRef] [PubMed]
- Carvalho-Costa, T.M.; Tiveron, R.D.R.; Mendes, M.T.; Barbosa, C.G.; Nevoa, J.C.; Roza, G.A.; Silva, M.V.; Figueiredo, H.C.P.; Rodrigues, V.; Soares, S.d.C.; et al. Salivary and Intestinal Transcriptomes Reveal Differential Gene Expression in Starving, Fed and Trypanosoma cruzi-Infected Rhodnius neglectus. Front. Cell. Infect. Microbiol. 2021, 11, 773357. [Google Scholar] [CrossRef] [PubMed]
- Buarque, D.S.; Gomes, C.M.; Araújo, R.N.; Pereira, M.H.; Ferreira, R.C.; Guarneri, A.A.; Tanaka, A.S. A New Antimicrobial Protein from the Anterior Midgut of Triatoma infestans Mediates Trypanosoma cruzi Establishment by Controlling the Microbiota. Biochimie 2016, 123, 138–143. [Google Scholar] [CrossRef]
- Riccillo, F.L.; Ronderos, J.R. Allatotropin Expression during the Development of the Fourth Instar Larvae of the Kissing-Bug Triatoma infestans (Klüg). Tissue Cell 2010, 42, 355–359. [Google Scholar] [CrossRef]
- Díaz-Garrido, P.; Cárdenas-Guerra, R.E.; Martínez, I.; Poggio, S.; Rodríguez-Hernández, K.; Rivera-Santiago, L.; Ortega-López, J.; Sánchez-Esquivel, S.; Espinoza, B. Differential Activity on Trypanosomatid Parasites of a Novel Recombinant Defensin Type 1 from the Insect Triatoma (Meccus) pallidipennis. Insect Biochem. Mol. Biol. 2021, 139, 103673. [Google Scholar] [CrossRef]
- Vieira, L.R.; Polomé, A.; Mesquita, R.D.; Salmon, D.; Braz, G.R.C.; Bousbata, S. Protein 2DE Reference Map of the Anterior Midgut of the Blood-Sucking Bug Rhodnius prolixus. Proteomics 2015, 15, 3901–3904. [Google Scholar] [CrossRef]
- Alves-Bezerra, M.; Majerowicz, D.; Grillo, L.A.M.; Tremonte, H.; Almeida, C.B.; Braz, G.R.C.; Sola-Penna, M.; Paiva-Silva, G.O.; Gondim, K.C. Serotonin Regulates an Acyl-CoA-Binding Protein (ACBP) Gene Expression in the Midgut of Rhodnius prolixus. Insect Biochem. Mol. Biol. 2010, 40, 119–125. [Google Scholar] [CrossRef]
- Soares, T.S.; Buarque, D.S.; Queiroz, B.R.; Gomes, C.M.; Braz, G.R.C.; Araújo, R.N.; Pereira, M.H.; Guarneri, A.A.; Tanaka, A.S. A Kazal-Type Inhibitor Is Modulated by Trypanosoma cruzi to Control Microbiota inside the Anterior Midgut of Rhodnius prolixus. Biochimie 2015, 112, 41–48. [Google Scholar] [CrossRef]
- Mury, F.B.; da Silva, J.R.; Ferreira, L.S.; dos Santos Ferreira, B.; de Souza-Filho, G.A.; de Souza-Neto, J.A.; Ribolla, P.E.M.; Silva, C.P.; do Nascimento, V.V.; Machado, O.L.T.; et al. Alpha-Glucosidase Promotes Hemozoin Formation in a Blood-Sucking Bug: An Evolutionary History. Public Libr. Sci. One 2009, 4, e6966. [Google Scholar] [CrossRef]
- Araújo, C.A.C.; Pacheco, J.P.F.; Waniek, P.J.; Geraldo, R.B.; Sibajev, A.; Dos Santos, A.L.; Evangelho, V.G.O.; Dyson, P.J.; Azambuja, P.; Ratcliffe, N.A.; et al. A Rhamnose-Binding Lectin from Rhodnius prolixus and the Impact of Its Silencing on Gut Bacterial Microbiota and Trypanosoma cruzi. Dev. Comp. Immunol. 2021, 114, 103823. [Google Scholar] [CrossRef]
- Zandawala, M.; Orchard, I. Identification and Functional Characterization of FGLamide-Related Allatostatin Receptor in Rhodnius prolixus. Insect Biochem. Mol. Biol. 2015, 57, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Waniek, P.J.; Castro, H.C.; Sathler, P.C.; Miceli, L.; Jansen, A.M.; Araújo, C.A.C. Two Novel Defensin-Encoding Genes of the Chagas Disease Vector Triatoma brasiliensis (Reduviidae, Triatominae): Gene Expression and Peptide-Structure Modeling. J. Insect Physiol. 2009, 55, 840–848. [Google Scholar] [CrossRef] [PubMed]
- Batista, K.K.S.; Vieira, C.S.; Figueiredo, M.B.; Costa-Latgé, S.G.; Azambuja, P.; Genta, F.A.; Castro, D.P. Influence of Serratia marcescens and Rhodococcus rhodnii on the Humoral Immunity of Rhodnius prolixus. Int. J. Mol. Sci. 2021, 22, 10901. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, J.M.C.; Genta, F.A.; Sorgine, M.H.F.; Logullo, R.; Mesquita, R.D.; Paiva-Silva, G.O.; Majerowicz, D.; Medeiros, M.; Koerich, L.; Terra, W.R.; et al. An Insight into the Transcriptome of the Digestive Tract of the Bloodsucking Bug, Rhodnius prolixus. Public Libr. Sci. Negl. Trop. Dis. 2014, 8, e2594. [Google Scholar] [CrossRef]
- Balczun, C.; Knorr, E.; Topal, H.; Meiser, C.K.; Kollien, A.H.; Schaub, G.A. Sequence Characterization of an Unusual Lysozyme Gene Expressed in the Intestinal Tract of the Reduviid Bug Triatoma infestans (Insecta). Parasitol. Res. 2008, 102, 229–232. [Google Scholar] [CrossRef]
- Vieira, C.S.; Figueiredo, M.B.; Moraes, C.d.S.; Pereira, S.B.; Dyson, P.; Mello, C.B.; Castro, D.P.; Azambuja, P. Azadirachtin Interferes with Basal Immunity and Microbial Homeostasis in the Rhodnius prolixus Midgut. Dev. Comp. Immunol. 2021, 114, 103864. [Google Scholar] [CrossRef]
- Alves-Bezerra, M.; Cosentino-Gomes, D.; Vieira, L.P.; Rocco-Machado, N.; Gondim, K.C.; Meyer-Fernandes, J.R. Identification of Uncoupling Protein 4 from the Blood-Sucking Insect Rhodnius prolixus and Its Possible Role on Protection against Oxidative Stress. Insect Biochem. Mol. Biol. 2014, 50, 24–33. [Google Scholar] [CrossRef]
- Ursic-Bedoya, R.J.; Nazzari, H.; Cooper, D.; Triana, O.; Wolff, M.; Lowenberger, C. Identification and Characterization of Two Novel Lysozymes from Rhodnius prolixus, a Vector of Chagas Disease. J. Insect Physiol. 2008, 54, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Gumiel, M.; de Mattos, D.P.; Vieira, C.S.; Moraes, C.S.; Moreira, C.J.d.C.; Gonzalez, M.S.; Teixeira-Ferreira, A.; Waghabi, M.; Azambuja, P.; Carels, N. Proteome of the Triatomine Digestive Tract: From Catalytic to Immune Pathways; Focusing on Annexin Expression. Front. Mol. Biosci. 2020, 7, 589435. [Google Scholar] [CrossRef]
- Vieira, C.S.; Waniek, P.J.; Mattos, D.P.; Castro, D.P.; Mello, C.B.; Ratcliffe, N.A.; Garcia, E.S.; Azambuja, P. Humoral Responses in Rhodnius prolixus: Bacterial Feeding Induces Differential Patterns of Antibacterial Activity and Enhances MRNA Levels of Antimicrobial Peptides in the Midgut. Parasites Vectors 2014, 7, 232. [Google Scholar] [CrossRef] [PubMed]
- Ursic-Bedoya, R.J.; Lowenberger, C.A. Rhodnius prolixus: Identification of Immune-Related Genes up-Regulated in Response to Pathogens and Parasites Using Suppressive Subtractive Hybridization. Dev. Comp. Immunol. 2007, 31, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Ouali, R.; Valentim de Brito, K.C.; Salmon, D.; Bousbata, S. High-Throughput Identification of the Rhodnius prolixus Midgut Proteome Unravels a Sophisticated Hematophagic Machinery. Proteomes 2020, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Waniek, P.J.; Araújo, C.A.C.; Momoli, M.M.; Azambuja, P.; Jansen, A.M.; Genta, F.A. Serine Carboxypeptidases of Triatoma brasiliensis (Hemiptera, Reduviidae): Sequence Characterization, Expression Pattern and Activity Localization. J. Insect Physiol. 2014, 63, 9–20. [Google Scholar] [CrossRef]
- Sangha, V.; Lange, A.B.; Orchard, I. Identification and Cloning of the Kinin Receptor in the Chagas Disease Vector, Rhodnius prolixus. Gen. Comp. Endocrinol. 2020, 289, 113380. [Google Scholar] [CrossRef]
- Buarque, D.S.; Braz, G.R.C.; Martins, R.M.; Tanaka-Azevedo, A.M.; Gomes, C.M.; Oliveira, F.A.A.; Schenkman, S.; Tanaka, A.S. Differential Expression Profiles in the Midgut of Triatoma infestans Infected with Trypanosoma cruzi. Public Libr. Sci. One 2013, 8, e61203. [Google Scholar] [CrossRef]
- Araujo, R.N.; Campos, I.T.N.; Tanaka, A.S.; Santos, A.; Gontijo, N.F.; Lehane, M.J.; Pereira, M.H. Brasiliensin: A Novel Intestinal Thrombin Inhibitor from Triatoma brasiliensis (Hemiptera: Reduviidae) with an Important Role in Blood Intake. Int. J. Parasitol. 2007, 37, 1351–1358. [Google Scholar] [CrossRef]
- Staniscuaski, F.; Paluzzi, J.-P.; Real-Guerra, R.; Carlini, C.R.; Orchard, I. Expression Analysis and Molecular Characterization of Aquaporins in Rhodnius prolixus. J. Insect Physiol. 2013, 59, 1140–1150. [Google Scholar] [CrossRef]
- Whitten, M.; Sun, F.; Tew, I.; Schaub, G.; Soukou, C.; Nappi, A.; Ratcliffe, N. Differential Modulation of Rhodnius prolixus Nitric Oxide Activities Following Challenge with Trypanosoma rangeli, T. cruzi and Bacterial Cell Wall Components. Insect Biochem. Mol. Biol. 2007, 37, 440–452. [Google Scholar] [CrossRef]
- González-Rete, B.; Salazar-Schettino, P.M.; Bucio-Torres, M.I.; Córdoba-Aguilar, A.; Cabrera-Bravo, M. Activity of the Prophenoloxidase System and Survival of Triatomines Infected with Different Trypanosoma cruzi Strains under Different Temperatures: Understanding Chagas Disease in the Face of Climate Change. Parasites Vectors 2019, 12, 219. [Google Scholar] [CrossRef]
- Sterkel, M.; Oliveira, P.L.; Urlaub, H.; Hernandez-Martinez, S.; Rivera-Pomar, R.; Ons, S. OKB, a Novel Family of Brain-Gut Neuropeptides from Insects. Insect Biochem. Mol. Biol. 2012, 42, 466–473. [Google Scholar] [CrossRef] [PubMed]
- Lovato, D.V.; Nicolau de Campos, I.T.; Amino, R.; Tanaka, A.S. The Full-Length CDNA of Anticoagulant Protein Infestin Revealed a Novel Releasable Kazal Domain, a Neutrophil Elastase Inhibitor Lacking Anticoagulant Activity. Biochimie 2006, 88, 673–681. [Google Scholar] [CrossRef] [PubMed]
- Favila-Ruiz, G.; Jiménez-Cortés, J.G.; Córdoba-Aguilar, A.; Salazar-Schettino, P.M.; Gutiérrez-Cabrera, A.E.; Pérez-Torres, A.; De Fuentes-Vicente, J.A.; Vences-Blanco, M.O.; Bucio-Torres, M.I.; Flores-Villegas, A.L.; et al. Effects of Trypanosoma cruzi on the Phenoloxidase and Prophenoloxidase Activity in the Vector Meccus pallidipennis (Hemiptera: Reduviidae). Parasites Vectors 2018, 11, 434. [Google Scholar] [CrossRef]
- Balczun, C.; Siemanowski, J.; Pausch, J.K.; Helling, S.; Marcus, K.; Stephan, C.; Meyer, H.E.; Schneider, T.; Cizmowski, C.; Oldenburg, M.; et al. Intestinal Aspartate Proteases TiCatD and TiCatD2 of the Haematophagous Bug Triatoma infestans (Reduviidae): Sequence Characterisation, Expression Pattern and Characterisation of Proteolytic Activity. Insect Biochem. Mol. Biol. 2012, 42, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Borges, E.C.; Machado, E.M.M.; Garcia, E.S.; Azambuja, P. Trypanosoma cruzi: Effects of Infection on Cathepsin D Activity in the Midgut of Rhodnius prolixus. Exp. Parasitol. 2006, 112, 130–133. [Google Scholar] [CrossRef] [PubMed]
- Sedra, L.; Paluzzi, J.-P.; Lange, A.B. Characterization and Expression of a Long Neuropeptide F (NPF) Receptor in the Chagas Disease Vector Rhodnius prolixus. Public Libr. Sci. One 2018, 13, e0202425. [Google Scholar] [CrossRef]
- Paim, R.M.; Pereira, M.H.; Di Ponzio, R.; Rodrigues, J.O.; Guarneri, A.A.; Gontijo, N.F.; Araújo, R.N. Validation of Reference Genes for Expression Analysis in the Salivary Gland and the Intestine of Rhodnius prolixus (Hemiptera, Reduviidae) under Different Experimental Conditions by Quantitative Real-Time PCR. BMC Res. Notes 2012, 5, 128. [Google Scholar] [CrossRef] [PubMed]
- Kwok, R.; Chung, D.; Brugge, V.T.; Orchard, I. The Distribution and Activity of Tachykinin-Related Peptides in the Blood-Feeding Bug, Rhodnius prolixus. Peptides 2005, 26, 43–51. [Google Scholar] [CrossRef]
- Vieira, C.S.; Moreira, O.C.; Batista, K.K.S.; Ratcliffe, N.A.; Castro, D.P.; Azambuja, P. The NF-ΚB Inhibitor, IMD-0354, Affects Immune Gene Expression, Bacterial Microbiota and Trypanosoma cruzi Infection in Rhodnius prolixus Midgut. Front. Physiol. 2018, 9, 1189. [Google Scholar] [CrossRef]
- Paluzzi, J.-P.; O’Donnell, M.J. Identification, Spatial Expression Analysis and Functional Characterization of a Pyrokinin-1 Receptor in the Chagas’ Disease Vector, Rhodnius prolixus. Mol. Cell. Endocrinol. 2012, 363, 36–45. [Google Scholar] [CrossRef]
- Kollien, A.H.; Fechner, S.; Waniek, P.J.; Schaub, G.A. Isolation and Characterization of a CDNA Encoding for a Lysozyme from the Gut of the Reduviid Bug Triatoma Infestans. Archives of Insect Biochemistry and Physiology. 2003, 53, 134–145. [Google Scholar] [CrossRef] [PubMed]
- Gandara, A.C.P.; Oliveira, J.H.M.; Nunes, R.D.; Goncalves, R.L.S.; Dias, F.A.; Hecht, F.; Fernandes, D.C.; Genta, F.A.; Laurindo, F.R.M.; Oliveira, M.F.; et al. Amino Acids Trigger Down-Regulation of Superoxide via TORC Pathway in the Midgut of Rhodnius prolixus. Biosci. Rep. 2016, 36, e00321. [Google Scholar] [CrossRef] [PubMed]
- Waniek, P.J.; Pacheco Costa, J.E.; Jansen, A.M.; Costa, J.; Araújo, C.A.C. Cathepsin L of Triatoma brasiliensis (Reduviidae, Triatominae): Sequence Characterization, Expression Pattern and Zymography. J. Insect Physiol. 2012, 58, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, N.R.S.; Tobe, S.S.; Orchard, I. The Distribution and Effects of Dippu-Allatostatin-like Peptides in the Blood-Feeding Bug, Rhodnius prolixus. Peptides 2003, 24, 1553–1562. [Google Scholar] [CrossRef] [PubMed]
- Vieira, C.S.; Waniek, P.J.; Castro, D.P.; Mattos, D.P.; Moreira, O.C.; Azambuja, P. Impact of Trypanosoma cruzi on Antimicrobial Peptide Gene Expression and Activity in the Fat Body and Midgut of Rhodnius Prolixus. Parasites Vectors 2016, 9, 119. [Google Scholar] [CrossRef]
- Buarque, D.S.; Spindola, L.M.N.; Martins, R.M.; Braz, G.R.C.; Tanaka, A.S. Tigutcystatin, a Cysteine Protease Inhibitor from Triatoma infestans Midgut Expressed in Response to Trypanosoma cruzi. Biochem. Biophys. Res. Commun. 2011, 413, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Kollien, A.; Billingsley, P. Differential Display of MRNAs Associated with Blood Feeding in the Midgut of the Bloodsucking Bug, Triatoma Infestans. Parasitology Research 2002, 88, 1026–1033. [Google Scholar] [CrossRef] [PubMed]
- Paim, R.M.M.; Araujo, R.N.; Leis, M.; Sant’anna, M.R.V.; Gontijo, N.F.; Lazzari, C.R.; Pereira, M.H. Functional Evaluation of Heat Shock Proteins 70 (HSP70/HSC70) on Rhodnius prolixus (Hemiptera, Reduviidae) Physiological Responses Associated with Feeding and Starvation. Insect Biochem. Mol. Biol. 2016, 77, 10–20. [Google Scholar] [CrossRef]
- Waniek, P.J.; Jansen, A.M.; Araújo, C.A.C. Trypanosoma cruzi Infection Modulates the Expression of Triatoma brasiliensis Def1 in the Midgut. Vector Borne Zoonotic Dis. 2011, 11, 845–847. [Google Scholar] [CrossRef]
- Lopez-Ordoñez, T.; Rodriguez, M.H.; de la Cruz Hernández-Hernández, F. Characterization of a cDNA encoding a cathepsin L-like protein of Rhodnius Prolixus Insect. Mol. Biol. 2001, 10, 505–511. [Google Scholar] [CrossRef]
- Alvarenga, E.S.L.; Mansur, J.F.; Justi, S.A.; Figueira-Mansur, J.; dos Santos, V.M.; Lopez, S.G.; Masuda, H.; Lara, F.A.; Melo, A.C.A.; Moreira, M.F. Chitin Is a Component of the Rhodnius prolixus Midgut. Insect Biochem. Mol. Biol. 2016, 69, 61–70. [Google Scholar] [CrossRef]
- Sterkel, M.; Urlaub, H.; Rivera-Pomar, R.; Ons, S. Functional Proteomics of Neuropeptidome Dynamics during the Feeding Process of Rhodnius prolixus. J. Proteome Res. 2011, 10, 3363–3371. [Google Scholar] [CrossRef]
- Villalobos-Sambucaro, M.J.; Diambra, L.A.; Noriega, F.G.; Ronderos, J.R. Allatostatin-C Antagonizes the Synergistic Myostimulatory Effect of Allatotropin and Serotonin in Rhodnius prolixus (Stal). Gen. Comp. Endocrinol. 2016, 233, 1–7. [Google Scholar] [CrossRef]
- Meiser, C.K.; Piechura, H.; Werner, T.; Dittmeyer-Schäfer, S.; Meyer, H.E.; Warscheid, B.; Schaub, G.A.; Balczun, C. Kazal-Type Inhibitors in the Stomach of Panstrongylus megistus (Triatominae, Reduviidae). Insect Biochem. Mol. Biol. 2010, 40, 345–353. [Google Scholar] [CrossRef]
- Mauri, M.; Elli, T.; Caviglia, G.; Uboldi, G.; Azzi, M. RAWGraphs: A Visualisation Platform to Create Open Outputs. In Proceedings of the Proceedings of the 12th Biannual Conference on Italian SIGCHI Chapter, Cagliari, Italy, 18–20 September 2017; pp. 1–5. [Google Scholar]
- Liebler, D.C. Introduction to Proteomics: Tools for the New Biology; Humana Press: Totowa, NJ, USA, 2002; p. 198. [Google Scholar]
- Alevi, K.C.C.; De Oliveira, J.; Da Silva Rocha, D.; Galvão, C. Trends in Taxonomy of Chagas Disease Vectors (Hemiptera, Reduviidae, Triatominae): From Linnaean to Integrative Taxonomy. Pathogens 2021, 10, 1627. [Google Scholar] [CrossRef]
- Hashimoto, K.; Schofield, C.J. Elimination of Rhodnius prolixus in Central America. Parasites Vectors 2012, 5, 45. [Google Scholar] [CrossRef]
- Terra, W.R.; Ferreira, C.; Garcia, E.S. Origin, Distribution, Properties and Functions of the Major Rhodnius prolixus Midgut Hydrolases. Insect Biochem. 1988, 18, 423–434. [Google Scholar] [CrossRef]
- González-Santoyo, I.; Córdoba-Aguilar, A. Phenoloxidase: A Key Component of the Insect Immune System: Biochemical and Evolutionary Ecology of PO. Entomol. Exp. Et Appl. 2012, 142, 1–16. [Google Scholar] [CrossRef]
- Flores-Villegas, A.L.; Salazar-Schettino, P.M.; Córdoba-Aguilar, A.; Gutiérrez-Cabrera, A.E.; Rojas-Wastavino, G.E.; Bucio-Torres, M.I.; Cabrera-Bravo, M. Immune Defence Mechanisms of Triatomines against Bacteria, Viruses, Fungi and Parasites. Bull. Entomol. Res. 2015, 105, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Schaub, G.A.; Meiser, C.K.; Balczun, C. Interactions of Trypanosoma cruzi and Triatomines. In Progress in Parasitology; Mehlhorn, H., Ed.; Springer: Berlin, Heidelberg, Germany, 2011; pp. 155–178. ISBN 978-3-642-21395-3. [Google Scholar]
- Castro, D.P.; Moraes, C.S.; Gonzalez, M.S.; Ratcliffe, N.A.; Azambuja, P.; Garcia, E.S. Trypanosoma cruzi Immune Response Modulation Decreases Microbiota in Rhodnius prolixus Gut and Is Crucial for Parasite Survival and Development. Public Libr. Sci. One 2012, 7, e36591. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Cabrera, A.E.; Alejandre-Aguilar, R.; Hernández-Martínez, S.; Espinoza, B. Development and Glycoprotein Composition of the Perimicrovillar Membrane in Triatoma (Meccus) pallidipennis (Hemiptera: Reduviidae). Arthropod Struct. Dev. 2014, 43, 571–578. [Google Scholar] [CrossRef]

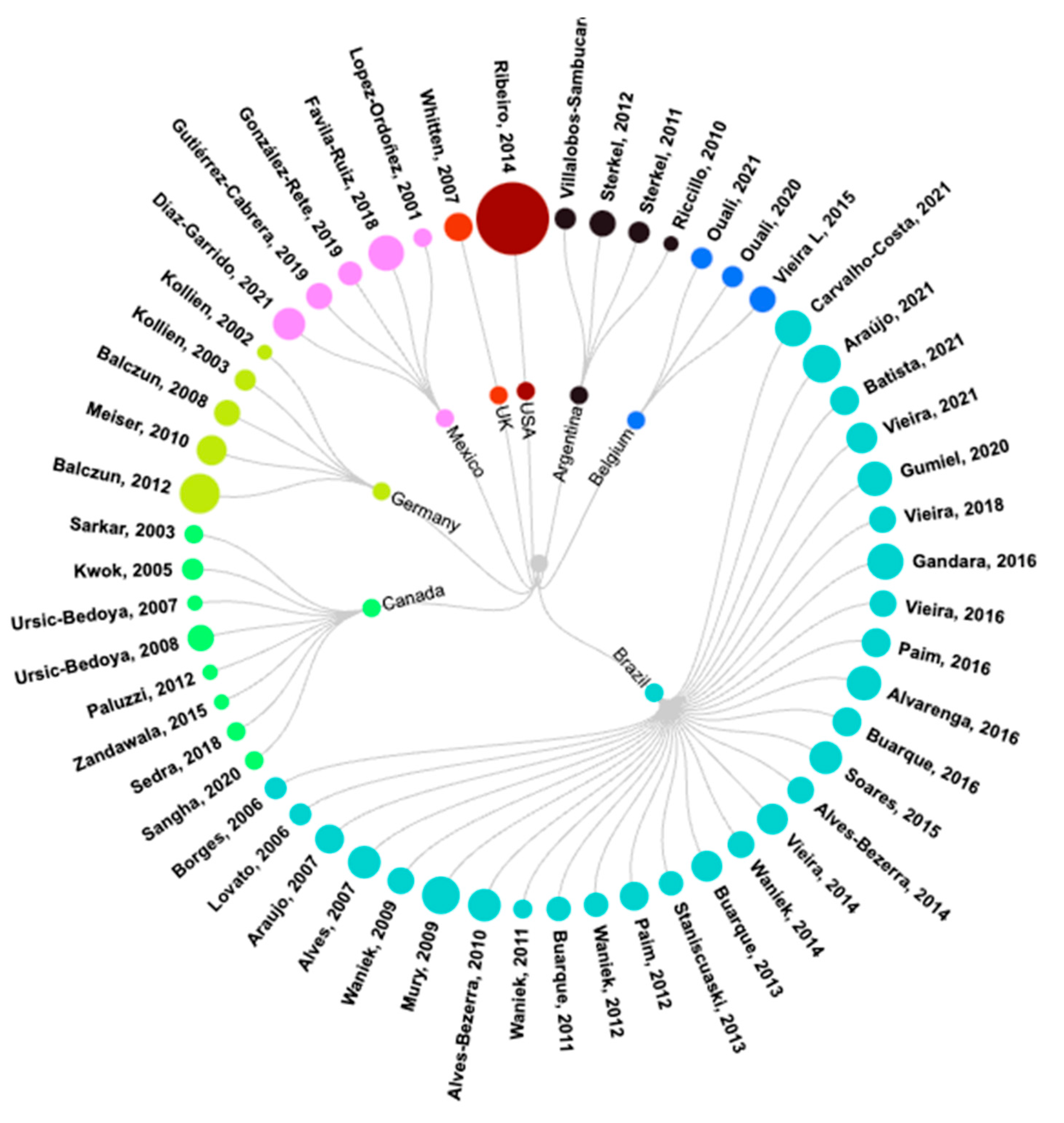
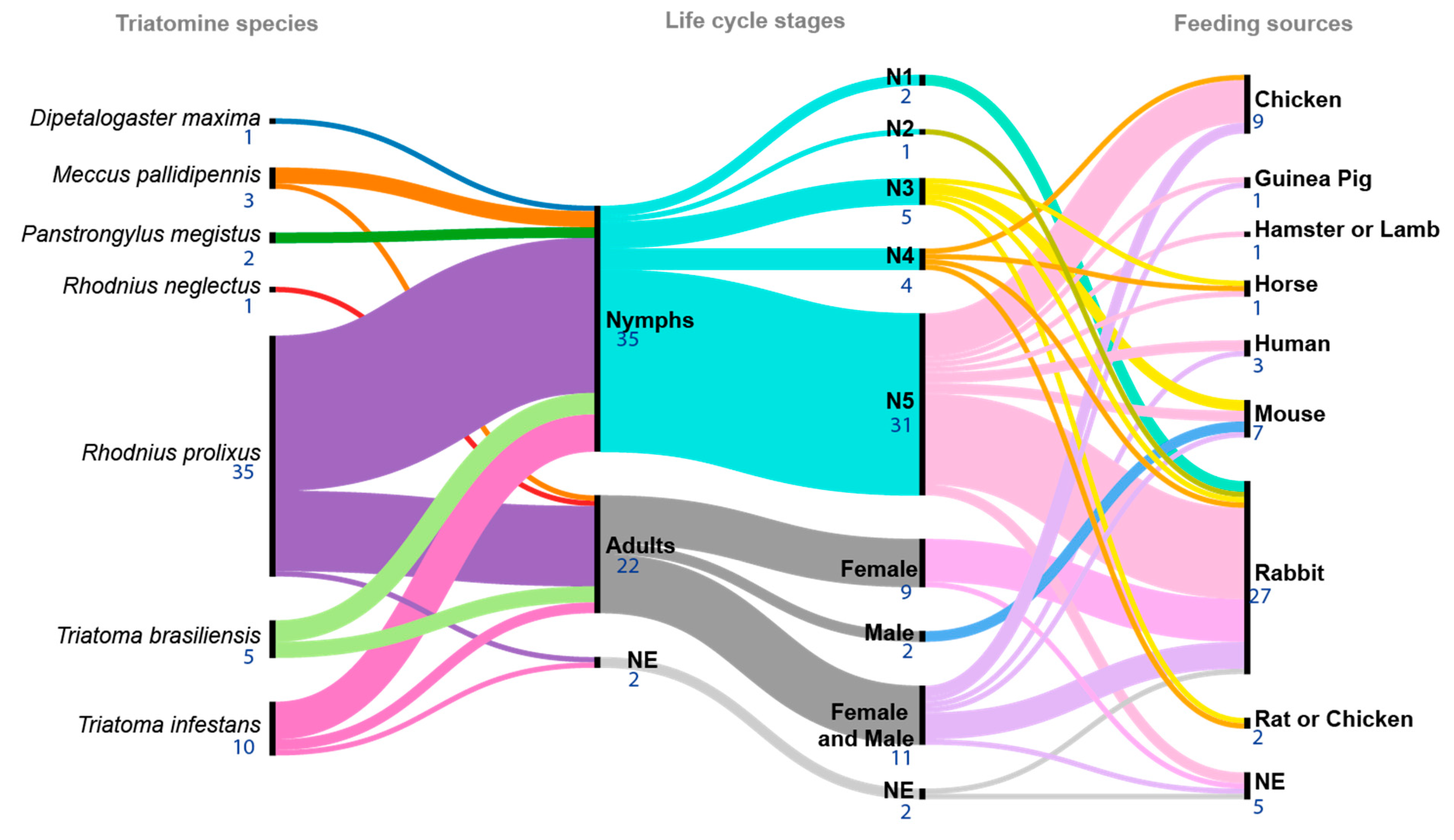
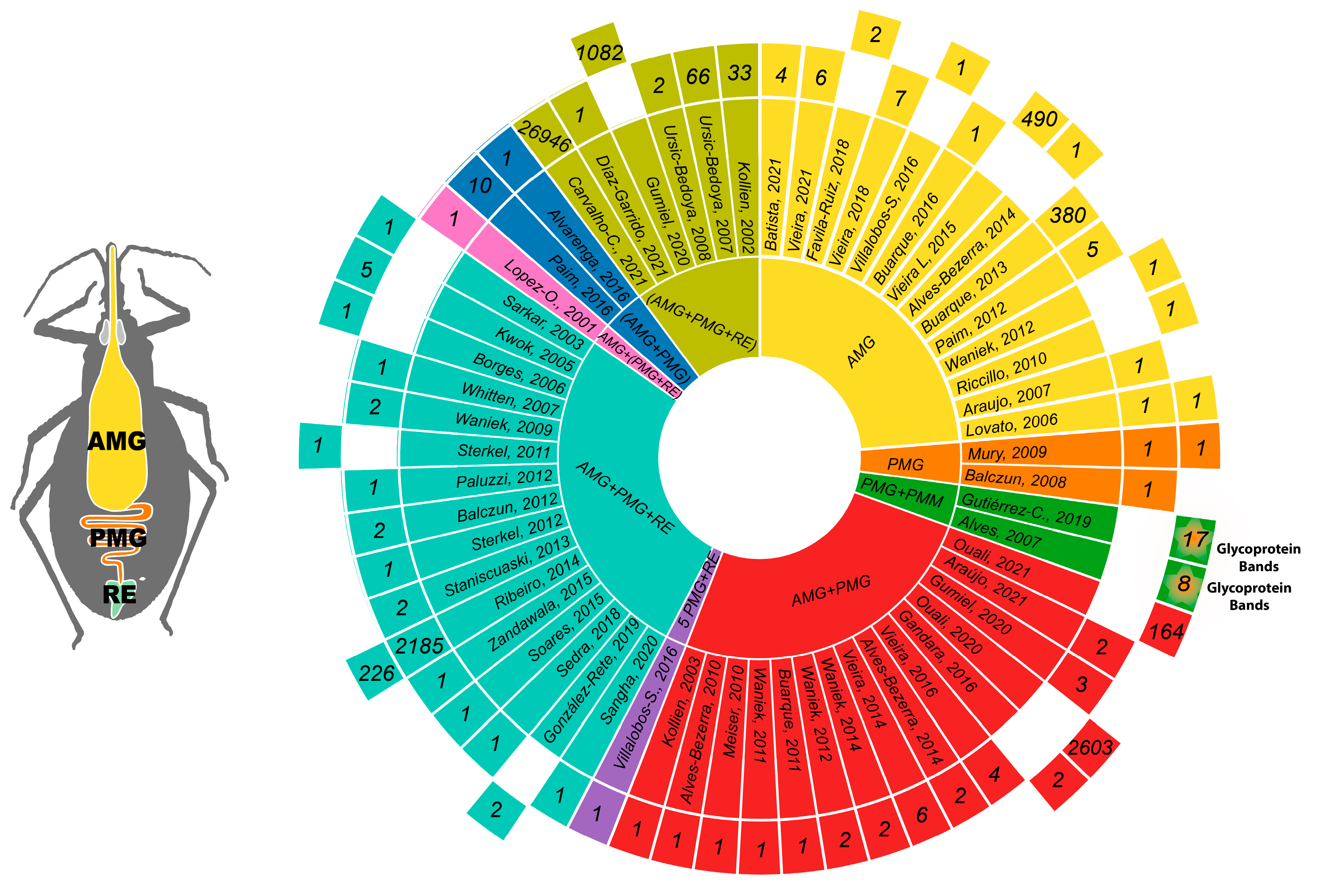
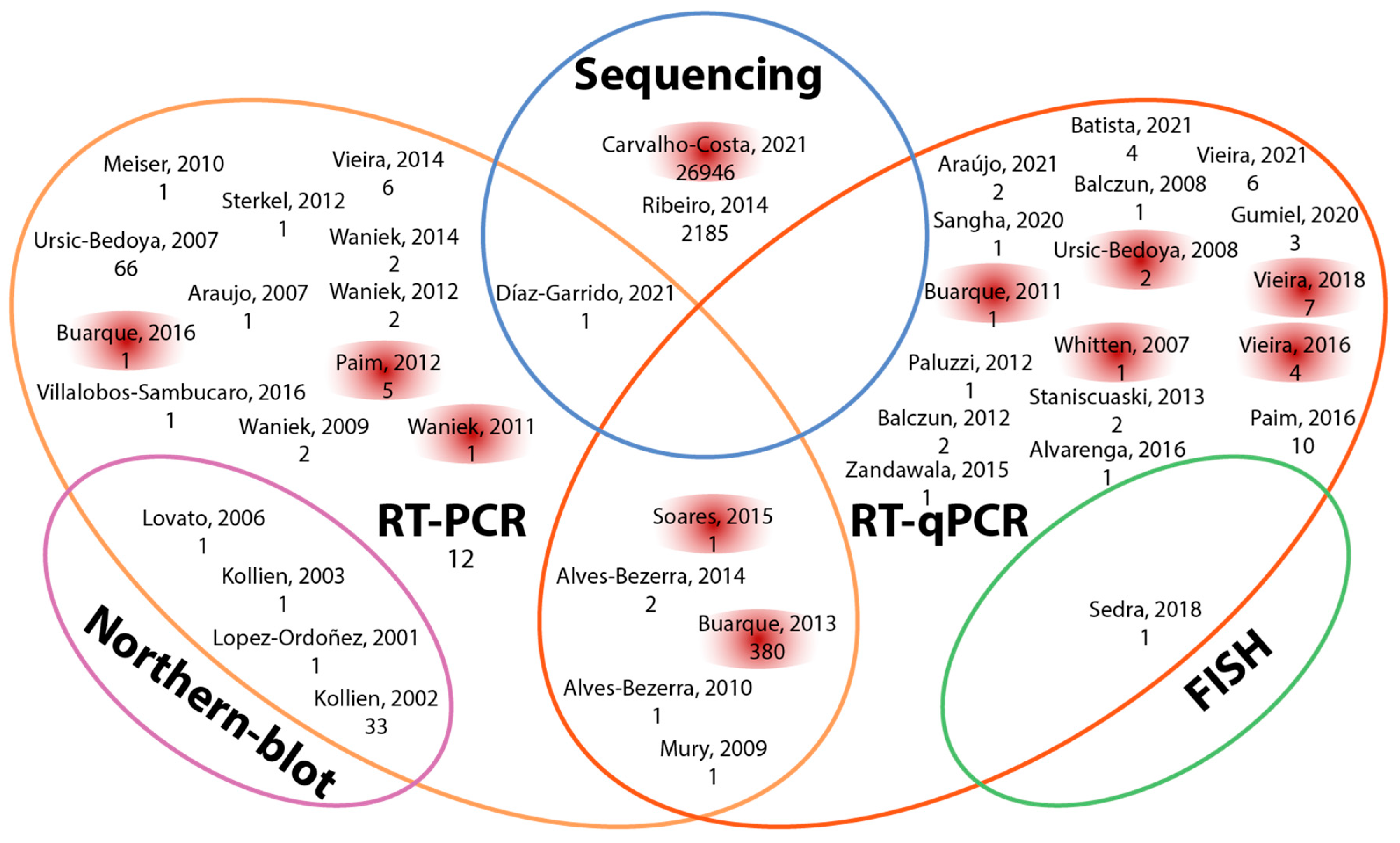
| # | Year | Author | T/P | Ref. | # | Year | Author | T/P | Ref. | # | Year | Author | T/P | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2021 | Carvalho-Costa, et al. | T | [16] | 20 | 2016 | Buarque, et al. | T | [17] | 39 | 2010 | Riccillo, et al. | P | [18] |
| 2 | 2021 | Díaz-Garrido, et al. | T | [19] | 21 | 2015 | Vieira L., et al. | P | [20] | 40 | 2010 | Alves-Bezerra, et al. | T | [21] |
| 3 | 2021 | Ouali, et al. | P | [7] | 22 | 2015 | Soares, et al. | T | [22] | 41 | 2009 | Mury, et al. | T&P | [23] |
| 4 | 2021 | Araújo, et al. | T | [24] | 23 | 2015 | Zandawala, et al. | T | [25] | 42 | 2009 | Waniek, et al. | T | [26] |
| 5 | 2021 | Batista, et al. | T | [27] | 24 | 2014 | Ribeiro, et al. | T&P | [28] | 43 | 2008 | Balczun, et al. | T | [29] |
| 6 | 2021 | Vieira C., et al. | T | [30] | 25 | 2014 | Alves-Bezerra, et al. | T&P | [31] | 44 | 2008 | Ursic-Bedoya, et al. | T | [32] |
| 7 | 2020 | Gumiel, et al. | T&P | [33] | 26 | 2014 | Vieira C., et al. | T | [34] | 45 | 2007 | Ursic-Bedoya, et al. | T | [35] |
| 8 | 2020 | Ouali, et al. | P | [36] | 27 | 2014 | Waniek, et al. | T | [37] | 46 | 2007 | Alves, et al. | P | [12] |
| 9 | 2020 | Sangha, et al. | T | [38] | 28 | 2013 | Buarque, et al. | T | [39] | 47 | 2007 | Araujo, et al. | T | [40] |
| 10 | 2019 | Gutiérrez-Cabrera, et al. | P | [15] | 29 | 2013 | Staniscuaski, et al. | T | [41] | 48 | 2007 | Whitten, et al. | T | [42] |
| 11 | 2019 | González-Rete, et al. | P | [43] | 30 | 2013 | Sterkel, et al. | T | [44] | 49 | 2006 | Lovato, et al. | T&P | [45] |
| 12 | 2018 | Favila-Ruiz, et al. | P | [46] | 31 | 2012 | Balczun, et al. | T | [47] | 50 | 2006 | Borges, et al. | P | [48] |
| 13 | 2018 | Sedra, et al. | T | [49] | 32 | 2012 | Paim, et al. | T | [50] | 51 | 2005 | Kwok, et al. | P | [51] |
| 14 | 2018 | Vieira C, et al. | T | [52] | 33 | 2012 | Paluzzi, et al. | T | [53] | 52 | 2003 | Kollien, et al. | T | [54] |
| 15 | 2016 | Gandara, et al. | P | [55] | 34 | 2012 | Waniek, P et al. | T&P | [56] | 53 | 2003 | Sarkar, et al. | P | [57] |
| 16 | 2016 | Vieira, C. et al. | T | [58] | 35 | 2011 | Buarque, et al. | T | [59] | 54 | 2002 | Kollien, et al. | T | [60] |
| 17 | 2016 | Paim, et al. | T&P | [61] | 36 | 2011 | Waniek, et al. | T | [62] | 55 | 2001 | Lopez-Ordoñez, et al. | T | [63] |
| 18 | 2016 | Alvarenga, et al. | T | [64] | 37 | 2011 | Sterkel, et al. | P | [65] | |||||
| 19 | 2016 | VillalobosSambucaro, et al. | T&P | [66] | 38 | 2010 | Meiser, et al. | X | [67] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reynoso-Ducoing, O.A.; González-Rete, B.; Díaz, E.; Candelas-Otero, F.N.; López-Aviña, J.A.; Cabrera-Bravo, M.; Bucio-Torres, M.I.; Torres-Gutiérrez, E.; Salazar-Schettino, P.M. Expression of Proteins, Glycoproteins, and Transcripts in the Guts of Fasting, Fed, and Trypanosoma cruzi-Infected Triatomines: A Systematic Review. Pathogens 2023, 12, 1124. https://doi.org/10.3390/pathogens12091124
Reynoso-Ducoing OA, González-Rete B, Díaz E, Candelas-Otero FN, López-Aviña JA, Cabrera-Bravo M, Bucio-Torres MI, Torres-Gutiérrez E, Salazar-Schettino PM. Expression of Proteins, Glycoproteins, and Transcripts in the Guts of Fasting, Fed, and Trypanosoma cruzi-Infected Triatomines: A Systematic Review. Pathogens. 2023; 12(9):1124. https://doi.org/10.3390/pathogens12091124
Chicago/Turabian StyleReynoso-Ducoing, Olivia A., Berenice González-Rete, Elsa Díaz, Frida N. Candelas-Otero, J. Antonio López-Aviña, Margarita Cabrera-Bravo, Martha I. Bucio-Torres, Elia Torres-Gutiérrez, and Paz María Salazar-Schettino. 2023. "Expression of Proteins, Glycoproteins, and Transcripts in the Guts of Fasting, Fed, and Trypanosoma cruzi-Infected Triatomines: A Systematic Review" Pathogens 12, no. 9: 1124. https://doi.org/10.3390/pathogens12091124
APA StyleReynoso-Ducoing, O. A., González-Rete, B., Díaz, E., Candelas-Otero, F. N., López-Aviña, J. A., Cabrera-Bravo, M., Bucio-Torres, M. I., Torres-Gutiérrez, E., & Salazar-Schettino, P. M. (2023). Expression of Proteins, Glycoproteins, and Transcripts in the Guts of Fasting, Fed, and Trypanosoma cruzi-Infected Triatomines: A Systematic Review. Pathogens, 12(9), 1124. https://doi.org/10.3390/pathogens12091124








