Putrescine Detected in Strains of Staphylococcus aureus
Abstract
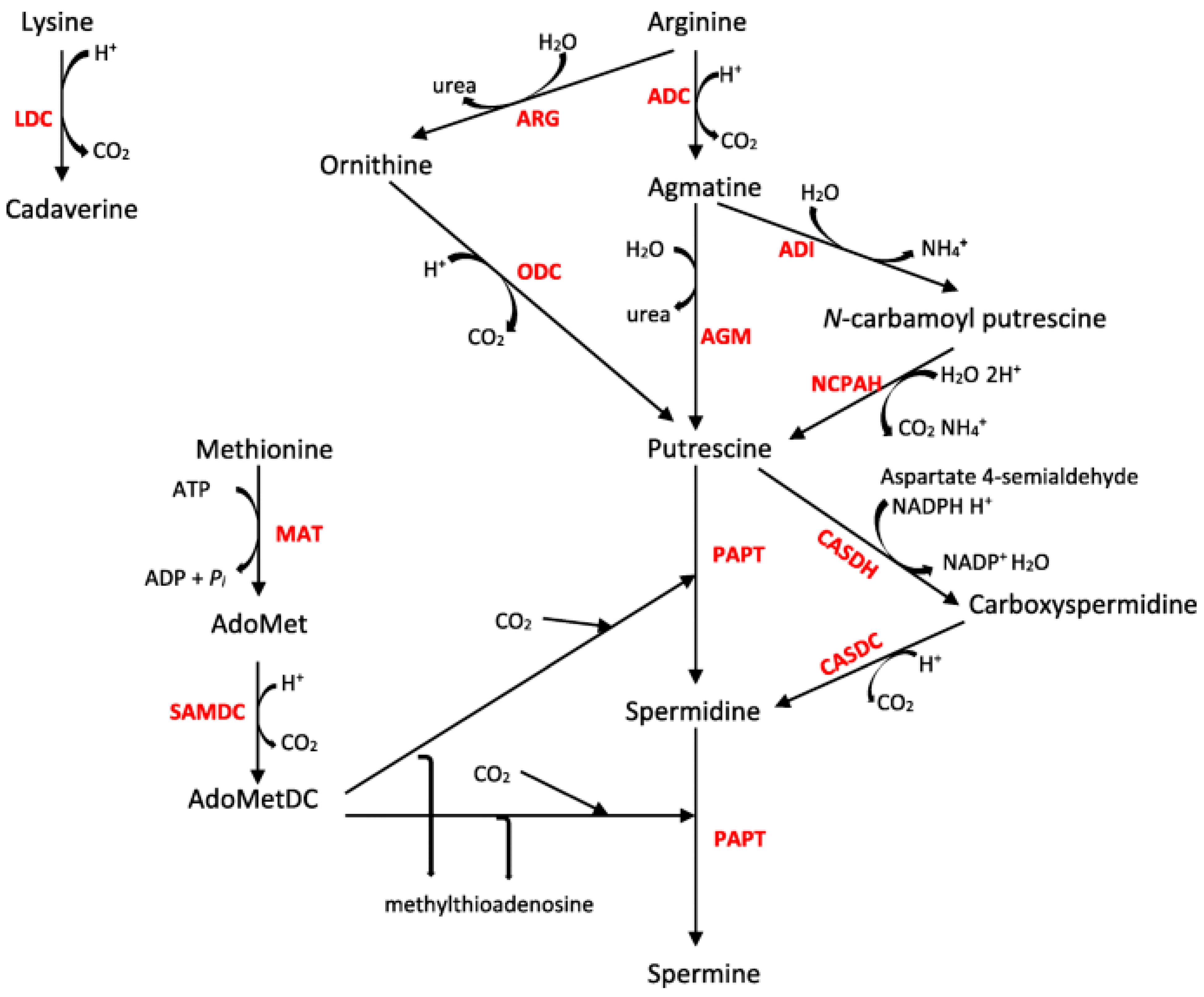
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Igarashi, K.; Kashiwag, K. The Functional Role of Polyamines in Eukaryotic Cells. Intl. J. Biochem. Cell Biol. 2018, 107, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Coton, E.; Mulder, N.; Coton, M.; Pochet, S.; Trip, H.; Lolkema, J.S. Origin of the Putrescine-Producing Ability of the Coagulase-Negative Bacterium Staphylococcus epidermis 2015B. Appl. Environ. Microbiol. 2010, 76, 5570–5576. [Google Scholar] [CrossRef] [PubMed]
- Joshi, G.S.; Spontak, J.S.; Klapper, D.G.; Richardson, A.R. ACME Encoded speG Abrogates the Unique Hypersensitivity of Staphylococcus aureus to Exogenous Polyamines. Mol. Microbiol. 2011, 82, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Maezato, Y.; Kim, S.H.; Kurihara, S.; Liang, J.; Michael, A.J. Polyamine Independent Growth and Biofilm Formation, and Functional Spermidine/Spermine N-acetyltransferases in Staphylococcus aureus and Enterococcus faecalis. Mol. Microbiol. 2018, 111, 159–175. [Google Scholar] [CrossRef]
- El-Halfawy, O.; Valvano, M.A. Putrescine Reduces Antibiotic-Induced Oxidative Stress as a Mechanism of Modulation of Antibiotic Resistance in Burkholderia cenocepacia. Antimicrob. Agents Chemother. 2014, 58, 4162–4171. [Google Scholar] [CrossRef]
- Banerji, R.; Kanojiya, P.; Patil, A.; Saroj, S.D. Polyamines in the Virulence of Bacterial Pathogens of Respiratory Tract. Mol. Oral Microbiol. 2021, 36, 1–11. [Google Scholar] [CrossRef]
- Miller, R.D.; Fung, D.Y.C. Amino Acid Requirements for the Production of Enterotoxin B by Staphylococcus aureus S-6 in a Chemically Defined Medium. Appl. Microbiol. 1973, 25, 800–806. [Google Scholar] [CrossRef]
- Carreau, A.; Hafny-Rahbi, B.E.; Matejuk, A.; Grillon, C.; Kleda, C. Why Is the Partial Oxygen Pressure of Human Tissues a Crucial Parameter? Small Molecules and Hypoxia. J. Cell. Mol. Med. 2011, 15, 1239–1253. [Google Scholar] [CrossRef]
- Yarwood, J.M.; Schlievert, P.M. Oxygen and Carbon Dioxide Regulation of Toxic Shock Syndrome Toxin 1 Production by Staphylococcus aureus MN8. J. Clin. Microbiol. 2000, 38, 1797–1803. [Google Scholar] [CrossRef]
- George, S.E.; Hrubesch, J.; Breuing, I.; Vetter, N.; Korn, N.; Hennemann, K.; Bleul, L.; Willmann, M.; Ebner, P.; Götz, F.; et al. Oxidative Stress Drives the Selection of Quorum Sensing Mutants in the Staphylococcus aureus population. Proc. Natl. Acad. Sci. USA 2019, 116, 19145–19154. [Google Scholar] [CrossRef]
- Wang, L.C. Polyamines in Soybeans. Plant Physiol. 1972, 50, 152–156. [Google Scholar] [CrossRef]
- Hanfrey, C.; Sommer, S.; Mayer, M.J.; Burtin, D.; Michael, A.J. Araidopsis Polyamine Biosynthesis: Absence of Ornithine Decarboxylase and the Mechanism of Arginine Decarboxylase Activity. Plant J. 2001, 27, 551–560. [Google Scholar] [CrossRef]
- Wong, C.; Santiago, J.C.; Rodriguez-Paez, L.; Ibanez, M.; Baeza, I. Synthesis of Putrescine Under Possible Primitive Earth Conditions. Origin Life Evol. Biosph. 1991, 21, 145–156. [Google Scholar] [CrossRef]
- Guerra, P.R.; Herrero-Fresno, A.; Ladero, V.; Pires dos Santos, T.; Spiegelhauer, M.R.; Jelsbak, L.; Olsen, J.E. Putrescine Biosynthesis and Export Genes are Essential for Normal Growth of Avian Pathogenic Escherichia coli. BMC Microbiol. 2018, 18, 226–237. [Google Scholar] [CrossRef]
- Antti, H.; Fahlgren, A.; Näsström, E.; Kouremenos, K.; Sundén-Cullberg, J.; Guo, Y.; Moritz, T.; Wolf-Watz, H.; Johansson, A.; Fallman, M. Metabolic Profiling for Detection of Staphylococcus aureus Infection and Antibiotic Resistance. PLoS ONE 2013, 8, e56971. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, S.; Chen, J.; Han, B. Metabolic Profiling of Staphylococcus aureus Cultivated Under Aerobic and Anaerobic Conditions with 1H NMR-based Nontargeted Analysis. Can. J. Microbiol. 2012, 58, 709–718. [Google Scholar] [CrossRef]
- Ammons, M.C.B.; Tripet, B.P.; Carlson, R.P.; Kirker, K.R.; Gross, M.A.; Stanisich, J.J.; Copie, V. Quantitative NMR Metabolite Profiling of Methicillin-Resistant and Methicillin-Susceptible Staphylococcus aureus Discriminates Between Biofilm and Planktonic Phenotypes. J. Proteome Res. 2014, 13, 2973–2985. [Google Scholar] [CrossRef]
- Freudenberg, R.A.; Wittemeier, L.; Einhaus, A.; Baier, T.; Kruse, O. Advanced Pathway Engineering for Phototrophic Putrescine Production. Plant Biotech. J. 2022, 20, 1968–1982. [Google Scholar] [CrossRef]
- Rosenthal, S.M.; Dubin, D.T. Metabolism of Polyamines by Staphylococcus. J. Bacteriol. 1962, 84, 859–863. [Google Scholar] [CrossRef]
- Michael, A.J. Polyamines in Eukaryotes, Bacteria, and Archaea. J. Biol. Chem. 2016, 291, 14896–14903. [Google Scholar] [CrossRef]
- Lodish, H.; Berk, A.; Zipursky, S.L.; Matsudaira, P.; Baltimore, D.; Darnell, J. Molecular Cell Biology, 4th ed.; W.H. Freeman and Company: New York, NY, USA, 2000. [Google Scholar] [CrossRef]
- Kong, X.; Wang, X.; Yin, Y.; Li, X.; Gao, H.; Bazer, F.W.; Wu, G. Putrescine Stimulates the mTOR Signaling Pathway and Protein Synthesis in Porcine Trophectoderm Cells. Biol. Reprod. 2014, 91, 106. [Google Scholar] [CrossRef] [PubMed]
- Pollitt, E.J.G.; Szkuta, P.T.; Burns, N.; Foster, S.J. Staphylococcus aureus infection dynamics. PLoS Pathog. 2018, 14, e1007112. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Wang, Q.; Li, Y.; Liang, Z.; Xu, L.; Zhou, J.; Cui, Z.; Zhang, L.-H. Putrescine Is an Intraspecies and Interkingdom Cell-Cell Communication Signal Modulating the Virulence of Dickeya zeae. Front. Microbiol. 2019, 10, 1950. [Google Scholar] [CrossRef] [PubMed]
- Pegg, A.E.; Casero, R.A. Current Status of the Polyamine Research Field. Methods Mol. Biol. 2011, 720, 3–35. [Google Scholar] [CrossRef]
- Mandal, S.; Mandal, A.; Johansson, H.E.; Orjalo, A.V.; Park, M.H. Depletion of Cellular Polyamines, Spermidine, and Spermine, Cause a Total Arrest in Translation and Growth in Mammalian Cells. Proc. Natl. Acad. Sci. USA 2013, 110, 2169–2174. [Google Scholar] [CrossRef]
- Nanduri, B.; Swiatlo, E. The expansive effects of polyamines on the metabolism and virulence of Streptococcus pneumoniae. Pneumonia 2021, 13, 4. [Google Scholar] [CrossRef]

| Strain | Putrescine | Putrescine (nM) | N-Acetyl-Putrescine | Acetyl Putrescine (nM) |
|---|---|---|---|---|
| ATCC 25923 | 4.17 ± 3.03 | 0.4 | 17.2 ± 15.14 | 1.65 |
| CDC AR1454 | 2.66 ± 0.91 b | 6 | 12.59 ± 11.09 b | 28.4 |
| CDC AR3175 | 1.78 ± 0.58 | 3.95 | --- | |
| CDC AR7067 | --- | --- | 149.98 ± 123.37 a | 14.4 |
| Strain | Antibiotic Resistance | Hemolytic Activity (24 h) | Concentration CFU/mL |
|---|---|---|---|
| ATCC BAA-44 | Multiple 1 | None | 3.69 × 109 |
| NCBI COL-S | None | None | 1.7 × 109 |
| ATCC 43300 | Methicillin, Oxacillin | None | 2 × 109 |
| ATCC 25923 | Cefoxitin, Penicillin, Mupirocin | None | 1.8 × 109 |
| CDC AR1454 | Oxacillin, Penicillin, Erythromycin | None | 4.35 × 109 |
| CDC AR2789 | Oxacillin, Penicillin, Clindamycin, Erythromycin, Levofloxacin | None | 2.9 × 109 |
| CDC AR3175 | Oxacillin, Penicillin, Clindamycin, Erythromycin | None | 2.87 × 109 |
| CDC AR42208 | Oxacillin, Penicillin, Erythromycin, Levofloxacin | Some | 1.3 × 109 |
| CDC AR 5738 | Oxacillin, Penicillin, Erythromycin | Some | 2.21 × 109 |
| CDC AR6498 | Oxacillin, Penicillin, Clindamycin, Erythromycin, Levofloxacin | None | 1.9 × 109 |
| CDC AR7067 | Oxacillin, Penicillin, Erythromycin, Levofloxacin | Some | 2.23 × 109 |
| Baylor MRSA USA300 | Erythromycin, Vancomycin, Methicillin | Strong | 2.7 × 109 |
| GENE | 25923 | 43300 |
|---|---|---|
| Hypothetical protein 132469..132870 | 1 | 0 |
| Hypothetical protein 135487..136044 | 1 | 2 |
| Hypothetical protein 156985..157179 | 2 | 0 |
| Hypothetical protein 157404..158372 | 2 | 0 |
| Hypothetical protein 158649..159218 | 3 | 2 |
| Hypothetical protein 159559..160086 | 4 | 3 |
| Hypothetical protein 160137..160355 | 2 | 1 |
| Hypothetical protein 160356..160952 | 3 | 2 |
| Hypothetical protein 160964..161305 | 1 | 0 |
| Hypothetical protein 161652..162293 | 3 | 2 |
| Hypothetical protein 162290..162574 | 1 | 2 |
| Hypothetical protein 162576..162938 | 2 | 3 |
| Hypothetical protein 164721..165590 | 1 | 0 |
| Hypothetical protein 165974..166183 | 2 | 0 |
| Hypothetical protein 166176..166322 | 1 | 0 |
| Hypothetical protein 166650..167291 | 1 | 0 |
| Hypothetical protein 167295..167507 | 3 | 2 |
| Hypothetical protein 167661..168323 | 1 | 0 |
| Hypothetical protein 177188..177328 | 3 | 4 |
| Hypothetical protein 434789..435115 | 1 | 2 |
| Hypothetical protein 460317..460508 | 1 | 0 |
| Hypothetical protein 490900..491820 | 1 | 2 |
| Hypothetical protein 513477..515150 | 2 | 3 |
| Hypothetical protein 637833..638594 | 1 | 2 |
| Hypothetical protein 689131..690099 | 2 | 1 |
| Hypothetical protein 690618..691028 | 1 | 3 |
| Hypothetical protein 734010..734387 | 2 | 6 |
| Hypothetical protein 734404..735225 | 2 | 3 |
| Hypothetical protein 945446..945742 | 1 | 0 |
| Hypothetical protein 992307..993047 | 1 | 2 |
| Hypothetical protein 1004508..1005284 | 3 | 2 |
| Hypothetical protein 1182854..1183327 | 2 | 1 |
| Hypothetical protein 1183342..1183716 | 2 | 1 |
| Hypothetical protein 1183734..1184123 | 2 | 1 |
| Hypothetical protein 1184125..1184517 | 1 | 0 |
| Hypothetical protein 1750363..1750452 | 5 | 4 |
| Hypothetical protein 1254171..1255004 | 1 | 0 |
| Hypothetical protein 1256675..1257370 | 1 | 0 |
| Hypothetical protein 1257514..1257726 | 3 | 2 |
| Hypothetical protein 1257727..1257999 | 1 | 0 |
| Hypothetical protein 1258150..1258359 | 2 | 0 |
| Hypothetical protein 1258750..1259619 | 2 | 3 |
| Hypothetical protein 1259633..1261342 | 1 | 0 |
| Hypothetical protein 1262031..1262672 | 3 | 2 |
| Hypothetical protein 1263189..1263530 | 2 | 3 |
| Hypothetical protein 1263542..1264138 | 3 | 2 |
| Hypothetical protein 1264139..1264357 | 2 | 1 |
| Hypothetical protein 1264408..1264935 | 4 | 3 |
| Hypothetical protein 1265276..1265845 | 3 | 2 |
| Hypothetical protein 1266122..1267090 | 2 | 0 |
| Hypothetical protein 1267386..1267667 | 1 | 0 |
| Hypothetical protein 1267785..1268297 | 1 | 0 |
| Hypothetical protein 1274829..1275455 | 1 | 2 |
| Hypothetical protein 1488328..1488564 | 2 | 1 |
| Hypothetical protein 1521968..1522474 | 3 | 4 |
| Hypothetical protein 1587960..1588349 | 4 | 6 |
| Hypothetical protein 1653173..1654051 | 4 | 3 |
| Hypothetical protein 1744950..1745777 | 2 | 1 |
| Hypothetical protein 1850234..1850428 | 1 | 0 |
| Hypothetical protein 1975041..1976078 | 4 | 6 |
| Hypothetical protein 2009292..2009777 | 1 | 0 |
| Hypothetical protein 2020584..2021309 | 3 | 2 |
| Hypothetical protein 2021419..2022144 | 2 | 1 |
| Hypothetical protein 2187375..2187581 | 2 | 3 |
| Hypothetical protein 2197213..2197338 | 1 | 0 |
| Hypothetical protein 2268153..2268722 | 1 | 0 |
| Hypothetical protein 2292291..2292692 | 1 | 2 |
| Hypothetical protein 2314681..2314902 | 2 | 3 |
| Hypothetical protein 2321684..2322388 | 6 | 5 |
| Hypothetical protein 2413521..2414975 | 2 | 1 |
| Hypothetical protein 2414986..2415288 | 2 | 1 |
| Hypothetical protein 2415414..2415788 | 1 | 0 |
| Hypothetical protein 2420121..2421611 | 2 | 1 |
| Hypothetical protein 2421611..2426260 | 1 | 0 |
| Hypothetical protein 2426498..2426944 | 1 | 0 |
| Hypothetical protein 2427009..2427962 | 5 | 3 |
| Hypothetical protein 2427963..2428343 | 2 | 1 |
| Hypothetical protein 2428340..2428717 | 1 | 0 |
| Hypothetical protein 2428717..2429052 | 1 | 0 |
| Hypothetical protein 2429039..2429371 | 1 | 0 |
| Hypothetical protein 2429380..2429538 | 2 | 1 |
| Hypothetical protein 2429574..2430821 | 2 | 1 |
| Hypothetical protein 2430909..2431493 | 1 | 0 |
| Hypothetical protein 2431486..2432736 | 1 | 0 |
| Hypothetical protein 2432742..2432981 | 1 | 0 |
| Hypothetical protein 2432956..2434650 | 1 | 0 |
| Hypothetical protein 2434653..2435120 | 2 | 1 |
| Hypothetical protein 2435250..2435603 | 1 | 0 |
| Hypothetical protein 2435610..2436062 | 1 | 0 |
| Hypothetical protein 2436177..2436611 | 1 | 0 |
| Hypothetical protein 2437643..2437795 | 2 | 1 |
| Hypothetical protein 2437795..2437995 | 1 | 0 |
| Hypothetical protein 2438155..2438400 | 1 | 0 |
| Hypothetical protein 2439228..2439476 | 2 | 1 |
| Hypothetical protein 2439477..2439836 | 1 | 0 |
| Hypothetical protein 2439837..2440022 | 1 | 0 |
| Hypothetical protein 2440027..2440431 | 1 | 0 |
| Hypothetical protein 2440441..2440662 | 1 | 0 |
| Hypothetical protein 2440675..2440833 | 2 | 1 |
| Hypothetical protein 2440827..2441606 | 1 | 0 |
| Hypothetical protein 2441616..2442386 | 1 | 0 |
| Hypothetical protein 2442452..2442733 | 1 | 0 |
| Hypothetical protein 2442872..2443543 | 1 | 0 |
| Hypothetical protein 2444140..2444919 | 1 | 0 |
| Hypothetical protein 2444912..2445133 | 1 | 0 |
| Hypothetical protein 2445143..2445403 | 1 | 0 |
| Hypothetical protein 2445814..2445975 | 1 | 0 |
| Hypothetical protein 2446877..2447104 | 1 | 0 |
| Hypothetical protein 2447106..2447297 | 1 | 0 |
| Hypothetical protein 2447354..2447893 | 1 | 0 |
| Hypothetical protein 2450801..2450926 | 1 | 0 |
| Strain | Transport | Ornithine Decarboxylase | Agmatinase | Agmatine Deiminase | N-Carbamoyl- Putrescine Amidohydrolase | Arginine Decarboxylase |
|---|---|---|---|---|---|---|
| Streptococcus pyogenes | ||||||
| NCTC12064 | Spermidine/putrescine transport permease | No | No | No | No | No |
| HKU488 | Spermidine/putrescine transport permease | No | No | No | No | No |
| MGAS29326 | Spermidine/putrescine transport permease | No | No | No | No | No |
| TJ11-001 | No transport | No | No | No | No | No |
| BSAC_bs1388 | ABC transporter permease | No | No | No | No | No |
| ABC221 | ABC transporter permease | No | No | No | No | No |
| 37-97S | ABC transporter permease | No | No | No | No | No |
| NCTC8324 | Spermidine/putrescine transport permease | No | No | No | No | No |
| HKU419 | No transport | No | No | No | No | No |
| MGAS10786 | ABC transporter permease | No | No | No | No | No |
| 21SPY7071 | ABC transporter permease | No | No | No | No | No |
| PS003 | No transport | No | No | No | No | No |
| Pseudomonas aeruginosa | ||||||
| Pa58 | Spermidine/putrescine ABC transporter | Yes | Yes | No | Yes | Yes |
| AR_0353 | Polyamine antiporter | No | Yes | Yes | Yes | Yes |
| AG1 | Putrescine ABC transporter | No | No | Yes | Yes | Yes |
| PALA9 | Spermidine/putrescine transport system permease | Yes | No | Yes | No | Yes |
| WTJH36 | Spermidine/putrescine ABC transporter | No | Yes | Yes | Yes | Yes |
| H05 | Spermidine/putrescine ABC transporter | No | Yes | Yes | Yes | Yes |
| Bacillus subtilis | ||||||
| SRCM102754 | Putative ABC transporter ATP-binding protein, ABC transporter permease | No | Yes | No | No | Yes |
| PRO112 | ABC transporter permease, ABC transporter ATP-binding protein | No | Yes | No | No | Yes |
| ms-2 | No transport | No | No | No | No | No |
| SRCM103581 | ABC transporter permease | No | Yes | No | No | No |
| 29R7-12 | Spermidine/putrescine ABC transporter | No | Yes | No | No | Yes |
| TR21 | ABC transporter permease | No | Yes | No | No | Yes |
| ATC-3 | ABC transporter permease | No | Yes | No | No | Yes |
| MB9_B4 | ABC transporter permease | No | Yes | No | No | Yes |
| Staphylococcus aureus | ||||||
| ATCC BAA-44 | Putrescine export system ATP-binding protein | No | No | No | No | Yes |
| ATCC 6538 | Putrescine export system ATP-binding protein | No | No | No | No | Yes |
| ATCC 9144 | Putrescine export system ATP-binding protein | No | No | No | No | Yes |
| ATCC 12600 | Putrescine export system ATP-binding protein | No | No | No | No | Yes |
| ATCC BAA-1680 | Putrescine export system ATP-binding protein | No | No | No | No | Yes |
| ATCC 27660 | Putrescine export system ATP-binding protein | No | No | No | No | Yes |
| ATCC25923 | Putrescine export system ATP-binding protein | No | No | No | No | Yes |
| ATCC43300 | Putrescine export system ATP-binding protein | No | No | No | No | Yes |
| KG-22 | Spermidine/putrescine ABC transporter | No | No | No | No | No |
| JK3137 | Spermidine/putrescine ABC transporter | No | No | No | No | No |
| MRSA—AMRF 5 | Spermidine/putrescine ABC transporter | No | No | No | No | No |
| AR_0216 | Spermidine/putrescine import ATP-binding protein | No | No | No | No | No |
| USA300-SUR16 | Spermidine/putrescine ABC transporter | No | No | No | No | No |
| 110900 | Spermidine/putrescine ABC transporter | No | No | No | No | No |
| CMRSA-3 | Spermidine/putrescine ABC transporter | No | No | No | No | No |
| Escherichia coli | ||||||
| 2010C-3347 | No transport | No | No | No | No | No |
| W3110 | Putrescine/proton symporter, putrescine transport protein | Yes | Yes | No | No | Yes |
| K-12 (MG1655) | Putrescine-ornithine antiporter, putrescine transporter, putative amine transport | Yes | Yes | No | No | Yes |
| 117 | Spermidine/putrescine ABC transporter, putrescine ABC transporter permease, putrescine-ornithine antiporter | Yes | Yes | No | No | Yes |
| DSM 103246 | Spermidine/putrescine ABC transporter | Yes | Yes | No | No | Yes |
| CV261 | Putrescine-ornithine antiporter, spermidine/putrescine ABC transporter, putrescine ABC transporter permease | Yes | Yes | No | No | Yes |
| STEC2018-553 | Putrescine/proton symporter, putrescine/proton symporter, putrescine ABC transporter permease, putrescine-ornithine antiporter | Yes | Yes | No | No | Yes |
| 2013C-3277 | No transport | No | No | No | No | No |
| CFSAN027350 | Putrescine-ornithine antiporter, putrescine/proton symporter, putrescine/proton symporter, spermidine/putrescine ABC transporter, putrescine ABC transporter permease | Yes | Yes | No | No | Yes |
| Ecol_AZ146 | Putrescine-ornithine antiporter | Yes | Yes | No | No | Yes |
| 2014C-3655 | No transport | No | No | No | No | No |
| Mycobacterium tuberculosis | ||||||
| SEA02010036P6C4 | ABC transporter permease | No | No | No | No | No |
| 1-0110P6c4 | ABC transporter permease | No | No | No | No | No |
| MTB2 | ABC transporter permease | No | No | No | No | No |
| H54 | Spermidine/putrescine ABC transporter, ABC transporter permease | No | No | No | No | No |
| BLR-31d | ABC transporter permease, ABC transporter permease | No | No | No | No | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seravalli, J.; Portugal, F. Putrescine Detected in Strains of Staphylococcus aureus. Pathogens 2023, 12, 881. https://doi.org/10.3390/pathogens12070881
Seravalli J, Portugal F. Putrescine Detected in Strains of Staphylococcus aureus. Pathogens. 2023; 12(7):881. https://doi.org/10.3390/pathogens12070881
Chicago/Turabian StyleSeravalli, Javier, and Frank Portugal. 2023. "Putrescine Detected in Strains of Staphylococcus aureus" Pathogens 12, no. 7: 881. https://doi.org/10.3390/pathogens12070881
APA StyleSeravalli, J., & Portugal, F. (2023). Putrescine Detected in Strains of Staphylococcus aureus. Pathogens, 12(7), 881. https://doi.org/10.3390/pathogens12070881







