Abstract
A vaginal microbiota dominated by certain Lactobacillus species may have a protective effect against Chlamydia trachomatis infection. One of the key antimicrobial compounds produced is lactic acid, which is believed to play a central role in host defense. Lactobacillus strains producing the D(−)-lactic acid isomer are known to exert stronger protection. However, the molecular mechanisms underlying this antimicrobial action are not well understood. The aim of this study was to investigate the role of D(−)-lactic acid isomer in the prevention of C. trachomatis infection in an in vitro HeLa cell model. We selected two strains of lactobacilli belonging to different species: a vaginal isolate of Lactobacillus crispatus that releases both D(−) and L(+) isomers and a strain of Lactobacillus reuteri that produces only the L(+) isomer. Initially, we demonstrated that L. crispatus was significantly more effective than L. reuteri in reducing C. trachomatis infectivity. A different pattern of histone acetylation and lactylation was observed when HeLa cells were pretreated for 24 h with supernatants of Lactobacillus crispatus or L. reuteri, resulting in different transcription of genes such as CCND1, CDKN1A, ITAG5 and HER-1. Similarly, distinct transcription patterns were found in HeLa cells treated with 10 mM D(−)- or L(+)-lactic acid isomers. Our findings suggest that D(−) lactic acid significantly affects two non-exclusive mechanisms involved in C. trachomatis infection: regulation of the cell cycle and expression of EGFR and α5β1-integrin.
1. Introduction
Chlamydia trachomatis (CT), an obligate intracellular pathogen, is the causative agent of the most common bacterial sexually transmitted infection (STI) worldwide with significant clinical and economic impact [1]. During its developmental cycle, CT alternates between two functionally and morphologically distinct forms: the extracellular form known as the infectious elementary body (EB) and the intracellular, noninfectious and replicating form known as the reticular body (RB). Since a large proportion of urogenital chlamydial infections in women are asymptomatic, they are at high risk of not being treated, leading to various complications and sequelae such as pelvic inflammatory disease, infertility and ectopic pregnancy [2].
A vaginal microbiota dominated by lactobacilli is crucial for the prevention of CT infections [3,4,5], although there are large differences depending on the species [6,7,8]. The protective role of lactobacilli against CT is exerted by several mechanisms acting on both the extracellular and intracellular steps of the cycle [9,10,11], including changes in lipid composition and exposure of the α5-integrin subunit in the plasma membrane of epithelial cells [12].
Lactobacilli provide protection against pathogens in several ways, including lactic acid (LA), short-range bacteriocins and long-range hydrogen peroxide (H2O2) production. It is believed that LA plays a central role in host defense, and it is important to emphasize that there are two isomeric forms of LA, D(−) and L(+) (D(−)-LA and L(+)-LA) [9,13,14]. The presence of all these compounds, whether independently or in combination, influences the host’s susceptibility to CT infection, highlighting the significance of the host–microbiota relationship.
Since both D(−) and L(+) isomers are present in vaginal fluid and their relative amounts depend on which lactobacillus species predominates [15,16,17], we decided to conduct a comprehensive study to investigate the role of the two LA isomers in the reduction in CT infectivity.
To this end, using an in vitro HeLa cell model, we investigated the protective role of two different isolates of vaginal lactobacilli, namely Lactobacillus crispatus (L. crispatus), which releases both D(−) and L(+) isomers, and Lactobacillus reuteri (L. reuteri), which produces only the L(+) isomer. Our goal was to investigate the effects of LA in the context of several non-exclusive mechanisms, such as host energy resources, epigenetic changes and cell-cycle regulation. The depletion of energy resources in host cells is critical for both EB infection and subsequent transformation into RBs. In addition, epigenetic changes can trigger expression of various genes involved in both cell-cycle regulation and surface plasma membrane protein modification. To investigate these points, we analyzed the effect of cell-free supernatants of lactobacilli supplemented with different amounts of D(−)-LA or L(+)-LA on the viability of HeLa cells. Subsequently, cellular metabolic status was assessed by oxygen consumption rate (OCR). Finally, we analyzed both the acetylation and lactylation status of histones from HeLa cells treated with L. crispatus or L. reuteri supernatants.
2. Materials and Methods
2.1. Lactobacilli Strains and Growth Conditions
Lactobacilli were isolated during routine diagnostic procedures from vaginal samples submitted to the Microbiology Unit of Sant’Orsola-Malpighi Hospital of Bologna (Italy), using de Man, Rogosa and Sharpe (MRS) agar plates (Difco, Detroit, MI, USA) in anaerobic conditions. For bacterial identification, MALDI-TOF MS analysis (Bruker Daltonics, Bremen, Germany) was performed by using direct analysis of bacterial colonies as previously detailed [18]. For further experiments, two different isolates were used, one belonging to L. crispatus species and the other to L. reuteri species, identified by MALDI-TOF MS with scores > 2.0, and glycerol stock preparations were stored at −80 °C until use. L. crispatus and L. reuteri used throughout all experiments were grown in MRS medium supplemented with 0.05% L-cysteine, at 37 °C for 24 h in anaerobic jars supplemented with GasPak EZ (Sparks, MD, USA). The turbidity of 24 h lactobacilli cultures was adjusted to an optical density (OD600nm) of 2, corresponding to a cell concentration of 5 × 108 colony-forming units (CFU)/mL. Cell suspensions were centrifuged at 5000× g for 10 min at 4 °C, then supernatants were filtered through a 0.2 μm membrane filter to obtain cell-free supernatants.
2.2. Cell Culture
HeLa cells, a human cervical adenocarcinoma epithelial cell line (ATCC® CCL-2), were seeded at 2 × 104 cells/cm2 in a petri plastic plate (Orange Scientific, Braine-l’Alleud, Belgium) and cultured in 5% CO2 at 37 °C in Dulbecco’s modified Eagle medium (D-MEM) (Labtek Eurobio, Milan, Italy), supplemented with 10% FBS (Euroclone, Milan, Italy) and 2 mM L-glutamine (Sigma-Aldrich, Milan, Italy).
2.3. CT Propagation and Titration
CT GO/86 (serovar D), isolated during routine diagnostic procedures in the Microbiology Unit of Sant’Orsola-Malpighi Hospital of Bologna (Italy) and belonging to the laboratory collection, was used throughout all experiments. CT was propagated in HeLa cells, cultured in Dulbecco’s minimal essential medium (DMEM), and supplemented with 10% fetal bovine serum, 1% L-glutamine 200 mM and antibiotics (vancomycin 10 mg/L, gentamicin 10 mg/L and amphotericin B 0.3 mg/L). Purification of EBs and evaluation of the infectivity titer (expressed as inclusion-forming units, IFU/mL) have been described in detail, elsewhere [19].
2.4. Effect of Lactobacilli on CT Infectivity
HeLa cells were seeded at 2 × 104 cells/cm2 in plastic wells (Orange Scientific, Braine-l’Alleud, Belgium) or on sterile glass coverslips for 48 h and treated with L. crispatus or L. reuteri at a ratio of 1:100 or 1:200 (HeLa cells: Lactobacillus) for 1 h at 37 °C and 5% CO2 atmosphere. Afterwards, HeLa bacteria co-cultures were washed 3 times with PBS to remove unbound bacteria and then were treated with 5 × 104 CT EBs, corresponding to multiplicity of infection (MOI) 1 for 1 h at 37 °C and 5% CO2 atmosphere. Samples were washed 3 times with PBS, fixed in 3% paraformaldehyde for 10 min and marked with a monoclonal antibody against the chlamydial membrane lipopolysaccharide antigen conjugated with fluorescein (Meridian, Cincinnati, OH, USA) for 30 min at RT. The bacterial cells were centrifuged at 10,000× g, and the 1:100 diluted supernatant was sterile-filtered through a 0.2 µm filter and stored at −20 °C until use. The D(−)-LA or L(+)-LA enriched L. reuteri supernatant was prepared by adding 10 mM of D(−)-LA or L(+)-LA. HeLa cells were incubated with the enriched supernatant solutions for 1 h at 37 °C with 5% CO2. After incubation, the different samples cells were infected with CT EB for 48 h.
The number of IFUs was counted in 60 randomly chosen 100× microscopic fields. The results were expressed as a percentage (median percentage ± median absolute deviation) of CT infectivity, comparing the number of IFUs in the individual samples with the control slides (set at 100%).
2.5. Lactate Dehydrogenase Activity
Bacterial lysates for lactate dehydrogenase activity measurement were prepared diluting 5 × 108 CFU/mL of the sub-cultured bacterial culture of lactobacilli with PBS to OD600nm = 0.1, centrifuged at 4 °C and 12,000× g for 10 min and washed once using PBS buffer. Cell pellets were lysed in 500 μL of enzymatic lysis buffer (20 mM Tris HCl pH 8, 2 mM sodium EDTA, 1.2% Triton X-100, 20 mg/mL lysozyme), incubated at 37 °C for 30 min and then ultrasonicated to release the intracellular organelles and enzymes. The disrupted cells were then centrifuged (12,000× g for 10 min at 4 °C) and the supernatant collected. The protein content of the supernatant was determined by the Lowry method [20]. For the D-lactate dehydrogenase activity, 50 µg of protein lysate was transferred in a 1 mL cuvette and added to 20 µL of D(−)-lactic acid 100 mM and 10 µL of NAD+ 200 mM, and the final volume was adjusted to 1 mL, adding phosphate buffer 100 mM pH = 7.8. The enzyme activity was evaluated spectrophotometrically with the NADH absorbance at 340 nm (ε340nm = 6220 M−1 cm−1) using a spectrophotometer (Jasco V-550) equipped with a thermostatic control and stirring device.
2.6. Lactate Determination
Lactobacilli cultures were adjusted to an OD600nm of 2.0 with sterile phosphate buffer solution (PBS) (cell concentration: 5 × 108 CFU/mL) and centrifuged at 5000× g for 10 min at 4 °C. Supernatants were filtered through a 0.2 μm membrane filter to obtain stock cell-free supernatants.
Prior to injection, the supernatants were diluted 1:10 in mobile phase and centrifuged at 14,000× g for 5 min at 4 °C. The supernatant was then injected manually into the HPLC system (Agilent 1100 series). Metabolites were separated on a C18 column (Agilent ZORBAX SB-Phenyl, 5 μm, 250 Å, 4.6 mm), using a mobile phase consisting of 50 mM KH2PO4, pH 2.9, at a flow rate of 0.8 mL/min. Lactate was detected using an Agilent UV detector set to 210 nm and quantified using Agilent Chem Station software (version LTS 01.11).
2.7. Energetic Profile
To assess the energetic profile of HeLa cells after addition of diluted supernatants or addition of D(−)- or L(+)-LA at the final concentration of 10 mM, around 7.5 × 103 cells were seeded in every well of a 96-well plate. Treatment for 1 h with supernatant or LA solutions was carried out 24 h after the seeding. Cells were then washed twice with PBS, and 180 μL of fresh medium were added, then the 96-well plate was placed immediately in the Seahorse Agilent (Agilent Technologies, Santa Clara, CA, USA), and the ATPase inhibitor oligomycin A was added to evaluate the proton leak, while the uncoupler FCCP was added to obtain the maximal respiration rate.
Evaluation of the energetic profile was performed following the standard protocol of the Agilent Seahorse XF Cell Mito Stress Test.
2.8. Histone Post-Translational Modification
The level of histone acetylation/lactylation was determined by immunoblot analysis, a semi-quantitative technique widely used in molecular biology and biochemistry research. HeLa cells were seeded in a dish, and after 48 h they were treated for 24 h with L. crispatus or L. reuteri supernatant diluted 1:100 in culture medium. Cells were harvested and washed with 10 mM sodium butyrate in PBS, and nuclei were isolated in according to Amellem et al. [21]. The nuclear histones were extracted as previously described [22]. Histones were detected resolving samples on a 10% gel in MES buffer at 200 V for 40 min. Western Blotting was performed in transfer buffer at 100 V for 1 h. The nitrocellulose membrane was incubated with primary antibody specific for anti-acetylated lysine (Millipore, Billerica, MA, USA) for 1 h. After washes with PBS-TWEEN 20 0.1%, the membrane was incubated as before with secondary horseradish-peroxidase-conjugated antibody (GE Healthcare, Milan, Italy). After washes with PBS-TWEEN 20 0.1%, antibody binding was detected using an Amersham ECL Plus Western Blotting Detection System (GE Healthcare, Milan, Italy). Densitometry analysis was performed with a Fluor-S Max MultiImager (Bio-Rad, Hercules, CA, USA), and relative quantification of histone acetylation signals was carried out by using densitometry and normalized on H1 signal as a control.
2.9. Immunoblot
The level of protein expression was determined by immunoblot analysis, a semi-quantitative technique widely used in molecular biology and biochemistry research. HeLa cells were treated with L. crispatus or L. reuteri supernatant diluted 1:100 in culture medium for 48 h. Cells were washed twice with PBS and lysed for 1 h in lysis buffer (HEPES, pH 7.4 40 mM, glycerophosphate 60 mM, p-nitrophenylphosphate 20 mM, Na3PO4 0.5 mM, NaCl 250 mM, Triton X-100 1%, PMSF 0.5 mM and 10 mg/mL each of aprotinin, leupeptin, pepstatin and antipain (Sigma-Aldrich)) at 0 °C. Cell lysates were centrifuged at 12,000× g for 20 min. Supernatants were collected and protein concentration determined by using the Bio-Rad protein assay method (Bio-Rad, Hercules, CA, USA). The proteins were resolved on a 7.5% or 10% polyacrylamide gel and immunoblotted with a rabbit anti-human α5 integrin subunit (Cell Signaling Technology, Danvers, MA, USA), rabbit anti-EGFR (Biorbyt, Cowley Road, Cambridge, UK), rabbit anti-cyclin D1 (Millipore, Billerica, MA, USA), rabbit anti-p21 (Millipore, Billerica, MA, USA) or mouse anti-α-tubulin (Sigma-Aldrich) antibodies. Detection of immunoreactive bands was performed by using a rabbit or mouse HRP-conjugated secondary antibody (GE Healthcare, Milan, Italy) followed by the Amersham ECL Plus Western Blotting Detection System (GE Healthcare, Milan, Italy). Densitometry analysis of immunoreactive bands was conducted with a Fluor-S Max MultiImager (Bio-Rad). Relative quantification of bands was performed by using α-tubulin signal as control. Alternatively, HeLa cells were treated with a 10 mM solution od D(−)-LA or L(+)-LA. Protein extraction and immunoblot analysis were performed as described above.
3. Results
3.1. Lactate Determination and Lactate Dehydrogenase Activity
Two strains of lactobacilli belonging to two species, L. crispatus and L. reuteri, were selected for this in vitro study. The amount of LA released by the lactobacilli was determined in the supernatants of L. crispatus and L. reuteri after 24 h of culture by HPLC. Figure 1A shows that the supernatant of L. crispatus contains a higher amount of LA than that of L. reuteri. Because we were unable to determine the relative concentrations of the two LA isomers with our HPLC system, we tested the presence of the D-lactate dehydrogenase (LDHD) enzyme with kinetic analysis. To this end, we measured the activity of LDHD in the bacterial lysates in the presence of a saturating concentration of D(−)-LA, which can provide an approximate indication of the amount of the enzyme. The results shown in Figure 1B confirm the presence of LDHD in the L. crispatus lysate with a maximum specific activity of 1.46 nmol min−1/108 CFU, whereas it is almost completely absent in the L. reuteri lysate, where the maximal specific activity was 0.08 nmol min−1/108 CFU.
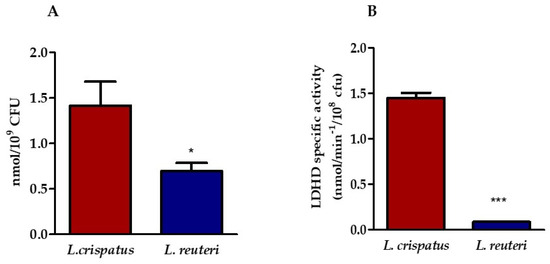
Figure 1.
Determination of the amount of LA in lactobacilli supernatant. (A) HPLC analysis of LA content in the supernatants of L. crispatus and L. reuteri. Statistical significance was calculated vs. control (* p ≤ 0.01). (B) Specific activity of LDHD in bacterial lysates (*** p ≤ 0.0001). Results are expressed as nanomoles (nmol) of LA/mL of supernatant containing 109 CFU of lactobacilli (panel A) and as nanomoles (nmol) of lactic acid min−1/109 CFU (panel B). Bars represent mean values, and error bars represent standard deviation. All measurements were performed in duplicate on six separate occasions.
These data were confirmed by the analysis performed using the liquid chromatography–mass spectrometry technique (LC-MS). The LC-MS analysis of peptides obtained by digestion of L. crispatus proteins led to the identification of 114 proteins, among which was D-lactate dehydrogenase. The identified peptides are listed in the supporting information (File S1) along with the other parameters obtained by searching the SwissProt database. The same analysis performed for proteins from L. reuteri resulted in the identification of 92 proteins, but D-lactate dehydrogenase was not found.
3.2. Evaluation of Lactobacilli Cell Pellets and Supernatant’s Protective Effect against CT Infection
To evaluate the role of lactobacilli in preventing CT infection, two different lactobacilli fractions (cell pellets and supernatant) were tested. First, HeLa cells were incubated with L. crispatus or L. reuteri at a ratio of 1:100 or 1:200 (HeLa cells: lactobacilli) for 1 h and then treated with 5 × 104 CT EBs for 1 h (exclusion mechanism). As shown in Figure 2A, treatment with L. crispatus reduced the infectivity of CT by 36.4% ± 1.65 (1:100 ratio) and 72.7% ± 0.36 (1:200 ratio), respectively, whereas L. reuteri was less active in protecting against infection.
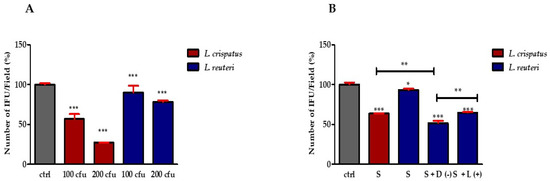
Figure 2.
Effect of lactobacilli on CT infectivity. (A) Effect of lactobacilli cell pellets on CT infectivity. Quantification of HeLa cells positive to CT inclusions. The results are expressed as a percentage of CT infectivity, comparing the number of IFUs with the control (set at 100%). Bars represent mean values, and red error bars represent standard deviation. (B) Percentage of CT-infected HeLa cells pre-exposed to L. crispatus or L. reuteri culture supernatant (S) (diluted 1:100) and L. reuteri culture supernatant supplemented with 10 mM D(−)- or L(+)-LA. Results are from three independent experiments. Statistical significance is calculated vs. control (* p ≤ 0.01; ** p ≤ 0.001; *** p ≤ 0.0001). All measurements were performed in duplicate on six separate occasions.
To distinguish the effect of lactobacilli from the effect of secreted molecules produced by bacteria during growth and to evaluate the effect of the two LA isomers, we tested the effect of bacterial supernatants and the additive effect of D(−)- or L(+)-LA on the infectivity of CT in HeLa cells. Before investigating the protective effect of Lactobacillus supernatants and LA isomers, we performed preliminary experiments to determine the concentrations to be used. For this purpose, we treated HeLa cells with different dilutions of supernatants from L. crispatus and L. reuteri cultures. The results shown in Figure S2A,B (File S2) indicate that the 1:100 dilution of the two bacterial supernatants is safe for HeLa cells. At the same time, we tested the effect of the two LA isomers on cell viability. The results shown in Figure S2C (File S2) indicate that both isomers of LA have very low cytotoxicity at a concentration of 10 mM for 24 h of treatment. Finally, we tested the effect of 24 h treatment with 1:100 diluted L. reuteri supernatant enriched with different concentrations of LA isomers (1, 5 and 10 mM) on HeLa cell viability. Figure S2D (File S2) shows that 10 mM of D(−)-LA has no significant effect on cell viability, while L(+)-LA reduces it by about 20%.
Based on these results, we treated HeLa cells with 1:100 diluted supernatants for 1 h, then the medium was removed, and the cells were treated with 5 × 104 CT EBs for 1 h. The results shown in Figure 2B indicate that the protective effect of L. crispatus supernatants was maintained. In particular, treatment with L. crispatus supernatant reduced the infectivity of CT by 37.08% ± 0.35, whereas treatment with L. reuteri had a much lower inhibitory effect against CT (the reduction only being equal to 7.01% ± 1.54). On the other hand, the addition of 10 mM D(−)-LA to the L. reuteri supernatant increased its resistance against infection (infectivity reduction rose to 48.33% ± 2.89), while the addition of 10 mM L(+)-LA led to a significantly lower infectivity reduction (35.16% ± 0.81) (Figure 2B). These results suggest that D(−)-LA plays a central role in reducing CT infectivity.
To improve this point, we preloaded HeLa cells with 10 mM of D(−)-LA or L(+)-LA prior to incubation with 5 × 104 CT EBs. The results shown in Figure 3 indicate that infection of CT was greatly reduced by D(−)-LA treatment.
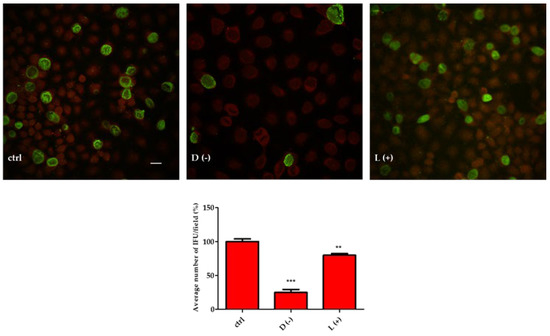
Figure 3.
D(−)-LA prevents CT infection in HeLa cells. (Above). HeLa cells were incubated or not with 10 mM of D(−)- or L(+)-LA for 1 h and then with CT EBs for an additional hour. Specimens were stained for chlamydial membrane lipopolysaccharide antigen (green fluorescence). Representative micrographs are shown. Bar: 20 µm. (Below). Percentage of CT infected HeLa cells. Results are from three independent experiments compared to control. (** p ≤ 0.01; *** p < 0.0001).
3.3. Oxygen Consumption Rate in HeLa Cells Treated with L. crispatus or L. reuteri Supernatants
The oxygen consumption rate (OCR) is an indicator of the metabolic state of the cell: cells with a more oxidative metabolism are characterized by a higher oxygen consumption rate, whereas a low oxygen consumption rate indicates a more glycolytic metabolism. To investigate the effect of D(−)- or L(+)-LA on the metabolic status of HeLa cells, we measured the OCR of cells treated with 10 mM D(−)- or L(+)-LA compared with the effect of treatment with L. crispatus or L. reuteri supernatants. The results shown in Figure 4 indicate that OCR in HeLa cells was stimulated by supplementation with D(−)-LA. On the other hand, addition of L. crispatus supernatant caused a slight decrease in OCR of HeLa, whereas addition of the L(+)-LA or addition of L. reuteri supernatant caused a strong decrease in both native and uncoupled OCR. This result suggests that HeLa cells exhibit a more oxidative metabolism in the presence of the D(−)-LA isomer, whereas the L(+)-LA isomer stimulates a more glycolytic metabolism.
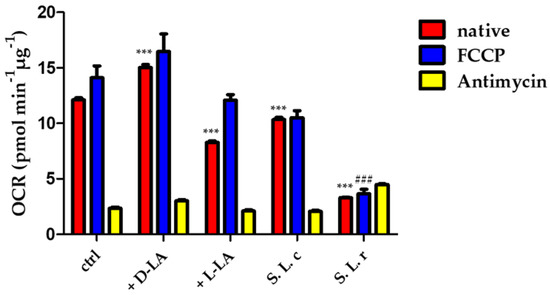
Figure 4.
Oxygen consumption rate (OCR) was measured in HeLa cells (ctrl) and in HeLa incubated with 10 mM of D(−) or L(+)-LA or treated for 1 h with L. crispatus (S L.c.) and L. reuteri (S L.r.) supernatant following the addition of the OXPHOS uncoupler FCCP (0.5 µM) and the electron transport inhibitor antimycin A (AA) (2 µM). Statistical significance of native OCR (red bars) is calculated vs. control (ctrl) (*** p ≤ 0.0001); the uncoupled respiration rate (blue bars) is significantly decreased by the treatment with L. reuteri supernatant (### p ≤ 0.0001). Data are mean ± SD.
3.4. Lactobacilli Culture Supernatants Modulate Histone Modification State and Gene Expression
Histones can be modified by cellular enzymes that add chemical marks such as methyl, acetyl and phosphate groups; these epigenetic modifications of the genome affect processes such as gene expression and DNA replication and repair [23]. Lactic acid can promote histone acetylation by inhibiting histone deacetylase (HDAC), thereby regulating gene expression [24]. In addition, it has recently been shown that LA can induce a particular type of histone modification called “lactylation” [25]. In this section, we analyzed the effect of the two isomers LA and the supernatants of L. crispatus and L. reuteri on histone modifications and gene expression in HeLa cells. HeLa cells exposed to culture supernatants of L. crispatus or L. reuteri for 24 h showed a significant increase in histone acetylation. Specifically, treatment with L. crispatus supernatant induced a greater increase in H2/H3 histone acetylation (~6-fold compared to a less than 4-fold increase induced by the addition of L. reuteri supernatant) (p < 0.0001) and a strong acetylation of histone H4 (+72.6%). Treatment with L. reuteri supernatant had no effect (Figure 5A). Interestingly, supernatant of both lactobacilli led to changes in the state of histone lactylation: supernatant of L. crispatus decreased lactylation of H2/H3 by 41.7% ± 12.76% and did not lead to changes in H4 histones. In contrast, the supernatant of L. reuteri significantly increased the lactylation of H2/H3 and H4 histones by approximately 26.1% ± 15.37 and 52.5% ± 34.59, respectively (Figure 5B).
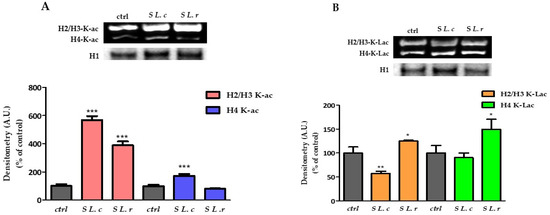
Figure 5.
Changes in the state of histone acetylation and lactylation. (A) (Above). Western blot of acetylated histones. (Below). Relative quantification of H2/H3 acetylated histones in HeLa control and exposed for 24 h to culture supernatants of L. crispatus (S L.c) or L. reuteri (S L.r). (B) (Above). Western blot of lactylated histones. (Below). Relative quantification of H4 lactylated histones in HeLa control and treated with culture supernatants of L. crispatus or L. reuteri for 24 h. Densitometry arbitrary units (A.U.) are normalized by H1 histone. Results are given as means ± SD of three independent experiments and are compared to controls, taken as 100%. (* p < 0.05; ** p < 0.001; *** p < 0.0001).
These results suggest that L. crispatus and L. reuteri induce different epigenetic alteration patterns in HeLa cells via their metabolites, which could lead to differential regulation of gene expression. Based on this observation, we performed qtRT-PCR analysis of genes involved in cell-cycle regulation and genes such as HER.1 and ITAG5, which encode membrane proteins required for the internalization of chlamydial EBs into host cells. The qtRT-PCR results were confirmed by immunoblot analysis of the corresponding host proteins.
It is known that proteins on the cell membrane, such as integrin α5β1, are important for CT entry into the cell or development of infection [12]. In addition, downregulation of cell-cycle regulators appears to be involved in C. trachomatis infection [26]. The transcription levels of CCND1 (cyclin D1), CDKN1A (p21), HER-1(EGFR) and ITAG5 (α5β1) were analyzed by qtRT-PCR on cDNA of HeLa control cells treated with diluted supernatants of L. crispatus or L. reuteri for 24 h. The results presented in Figure S3 (File S3) show that the supernatant of L. crispatus caused a strong downregulation of CCND1, Her-1 and ITAG5 genes (0.47 ± 0.01; 0.29 ± 0.01 and 0.53 ± 0.05, respectively) (p ≤ 0.001 compared to control); on the other hand, CDKN1A transcription was significantly more upregulated in cells treated with L. reuteri supernatant by a factor of 2.55 ± 0.063 (p ≤ 0.001 compared to control).
To validate the qtRT-PCR results, we examined the expression of the corresponding proteins by immunoblot analysis. The results shown in Figure 6 demonstrated a decrease in the expression of EGFR, cy-clin D1 and α5β1-integrin in HeLa cells treated with the supernatant of L. crispatus (Figure 6A) or 10 mM of D(−)-LA (Figure 6B). EGFR expression was greatly decreased by treatment with 10 mM of D(−)-LA (Figure 6B). On the other hand, the expression of p21 was increased only in cells treated with the supernatant of L. reuteri and was significantly increased in HeLa cells treated with 10 mM of L(+)-LA (Figure 6B).
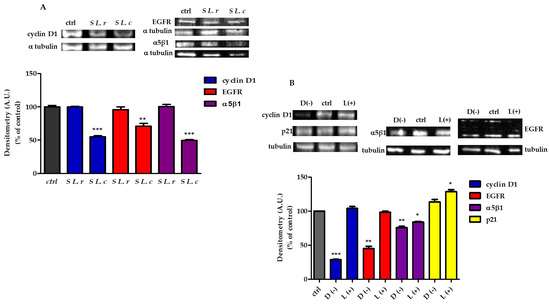
Figure 6.
Protein expression modifications by supernatants of L. crispatus (S L.c) or L. reuteri (S L.r) modified protein expression. (A) (Above). (sx) Immunoblot analysis of cyclin D1 protein and (dx) Western blot of EGFR and α5β1 subunit of integrin 1. (Below). Relative quantification of proteins. Densitometry arbitrary units (A.U.) were normalized by α-tubulin. Results are given as means ± SD of three independent experiments and are compared to controls (statistical significance: ** p ≤ 0.001; *** p ≤ 0.0001), taken as 100%. (B) HeLa treated with 10 mM of D(−)- or L(+)-LA. (Above). (sx) Immunoblot analysis of cyclin D1 and p21 proteins and (dx) Western blot of EGFR and α5β1 subunit of integrin 1. (Below). Relative quantification of proteins. Densitometry arbitrary units (A.U.) were normalized by α-tubulin. Results are given as means ± SD of three independent experiments and are compared to controls (statistical significance: * p ≤ 0.01; ** p ≤ 0.001; *** p ≤ 0.0001), taken as 100%.
4. Discussion
In this work, two different species of lactobacilli (L. crispatus and L. reuteri) were tested to investigate their protective effect against CT infection.
The differences that characterize these two bacterial strains include the amount and type of LA produced, which is the focus of this study. L. crispatus was chosen because it is known to be able to produce both isomers of LA. In contrast, L. reuteri, which is known to be very abundant in the gut, is a far less effective producer of D(−)-LA [15]. In particular, the isolate used in the present study produces only L(+)-LA isomer. Although L. reuteri is less abundant in vaginal fluids, it is more stable and easier to culture than the other lactobacilli species that colonize the vaginal environment.
LA is one of the most important antimicrobial compounds produced by vaginal lactobacilli [9], so we first investigated its content in the free lactobacilli supernatants using HPLC. The results showed that the amount of LA produced by L. crispatus was higher than the amount produced by L. reuteri.
These results are consistent with the stronger protective function of L. crispatus reported in the literature [10,11,12,16,17,27]. To confirm the ability of L. crispatus and L. reuteri to produce the D(−)-LA isomer, we assessed the presence of D-lactate dehydrogenase by LC-MS analysis and performed a kinetic assay to measure the maximum activity of the enzyme. We were able to demonstrate that only L. crispatus can produce the D(−)-LA isomer.
The mechanisms underlying the protective properties of LA are multiple: (i) the protonated form of LA can penetrate cell membranes and acidify the cytosol; (ii) LA can also weaken the bacterial cell wall or affect host cell membranes, reducing their susceptibility to infection and finally (iii) LA can induce epigenetic changes that could affect target cell gene expression [9,13,14]. These mechanisms independently or together determine host susceptibility and consequently the relationship between host and microbiota.
In terms of activity against CT infection, vaginal lactobacilli can be classified into strains with high and low protection. In the work of Edwards et al. [28] it is reported that lactobacilli producing only the L(+)-LA belong to the low activity group, suggesting that D(−)-LA producers provide higher protection against CT infection. To test this hypothesis, additional experiments were performed by adding 10 mM of D(−)-LA or L(+)-LA to L. reuteri supernatant, which was subsequently used to pretreat HeLa cells before CT infection. The addition of D(−)-LA resulted in enhanced protection against CT infection, comparable to the protection exerted by L. crispatus supernatant.
Since the acid-base properties of the two isomers are quite similar, the higher anti-chlamydial activity of D(−)-LA is likely due to the presence of specific targets. Edwards et al. suggested that D(−)-LA might modulate vaginal cell homeostasis through epigenetic modifications [28]. Therefore, we performed an in-depth analysis of epigenetic modifications of histone proteins and expression of genes involved in the regulation of cell proliferation.
HeLa cells exposed to L. crispatus supernatant for 24 h showed a significant increase in H2/H3 and H4 histone acetylation, whereas treatment with L. reuteri supernatant had only a minor effect on H2/H3 acetylation and no effect at all on H4 acetylation. It is well known that histone lysine acetylation plays an important role in epigenetic regulation of gene expression by decondensing nucleosome structure [26,29], which alters histone–DNA interactions [30], facilitating access and binding of transcription factors to genes [31,32].
Zhang et al. recently found that lactic acid can also alter the lysine residues of histones in a new epigenetic modification, lactylation, which directly stimulates gene transcription from chromatin [25]. The alteration of gene expression induced by epigenetic changes may provide further clues to explain the protective effect of D(−)-LA. For this reason, the supernatant-induced epigenetic changes were studied in relation to the state of acetylation and lactylation of histone proteins.
Lactate is an end product of anaerobic glycolysis, whose intracellular concentration increases during the Warburg effect and is responsible for several changes involved in cell survival under unfavorable environmental conditions. It has been suggested that the excess of lactate could be used to introduce epigenetic changes. Zhang et al. [25] showed that lysine histone lactylation stimulates gene transcription, suggesting a novel role of lactate in various pathological conditions such as infections or cancer. Recently, Moreno-Yruela et al. [33] reported that lysine histone lactylation is mediated by histone acetyltransferases such as p300, whereas HDAC 1-3 are responsible for lysine histone delactylation. Although both isomers are present in cells, the L(+)-LA isomer is preferred as a metabolic substrate and signaling molecule. The preferential use of the L(+)-LA isomer is mainly due to two factors: the concentration of the L(+)-LA is more than a thousand times higher than that of the D(−)-LA form [34], and L(+)-lactate is also recognized as a precursor for lysine lactylation [27]. The D(−) form of lactic acid may increase under certain conditions, such as the activation of the glyoxalase pathway observed in some tumor cells [35,36], or may be overproduced by certain microorganisms. Our hypothesis is that the presence of high levels of D(−)-LA influences epigenetic histone modifications that favor a cellular phenotype more resistant to CT infection.
Interestingly, we found that treatment of HeLa cells with the supernatants of both lactobacilli induced changes in the state of histone lactylation, suggesting that they may activate different epigenetic pathways. Indeed, treatment with the supernatant of L. crispatus resulted in downregulation of genes involved in cell-cycle regulation (CCND1, CDKN1A and HER-1) and significant downregulation of the ITGA5 gene encoding the α5β1 integrin. Immunoblot analysis of protein expressions of cyclin D1, p21, EGFR and α5β1 confirmed the results of RT-qPCR. Interestingly, treatment with 10 mM D(−)-LA induced decreased protein expression as observed in HeLa cells treated with L. crispatus supernatant. Edwards et al. [28] mentioned that the vaginal microbiota can counteract susceptibility to CT infection by controlling the cell cycle. Their results suggest that lactobacilli producing D(−)-LA downregulate the proliferation of vaginal epithelial cells, protecting them from CT infection. Other authors showed that culture supernatants of L. crispatus and L. jensenii modulated the expression of several genes related to cell proliferation, including a decrease in CCND1 and HER-1. In contrast, L. iners, which is unable to produce D(−)-LA, failed to downregulate these genes and protect against infection with CT [15,28]. Moreover, previous studies have shown that EGFR is required for the internalization of chlamydial EBs into host cells [37]. Parolin et al. demonstrated that infection of CT was greatly reduced when a specific blocking antibody masked the α5-integrin subunit or the corresponding gene expression was suppressed, supporting the hypothesis that the α5-subunit is critical for infection of CT in HeLa cells [12].
CT is an obligate intracellular bacterium with a very small (~1.04 Mb) genome [38] that lacks several genes for metabolic enzymes. For this reason, CT is strongly influenced by host metabolic conditions. Since CT lacks hexokinase, EBs and RBs require glucose-6 phosphate. Moreover, during the transition from EB to RB, CT is unable to meet the increased demand for ATP, making RB completely dependent on host ATP as an energy source [39,40]. Rother et al. [41] studied the changes in host metabolism following infection with CT and found that many glycolytic metabolites and TCA intermediates were upregulated in a manner similar to Warburg metabolism.
We examined the metabolic profile of HeLa cells before and after 24 h treatment with the lactobacilli supernatants using the Seahorse Flux Analyzer. Our results showed that HeLa cells treated with L. reuteri supernatant or 10 mM L(+)-LA for 24 h were characterized by a strong decrease in OCR profile, indicating a shift from oxidative to glycolytic metabolism that could promote CT infection. In contrast, both treatment with L. crispatus supernatant and with 10 mM D(−)-LA did not alter the metabolic profile of HeLa.
The lack of knowledge of the relative amounts of D(−)-LA and L(+)-LA present in the L. crispatus supernatant is a limitation of this work and may account for the differences in results observed when HeLa cells were treated with a 10 mM solution of the pure LA isomers. At the same time, another limitation concerns the use of two clinical isolates of Lactobacillus: we are fully aware that there are several strain-specific characteristics that can prevent a complete replication of the above listed results. However, treatment of the cells with the pure LA isomers replicated the effect of the supernatants of the two lactobacilli used.
In conclusion, our results demonstrate the stronger effect of D(−)-LA compared to L(+)-LA in protecting the host cell and provide new perspectives for the selection of probiotic lactobacilli strains for the prevention of chlamydia infections.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pathogens12070883/s1, File S1: LC-ESI-MS/MS analysis of protein from L. crispatus and L. reuteri; File S2: The effect on cell viability of pre-exposing host cells to lactic acid or Lactobacillus culture supernatants; File S3: The effect on gene expression of pre-exposing the host cells to Lactobacillus culture supernatants.
Author Contributions
Conceptualization, A.M., N.C., R.F. and C.F.; investigation, C.Z., N.R., G.M., S.M., M.E.D. and M.N.; writing—original draft preparation, N.C., R.F. and A.M; writing—review and editing, N.C., R.F., A.M., C.F. and M.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Alma Mater Studiorum—Università di Bologna grant RFO (to N.C., A.M. and R.F.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All relevant research data are presented in the text or in the Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chlamydia Infection—Annual Epidemiological Report for 2019. Available online: https://www.ecdc.europa.eu/en/publications-data/chlamydia-infection-annual-epidemiological-report-2019 (accessed on 20 March 2023).
- Price, M.J.; Ades, A.E.; De Angelis, D.; Welton, N.J.; Macleod, J.; Soldan, K.; Simms, I.; Turner, K.; Horner, P.J. Risk of Pelvic Inflammatory Disease Following Chlamydia Trachomatis Infection: Analysis of Prospective Studies with a Multistate Model. Am. J. Epidemiol. 2013, 178, 484–492. [Google Scholar] [CrossRef]
- Ceccarani, C.; Foschi, C.; Parolin, C.; D’Antuono, A.; Gaspari, V.; Consolandi, C.; Laghi, L.; Camboni, T.; Vitali, B.; Severgnini, M.; et al. Diversity of Vaginal Microbiome and Metabolome during Genital Infections. Sci. Rep. 2019, 9, 14095. [Google Scholar] [CrossRef] [PubMed]
- van Houdt, R.; Ma, B.; Bruisten, S.M.; Speksnijder, A.G.C.L.; Ravel, J.; de Vries, H.J.C. Lactobacillus Iners-Dominated Vaginal Microbiota Is Associated with Increased Susceptibility to Chlamydia Trachomatis Infection in Dutch Women: A Case-Control Study. Sex. Transm. Infect. 2018, 94, 117–123. [Google Scholar] [CrossRef]
- Valenti, P.; Rosa, L.; Capobianco, D.; Lepanto, M.S.; Schiavi, E.; Cutone, A.; Paesano, R.; Mastromarino, P. Role of Lactobacilli and Lactoferrin in the Mucosal Cervicovaginal Defense. Front. Immunol. 2018, 9, 376. [Google Scholar] [CrossRef] [PubMed]
- Di Pietro, M.; Filardo, S.; Simonelli, I.; Pasqualetti, P.; Sessa, R. Cervicovaginal Microbiota Composition in Chlamydia Trachomatis Infection: A Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2022, 23, 9554. [Google Scholar] [CrossRef] [PubMed]
- Tamarelle, J.; de Barbeyrac, B.; Hen, I.L.; Thiébaut, A.; Bébéar, C.; Ravel, J.; Delarocque-Astagneau, E. Vaginal Microbiota Composition and Association with Prevalent Chlamydia Trachomatis Infection: A Cross-Sectional Study of Young Women Attending a STI Clinic in France. Sex. Transm. Infect. 2018, 94, 616–618. [Google Scholar] [CrossRef]
- Armstrong, E.; Kaul, R. Beyond Bacterial Vaginosis: Vaginal Lactobacilli and HIV Risk. Microbiome 2021, 9, 239. [Google Scholar] [CrossRef]
- Gong, Z.; Luna, Y.; Yu, P.; Fan, H. Lactobacilli Inactivate Chlamydia Trachomatis through Lactic Acid but Not H2O2. PLoS ONE 2014, 9, e107758. [Google Scholar] [CrossRef]
- Rizzo, A.; Fiorentino, M.; Buommino, E.; Donnarumma, G.; Losacco, A.; Bevilacqua, N. Lactobacillus Crispatus Mediates Anti-Inflammatory Cytokine Interleukin-10 Induction in Response to Chlamydia Trachomatis Infection In Vitro. Int. J. Med. Microbiol. 2015, 305, 815–827. [Google Scholar] [CrossRef]
- Nardini, P.; Ñahui Palomino, R.A.; Parolin, C.; Laghi, L.; Foschi, C.; Cevenini, R.; Vitali, B.; Marangoni, A. Lactobacillus Crispatus Inhibits the Infectivity of Chlamydia Trachomatis Elementary Bodies, in Vitro Study. Sci. Rep. 2016, 6, 29024. [Google Scholar] [CrossRef]
- Parolin, C.; Frisco, G.; Foschi, C.; Giordani, B.; Salvo, M.; Vitali, B.; Marangoni, A.; Calonghi, N. Lactobacillus Crispatus BC5 Interferes With Chlamydia Trachomatis Infectivity Through Integrin Modulation in Cervical Cells. Front. Microbiol. 2018, 9, 2630. [Google Scholar] [CrossRef] [PubMed]
- O’Hanlon, D.E.; Moench, T.R.; Cone, R.A. In Vaginal Fluid, Bacteria Associated with Bacterial Vaginosis Can Be Suppressed with Lactic Acid but Not Hydrogen Peroxide. BMC Infect. Dis. 2011, 11, 200. [Google Scholar] [CrossRef] [PubMed]
- Hearps, A.C.; Tyssen, D.; Srbinovski, D.; Bayigga, L.; Diaz, D.J.D.; Aldunate, M.; Cone, R.A.; Gugasyan, R.; Anderson, D.J.; Tachedjian, G. Vaginal Lactic Acid Elicits an Anti-Inflammatory Response from Human Cervicovaginal Epithelial Cells and Inhibits Production of Pro-Inflammatory Mediators Associated with HIV Acquisition. Mucosal Immunol. 2017, 10, 1480–1490. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Min, S.; Wang, L.; Zhao, L.; Luo, F.; Lei, W.; Wen, Y.; Luo, L.; Zhou, Q.; Peng, L.; et al. Lactobacillus Modulates Chlamydia Infectivity and Genital Tract Pathology In Vitro and In Vivo. Front. Microbiol. 2022, 13, 1254. [Google Scholar] [CrossRef]
- Witkin, S.S.; Mendes-Soares, H.; Linhares, I.M.; Jayaram, A.; Ledger, W.J.; Forney, L.J. Influence of Vaginal Bacteria and D- and L-Lactic Acid Isomers on Vaginal Extracellular Matrix Metalloproteinase Inducer: Implications for Protection against Upper Genital Tract Infections. mBio 2013, 4, e00460-13. [Google Scholar] [CrossRef]
- Manhanzva, M.T.; Abrahams, A.G.; Gamieldien, H.; Froissart, R.; Jaspan, H.; Jaumdally, S.Z.; Barnabas, S.L.; Dabee, S.; Bekker, L.G.; Gray, G.; et al. Inflammatory and Antimicrobial Properties Differ between Vaginal Lactobacillus Isolates from South African Women with Non-Optimal versus Optimal Microbiota. Sci. Rep. 2020, 10, 6196. [Google Scholar] [CrossRef]
- Foschi, C.; Laghi, L.; Parolin, C.; Giordani, B.; Compri, M.; Cevenini, R.; Marangoni, A.; Vitali, B. Novel Approaches for the Taxonomic and Metabolic Characterization of Lactobacilli: Integration of 16S RRNA Gene Sequencing with MALDI-TOF MS and 1H-NMR. PLoS ONE 2017, 12, e0172483. [Google Scholar] [CrossRef]
- Foschi, C.; Bortolotti, M.; Marziali, G.; Polito, L.; Marangoni, A.; Bolognesi, A. Survival and Death of Intestinal Cells Infected by Chlamydia Trachomatis. PLoS ONE 2019, 14, e0215956. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein Measurement with the Folin Phenol Reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Amellem, O.; Stokke, T.; Sandvik, J.A.; Pettersen, E.O. The Retinoblastoma Gene Product Is Reversibly Dephosphorylated and Bound in the Nucleus in S and G2 Phases during Hypoxic Stress. Exp. Cell Res. 1996, 227, 106–115. [Google Scholar] [CrossRef]
- Calonghi, N.; Cappadone, C.; Pagnotta, E.; Boga, C.; Bertucci, C.; Fiori, J.; Tasco, G.; Casadio, R.; Masotti, L. Histone Deacetylase 1: A Target of 9-Hydroxystearic Acid in the Inhibition of Cell Growth in Human Colon Cancer. J. Lipid Res. 2005, 46, 1596–1603. [Google Scholar] [CrossRef]
- Zaib, S.; Rana, N.; Khan, I. Histone Modifications and Their Role in Epigenetics of Cancer. Curr. Med. Chem. 2022, 29, 2399–2411. [Google Scholar] [CrossRef] [PubMed]
- Latham, T.; Mackay, L.; Sproul, D.; Karim, M.; Culley, J.; Harrison, D.J.; Hayward, L.; Langridge-Smith, P.; Gilbert, N.; Ramsahoye, B.H. Lactate, a Product of Glycolytic Metabolism, Inhibits Histone Deacetylase Activity and Promotes Changes in Gene Expression. Nucleic Acids Res. 2012, 40, 4794–4803. [Google Scholar] [CrossRef]
- Zhang, D.; Tang, Z.; Huang, H.; Zhou, G.; Cui, C.; Weng, Y.; Liu, W.; Kim, S.; Lee, S.; Perez-Neut, M.; et al. Metabolic Regulation of Gene Expression by Histone Lactylation. Nature 2019, 574, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Loidl, P. Towards an Understanding of the Biological Function of Histone Acetylation. FEBS Lett. 1988, 227, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Foschi, C.; Parolin, C.; Giordani, B.; Morselli, S.; Luppi, B.; Vitali, B.; Marangoni, A. Lactobacillus Crispatus BC1 Biosurfactant Counteracts the Infectivity of Chlamydia Trachomatis Elementary Bodies. Microorganisms 2021, 9, 975. [Google Scholar] [CrossRef]
- Edwards, V.L.; Smith, S.B.; McComb, E.J.; Tamarelle, J.; Ma, B.; Humphrys, M.S.; Gajer, P.; Gwilliam, K.; Schaefer, A.M.; Lai, S.K.; et al. The Cervicovaginal Microbiota-Host Interaction Modulates Chlamydia Trachomatis Infection. mBio 2019, 10, e01548-19. [Google Scholar] [CrossRef]
- Garcia-Ramirez, M.; Rocchini, C.; Ausio, J. Modulation of Chromatin Folding by Histone Acetylation. J. Biol. Chem. 1995, 270, 17923–17928. [Google Scholar] [CrossRef] [PubMed]
- Hong, L.; Schroth, G.P.; Matthews, H.R.; Yau, P.; Bradbury, E.M. Studies of the DNA Binding Properties of Histone H4 Amino Terminus. Thermal Denaturation Studies Reveal That Acetylation Markedly Reduces the Binding Constant of the H4 “Tail” to DNA. J. Biol. Chem. 1993, 268, 305–314. [Google Scholar] [CrossRef]
- Lee, J.M.; Hwang, J.W.; Kim, M.J.; Jung, S.Y.; Kim, K.-S.; Ahn, E.H.; Min, K.; Choi, Y.-S. Mitochondrial Transplantation Modulates Inflammation and Apoptosis, Alleviating Tendinopathy Both In Vivo and In Vitro. Antioxidants 2021, 10, 696. [Google Scholar] [CrossRef]
- Vettese-Dadey, M.; Grant, P.A.; Hebbes, T.R.; Crane-Robinson, C.; Allis, C.D.; Workman, J.L. Acetylation of Histone H4 Plays a Primary Role in Enhancing Transcription Factor Binding to Nucleosomal DNA In Vitro. EMBO J. 1996, 15, 2508–2518. [Google Scholar] [CrossRef]
- Moreno-Yruela, C.; Zhang, D.; Wei, W.; Bæk, M.; Liu, W.; Gao, J.; Danková, D.; Nielsen, A.L.; Bolding, J.E.; Yang, L.; et al. Class I Histone Deacetylases (HDAC1–3) Are Histone Lysine Delactylases. Sci. Adv. 2022, 8, eabi6696. [Google Scholar] [CrossRef]
- Adeva-Andany, M.; López-Ojén, M.; Funcasta-Calderón, R.; Ameneiros-Rodríguez, E.; Donapetry-García, C.; Vila-Altesor, M.; Rodríguez-Seijas, J. Comprehensive Review on Lactate Metabolism in Human Health. Mitochondrion 2014, 17, 76–100. [Google Scholar] [CrossRef]
- Landro, J.A.; Brush, E.J.; Kozarich, J.W. Isomerization of (R)- and (S)-Glutathiolactaldehydes by Glyoxalase I: The Case for Dichotomous Stereochemical Behavior in a Single Active Site. Biochemistry 1992, 31, 6069–6077. [Google Scholar] [CrossRef]
- Sousa Silva, M.; Gomes, R.A.; Ferreira, A.E.N.; Ponces Freire, A.; Cordeiro, C. The Glyoxalase Pathway: The First Hundred Years… and Beyond. Biochem. J. 2013, 453, 1–15. [Google Scholar] [CrossRef]
- Patel, A.L.; Chen, X.; Wood, S.T.; Stuart, E.S.; Arcaro, K.F.; Molina, D.P.; Petrovic, S.; Furdui, C.M.; Tsang, A.W. Activation of Epidermal Growth Factor Receptor Is Required for Chlamydia Trachomatis Development. BMC Microbiol. 2014, 14, 277. [Google Scholar] [CrossRef] [PubMed]
- Stephens, R.S.; Kalman, S.; Lammel, C.; Fan, J.; Marathe, R.; Aravind, L.; Mitchell, W.; Olinger, L.; Tatusov, R.L.; Zhao, Q.; et al. Genome Sequence of an Obligate Intracellular Pathogen of Humans: Chlamydia Trachomatis. Science 1998, 282, 754–759. [Google Scholar] [CrossRef] [PubMed]
- Wyrick, P.B. Intracellular Survival by Chlamydia. Cell. Microbiol. 2000, 2, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Omsland, A.; Sixt, B.S.; Horn, M.; Hackstadt, T. Chlamydial Metabolism Revisited: Interspecies Metabolic Variability and Developmental Stage-Specific Physiologic Activities. FEMS Microbiol. Rev. 2014, 38, 779–801. [Google Scholar] [CrossRef]
- Rother, M.; Teixeira da Costa, A.R.; Zietlow, R.; Meyer, T.F.; Rudel, T. Modulation of Host Cell Metabolism by Chlamydia Trachomatis. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).