A Comparative Analysis of the Fecal Bacterial Communities of Light and Heavy Finishing Barrows Raised in a Commercial Swine Production Environment
Abstract
1. Introduction
2. Results
2.1. Animal Performance
2.2. Taxonomic Composition Analysis of Fecal Bacterial Communities
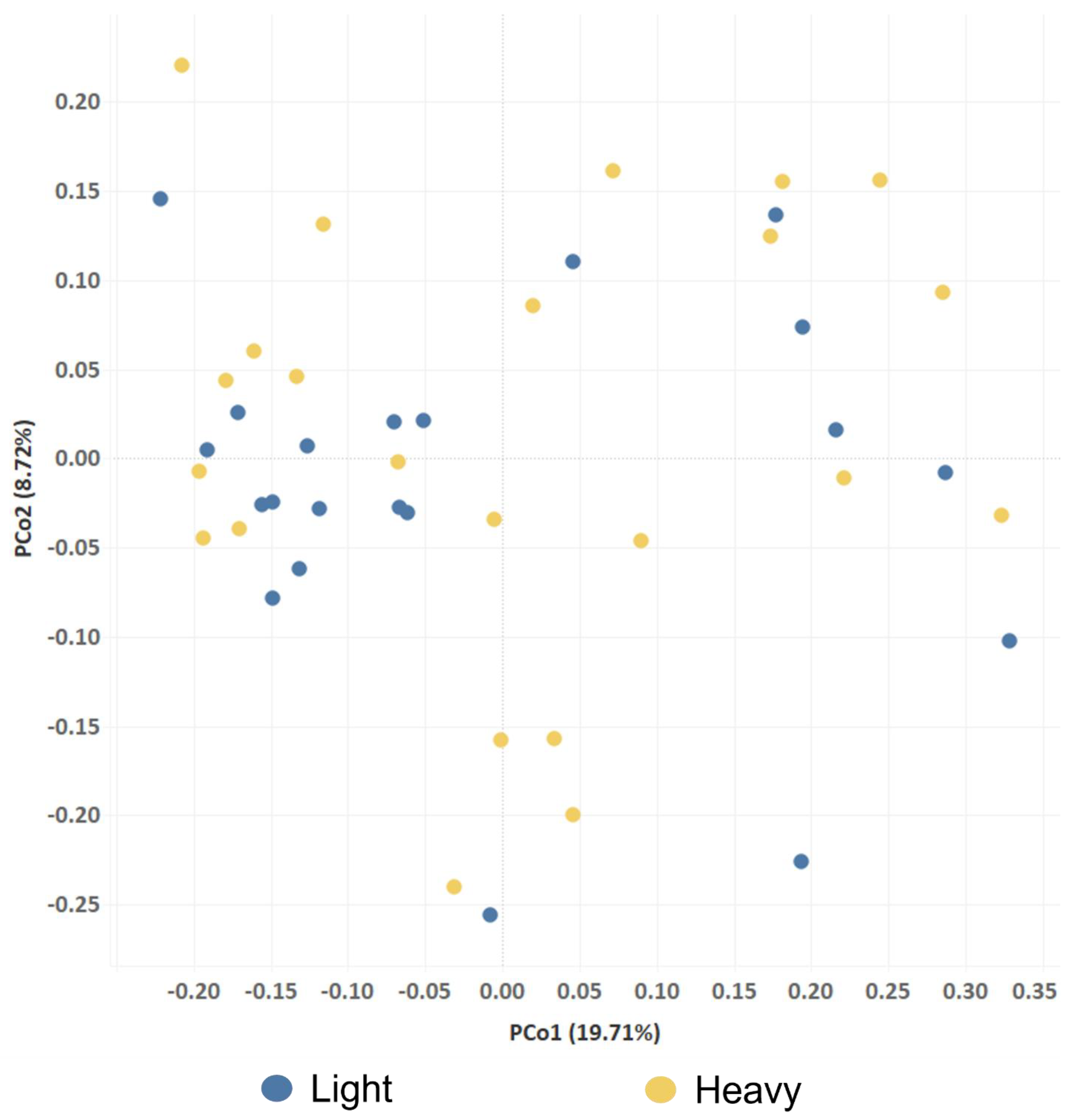
2.3. Analysis of Alpha and Beta Diversity of Fecal Bacterial Communities
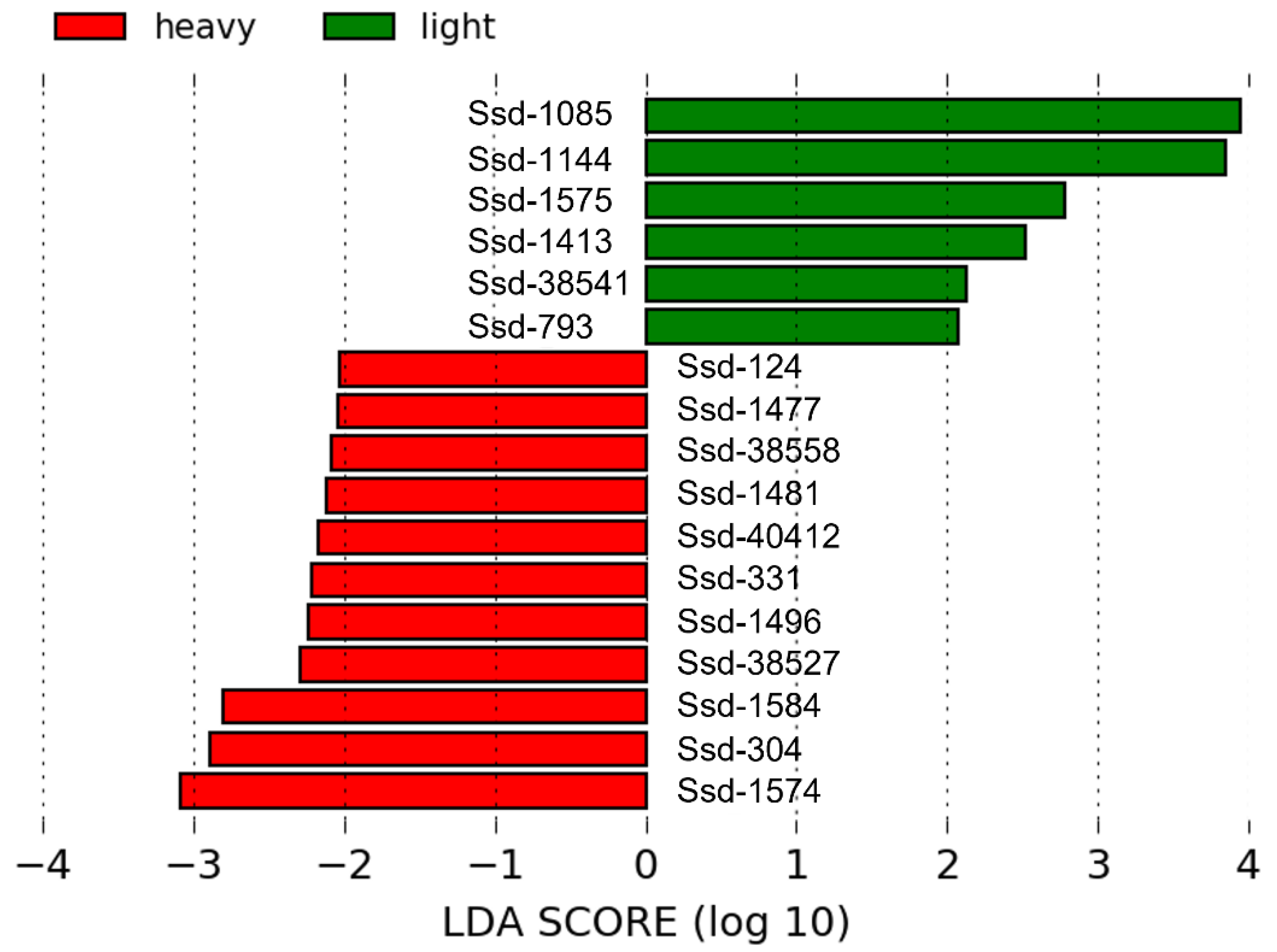
2.4. OTU Composition Analysis of Fecal Bacterial Communities
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Animals and Sample Collection
5.2. Isolation of Microbial Genomic DNA and Sequencing of 16S rRNA Gene Amplicons
5.3. Bacterial Composition Analyses
5.4. Statistical Analyses
5.5. Next Generation Sequencing Data Accessibility
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cook, H.; Schulz, L. The United States Pork Industry: Current Structure and Economic Importance; National Pork Producers Council: Des Moines, IA, USA, 2022. [Google Scholar]
- Thompson, B.K.; Fraser, D. Variation in piglet weights: Development of within-litter variation over a 5-week lactation and effect of farrowing crate design. Can. J. Anim. Sci. 1986, 66, 361–372. [Google Scholar] [CrossRef]
- Thompson, B.K.; Fraser, D. Variation in piglet weights: Weight gains in the first days after birth and their relationship with later performance. Can. J. Anim. Sci. 1988, 68, 581–590. [Google Scholar] [CrossRef]
- Milligan, B.N.; Fraser, D.; Kramera, D.L. Within-litter birth weight variation in the domestic pig and its relation to pre-weaning survival, weight gain, and variation in weaning weights. Livest. Prod. Sci. 2002, 76, 181–191. [Google Scholar] [CrossRef]
- O’Quinn, P.R.; Dritz, S.S.; Goodband, R.D.; Tokach, M.D.; Swanson, J.C.; Nelssen, J.L.; Musser, R.E. Sorting growing-finishing pigs by weight fails to improve growth performance or weight variation. J. Swine Health Prod. 2001, 9, 11–16. [Google Scholar]
- Marandu, N.; Halimani, T.E.; Chimonyo, M.; Shoniwa, A.; Mutibvu, T. Effect of within-litter birth weight variation on piglet survival and pre-weaning weight gain in a commercial herd. J. Agric. Rural. Dev. Trop. 2015, 116, 123–129. [Google Scholar]
- Wu, Y.; Zhao, J.; Xu, C.; Ma, N.; He, T.; Zhao, J.; Ma, X.; Thacker, P.A. Progress towards pig nutrition in the last 27 years. J. Sci. Food Agric. 2020, 100, 5102–5110. [Google Scholar] [CrossRef] [PubMed]
- Garrett, W.S.; Gordon, J.I.; Glimcher, L.H. Homeostasis and inflammation in the intestine. Cell 2010, 140, 859–870. [Google Scholar] [CrossRef]
- Isaacson, R.; Kim, H.B. The intestinal microbiome of the pig. Anim. Health Res. Rev. 2012, 13, 100–109. [Google Scholar] [CrossRef]
- Pluske, J.R.; Turpin, D.L.; Kim, J.C. Gastrointestinal tract (gut) health in the young pig. Anim. Nutr. 2018, 4, 187–196. [Google Scholar] [CrossRef]
- Fujisaka, S.; Watanabe, Y.; Tobe, K. The gut microbiome: A core regulator of metabolism. J. Endocrinol. 2023, 256, e220111. [Google Scholar] [CrossRef]
- Stokes, C.R. The development and role of microbial-host interactions in gut mucosal immune development. J. Anim. Sci. Biotechnol. 2017, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- McCormack, U.M.; Curiao, T.; Buzoianu, S.G.; Prieto, M.L.; Ryan, T.; Varley, P.; Crispie, F. Exploring a possible link between the intestinal microbiota and feed efficiency in pigs. Appl. Environ. Microbiol. 2017, 83, e00380-17. [Google Scholar] [CrossRef]
- Maltecca, C.; Bergamaschi, M.; Tiezzi, F. The interaction between microbiome and pig efficiency: A review. J. Anim. Breed. Genet. 2020, 137, 4–13. [Google Scholar] [CrossRef]
- Yu, M.; Li, Z.; Chen, W.; Rong, T.; Wang, G.; Ma, X. Hermetia illucens larvae as a potential dietary protein source altered the microbiota and modulated mucosal immune status in the colon of finishing pigs. J. Anim. Sci. Biotechnol. 2019, 10, 50. [Google Scholar] [CrossRef]
- He, W.; Gao, Y.; Guo, Z.; Yang, Z.; Wang, X.; Liu, H.; Sun, H.; Shi, B. Effects of fermented wheat bran and yeast culture on growth performance, immunity, and intestinal microflora in growing-finishing pigs. J. Anim. Sci. 2021, 99, skab308. [Google Scholar] [CrossRef]
- Smith, B.N.; Fleming, S.A.; Wang, M.; Dilger, R.N. Alterations of fecal microbiome characteristics by dietary soy isoflavone ingestion in growing pigs infected with porcine reproductive and respiratory syndrome virus. J. Anim. Sci. 2020, 98, skaa156. [Google Scholar] [CrossRef]
- Umu, Ö.C.O.; Mydland, L.T.; Øverland, M.; Press, C.M.; Sørum, H. Rapeseed-based diet modulates the imputed functions of gut microbiome in growing-finishing pigs. Sci. Rep. 2020, 10, 9372. [Google Scholar] [CrossRef]
- Pu, G.; Li, P.; Du, T.; Niu, Q.; Fan, L.; Wang, H.; Liu, H.; Li, K.; Niu, P.; Wu, C.; et al. Adding appropriate fiber in diet increases diversity and metabolic capacity of distal gut microbiota without altering fiber digestibility and growth rate of finishing pig. Front. Microbiol. 2020, 11, 533. [Google Scholar] [CrossRef]
- Niu, J.; Liu, X.; Xu, J.; Li, F.; Wang, J.; Zhang, X.; Yang, X.; Wang, L.; Ma, S.; Li, D.; et al. Effects of silage diet on meat quality through shaping gut microbiota in finishing pigs. Spectrum 2023, 11, e02416-22. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xie, J.; Fan, Z.; Ma, X.; Yin, Y. Effects of low protein diet with a balanced amino acid pattern on growth performance, meat quality and cecal microflora of finishing pigs. J. Sci. Food Agric. 2023, 103, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Sung, J.Y.; Johnson, T.A.; Ragland, D.R.; Adeola, O. Impact of ileal indigestible protein on fecal nitrogen excretion and fecal microbiota may be greater compared with total protein concentration of diets in growing pigs. J. Anim. Sci. 2022, 101, skac409. [Google Scholar] [CrossRef]
- Zhou, L.; Fang, L.; Sun, Y.; Su, Y.; Zhu, W. Effects of the dietary protein level on the microbial composition and metabolomic profile in the hindgut of the pig. Anaerobe 2016, 38, 61–69. [Google Scholar] [CrossRef] [PubMed]
- De Rodas, B.; Youmans, B.P.; Danzeisen, J.L.; Tran, H.; Johnson, T.J. Microbiome profiling of commercial pigs from farrow to finish. J. Anim. Sci. 2018, 96, 1778–1794. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tsai, T.; Deng, F.; Wei, X.; Chai, J.; Knapp, J.; Apple, J.; Maxwell, C.V.; Lee, J.A.; Li, Y.; et al. Longitudinal investigation of the swine gut microbiome from birth to market reveals stage and growth performance associated bacteria. Microbiome 2019, 7, 109. [Google Scholar] [CrossRef]
- Tao, X.; Deng, B.; Yuan, Q.; Men, X.; Wu, J.; Xu, Z. Low crude protein diet affects the intestinal microbiome and metabolome differently in barrows and gilts. Front. Microbiol. 2021, 12, 717727. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, H.; Wang, S.; Wang, Y.; Ji, H. Sex-dependent changes in the microbiota profile, serum metabolism, and hormone levels of growing pigs after dietary supplementation with Lactobacillus. Appl. Microbiol. Biotechnol. 2021, 105, 4775–4789. [Google Scholar] [CrossRef]
- Ma, J.; Chen, J.; Gan, M.; Chen, L.; Zhao, Y.; Zhu, Y.; Niu, L.; Zhang, S.; Zhu, L.; Shen, L. Gut microbiota composition and diversity in different commercial swine breeds in early and finishing growth stages. Animals 2022, 12, 1607. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Xiang, Y.; Robinson, K.; Wang, J.; Zhang, G.; Zhao, J.; Xiao, Y. Gut microbiota Is a major contributor to adiposity in pigs. Front. Microbiol. 2018, 9, 3045. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Yi, H.; Wu, Q.; Jiang, Z.; Wang, L. Effects of acute heat stress on intestinal microbiota in grow-finishing pigs, and associations with feed intake and serum profile. J. App Microbiol. 2019, 128, 840–852. [Google Scholar] [CrossRef]
- Tan, Z.; Yang, T.; Wang, Y.; Xing, K.; Zhang, F.; Zhao, X.; Ao, H.; Chen, J.; Liu, J.; Wang, C. Metagenomic Analysis of Cecal Microbiome Identified Microbiota and Functional Capacities Associated with Feed Efficiency in Landrace Finishing Pigs. Front. Microbiol. 2017, 8, 1546. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Kieu, H.T.; Garrigou, N.; Fadlane, A.; Brechard, L.; Armstrong, N.; Decloquement, P.; Yasir, M.; Azhar, E.I.; Al-Masaudi, S.B.; Lagier, J.-C.; et al. Clostridium culturomicium sp. nov. and Clostridium jeddahitimonense sp. nov., novel members of the Clostridium genus isolated from the stool of an obese Saudi Arabian. Curr. Microbiol. 2021, 78, 3586–3595. [Google Scholar] [CrossRef]
- Bertelsen, H.; Andersen, H.; Tvede, M. Fermentation of D-Tagatose by human intestinal bacteria and dairy lactic acid bacteria. Microb. Ecol. Health Dis. 2001, 13, 87–95. [Google Scholar] [CrossRef]
- Lobete, M.M.; Noriega, E.; Batalha, M.A.; De Beurme, S.; Van de Voorde, I.; Van Impe, J.F. Effect of tagatose on growth dynamics of Salmonella typhimurium and Listeria monocytogenes in media with different levels of structural complexity and in UHT skimmed milk. Food Control. 2017, 73, 31–42. [Google Scholar] [CrossRef]
- Li, M.; Guo, R.; Jiao, Y.; Jin, X.; Zhang, H.; Shi, L. Comparison of Salt Tolerance in Soja Based on Metabolomics of Seedling Roots. Front. Plant. Sci. 2017, 8, 1101. [Google Scholar] [CrossRef]
- Formanek, J.; Mackie, R.; Blaschek, H.P. Enhanced butanol production by Clostridium beijerinckii BA101 grown in semi defined P2 medium containing 6 percent maltodextrin or glucose. Appl. Environ. Microbiol. 1997, 63, 2306–2310. [Google Scholar] [CrossRef] [PubMed]
- López-Contreras, A.M.; Smidt, H.; van der Oost, J.; Claassen, P.A.; Mooibroek, H.; de Vos, W.M. Clostridium beijerinckii cells expressing Neocallimastix patriciarum glycoside hydrolases show enhanced lichenan utilization and solvent production. Appl. Environ. Microbiol. 2001, 67, 5127–5133. [Google Scholar] [CrossRef] [PubMed]
- Poudel, P.; Samuel, R.; Levesque, C.L.; St-Pierre, B. Investigating the effects of peptide-based, MOS and protease feed additives on the growth performance and fecal microbial composition of weaned pigs. J. Animal Sci. Biotechnol. 2022, 13, 25. [Google Scholar] [CrossRef] [PubMed]
- Poudel, P.; Levesque, C.L.; Samuel, R.; St-Pierre, B. Dietary inclusion of Peptiva, a peptide-based feed additive, can accelerate the maturation of the fecal bacterial microbiome in weaned pigs. BMC Vet. Res. 2020, 16, 60. [Google Scholar] [CrossRef]
- Fresno Rueda, A.; Samuel, R.; St-Pierre, B. Investigating the effects of a phytobiotics-based product on the fecal bacterial microbiome of weaned pigs. Animals 2021, 11, 1950. [Google Scholar] [CrossRef] [PubMed]
- Zeamer, K.M.; Samuel, R.S.; St-Pierre, B.; Thaler, R.C.; Woyengo, T.A.; Hymowitz, T.; Levesque, C.L. Effects of a low allergenic soybean variety on gut permeability, microbiota composition, ileal digestibility of amino acids, and growth performance in pigs. Livest. Sci. 2021, 243, 104369. [Google Scholar] [CrossRef]
- National Research Council (NRC). Nutrient Requirements of Swine; National Academy Press: Washington, DC, USA, 2012.
- Yu, Z.; Morrison, M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 2004, 36, 808–812. [Google Scholar] [CrossRef] [PubMed]
- Edwards, U.; Rogall, T.; Bloecker, H.; Emde, M.; Boettger, E.C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989, 17, 7843–7854. [Google Scholar] [CrossRef]
- Lane, D.; Pace, B.; Olsen, G.J.; Stahl, D.A.; Sogin, M.L.; Pace, N.R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 1985, 82, 6955–6959. [Google Scholar] [CrossRef]
- Kim, M.; Morrison, M.; Yu, Z. Evaluation of different partial 16S rRNA gene sequence regions for phylogenetic analysis of microbiomes. J. Microbiol. Methods 2011, 84, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucl. Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
| Pen Composition | |||||
|---|---|---|---|---|---|
| Week | Barrow | Gilt | Mixed | SEM | p-Value |
| 0 | 108.0 a | 103.7 b | 103.9 b | 0.5 | <0.01 |
| 1 | 115.5 a | 110.6 b | 111.2 b | 0.5 | <0.01 |
| 2 | 122.5 a | 116.8 b | 117.8 b | 0.5 | <0.01 |
| 3 | 129.0 a | 123.8 b | 124.5 b | 0.5 | <0.01 |
| 4 | 130.4 | 130.0 | 130.9 | 0.5 | 0.72 |
| 5 | 133.6 | 133.7 | 135.1 | 0.6 | 0.57 |
| 6 | - | 137.9 | 139.0 | 0.6 | 0.37 |
| Group | Mean BW (kg ± SEM) | Range BW (kg) | n |
|---|---|---|---|
| Light | 119.5 a ± 0.8 | 112.7–124.1 | 21 |
| Heavy | 145.9 b ± 1.1 | 139.1–160.0 | 23 |
| Pen mean | 133.0 ± 1.1 | 129.6–137.5 | 6 |
| Taxonomic Affiliation | Light | Heavy |
|---|---|---|
| Bacillota | 43.86 ± 2.61 | 45.60 ± 2.00 |
| Clostridiaceae 1 | 13.43 ± 1.36 | 10.79 ± 1.11 |
| Carnobacteriaceae | 5.51 ± 1.87 | 8.70 ± 2.34 |
| Ruminococcaceae | 4.23 ± 0.22 | 5.08 ± 0.37 |
| Peptostreptococcaceae | 2.58 ± 0.27 | 2.26 ± 0.22 |
| Unclassified Clostridiales & | 7.45 ± 0.70 | 8.85 ± 0.63 |
| Other Bacillota & | 10.66 ± 1.31 | 9.91 ± 0.84 |
| Planctomycetota | 24.75 ± 2.16 | 24.20 ± 2.16 |
| Unclassified Planctomycetacia & | 24.68 ± 2.16 | 24.13 ± 2.15 |
| Other Planctomycetota & | 0.07 ± 0.01 | 0.08 ± 0.01 |
| Bacteroidota | 14.79 ± 1.20 | 16.27 ± 1.05 |
| Prevotellaceae | 2.92 ± 0.40 | 3.75 ± 0.44 |
| Unclassified Bacteroidales & | 8.91 ± 0.90 | 9.23 ± 0.66 |
| Other Bacteroidota & | 2.96 ± 0.31 | 3.30 ± 0.33 |
| Spirochaetota | 8.37 ± 1.12 | 6.80 ± 0.77 |
| Spirochaetaceae | 8.29 ± 1.12 | 6.73 ± 0.76 |
| Other Spirochaetota & | 0.08 ± 0.01 | 0.07 ± 0.01 |
| Other Bacteria &$ | 8.23 ± 2.39 | 7.12 ± 0.97 |
| Index | Light (±SEM) | Heavy (±SEM) | p-Value |
|---|---|---|---|
| Observed OTUs | 811.4 ± 18.5 | 853.1 ± 18.0 | 0.1133 |
| Chao | 1750.8 ± 38.6 | 1880.9 ± 51.3 | 0.0521 |
| Ace | 2606.0 a ± 71.2 | 2866.7 b ± 86.0 | 0.0259 |
| Shannon | 3.89 ± 0.08 | 4.04 ± 0.08 | 0.2043 |
| Simpson | 0.11 ± 0.01 | 0.10 ± 0.01 | 0.5203 |
| OTUs | Light | Heavy | Closest Valid Relative (id% *) |
|---|---|---|---|
| Bacillota | |||
| Ssd-1085 1,3,4 | 5.88 a ± 0.67 | 4.16 b ± 0.37 | Clostridium jeddahitimonense (98.59%) |
| Ssd-1398 | 3.45 ± 1.27 | 5.25 ± 1.65 | Carnobacterium funditum (93.22%) |
| Ssd-1144 1,2,3,4 | 3.62 a ± 0.44 | 2.22 b ± 0.27 | Clostridium beijerinckii (97.80%) |
| Ssd-0675 1,2,3 | 2.19 ± 0.75 | 1.79 ± 0.48 | Christensenella massiliensis (84.54%) |
| Ssd-1566 | 1.12 ± 0.52 | 1.76 ± 0.69 | Carnobacterium funditum (93.42%) |
| Ssd-1079 1,3,4 | 1.44 ± 0.29 | 1.62 ± 0.26 | Mahella australiensi (83.01%) |
| Bacteroidota | |||
| Ssd-1048 1,3 | 3.09 ± 0.59 | 3.07 ± 0.36 | Caecibacteroides pullorum (86.93%) |
| Planctomycetota | |||
| Ssd-1095 1,2,3,4 | 24.52 ± 2.14 | 23.97 ± 2.09 | Lignipirellula cremea (80.91%) |
| Spirochaetota | |||
| Ssd-1115 1,4 | 4.58 ± 0.79 | 3.81 ± 0.45 | Treponema peruense (84.62%) |
| Ssd-1399 | 3.16 ± 0.72 | 2.34 ± 0.53 | Treponema bryantii (89.53%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fowler, E.C.; Samuel, R.S.; St-Pierre, B. A Comparative Analysis of the Fecal Bacterial Communities of Light and Heavy Finishing Barrows Raised in a Commercial Swine Production Environment. Pathogens 2023, 12, 738. https://doi.org/10.3390/pathogens12050738
Fowler EC, Samuel RS, St-Pierre B. A Comparative Analysis of the Fecal Bacterial Communities of Light and Heavy Finishing Barrows Raised in a Commercial Swine Production Environment. Pathogens. 2023; 12(5):738. https://doi.org/10.3390/pathogens12050738
Chicago/Turabian StyleFowler, Emily C., Ryan S. Samuel, and Benoit St-Pierre. 2023. "A Comparative Analysis of the Fecal Bacterial Communities of Light and Heavy Finishing Barrows Raised in a Commercial Swine Production Environment" Pathogens 12, no. 5: 738. https://doi.org/10.3390/pathogens12050738
APA StyleFowler, E. C., Samuel, R. S., & St-Pierre, B. (2023). A Comparative Analysis of the Fecal Bacterial Communities of Light and Heavy Finishing Barrows Raised in a Commercial Swine Production Environment. Pathogens, 12(5), 738. https://doi.org/10.3390/pathogens12050738









