Attenuation of In Vitro and In Vivo Virulence Is Associated with Repression of Gene Expression of AIG1 Gene in Entamoeba histolytica
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. Amoebiasis. Report on the WHO. Pan American Health Organization/UNESCO Expert Consultation Mexico City Geneva-WHO. Wkly. Epidemiol. Rec. 1997, 72, 97–100. [Google Scholar]
- Ali, I.K.M.; Clark, C.G.; Petri, W.A., Jr. Molecular Epidemiology of Amebiasis. Infect. Genet. Evol. 2008, 8, 698–707. [Google Scholar] [CrossRef]
- Diamond, L.S.; Clark, C.G. A Redescription of Entamoeba Histolytica Schaudinn, 1903 (Emended Walker, 1911) Separating It From Entamoeba Dispar Brumpt, 1925 1. J. Eukaryot. Microbiol. 1993, 40, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Ravdin, J.I.; Murphy, C.F.; Salata, R.A.; Guerrant, R.L.; Hewlett, E.L. N-Acetyl-D-Galactosamine-Inhibitable Adherence Lectin of Entamoeba Histolytica. I. Partial Purification and Relation to Amoebic Virulence in Vitro. J. Infect. Dis. 1985, 151, 804–815. [Google Scholar] [CrossRef]
- Petri, W., Jr. Amebiasis and the Entamoeba Histolytica Gal/GalNAc Lectin: From Lab Bench to Bedside. J. Investig. Med. 1996, 44, 24–36. [Google Scholar] [PubMed]
- Bruchhaus, I.; Jacobs, T.; Leippe, M.; Tannich, E. Entamoeba Histolytica and Entamoeba Dispar: Differences in Numbers and Expression of Cysteine Proteinase Genes. Mol. Microbiol. 1996, 22, 255–263. [Google Scholar] [CrossRef]
- Que, X.; Reed, S.L. The Role of Extracellular Cysteine Proteinases in Pathogenesis of Entamoeba Histolytica Invasion. Parasitol. Today 1997, 13, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Leippe, M. Amoebapores. Parasitol. Today 1997, 13, 178–183. [Google Scholar] [CrossRef]
- Tannicht, E. Entamoeba Histolytica and E. Dispar: Comparison of Molecules Considered Important for Host Tissue Destruction. Trans. R. Soc. Trop. Med. Hyg. 1998, 92, 593–596. [Google Scholar] [CrossRef]
- Mirelman, D.; Ankri, S.; Katz, U.; Padilla-Vaca, F.; Bracha, R. Pathogenesis of Entamoeba Histolytica Depends on the Concerted Action of Numerous Virulence Factors. Arch. Med. Res. 2000, 31, S214–S215. [Google Scholar] [CrossRef] [PubMed]
- Que, X.; Reed, S.L. Cysteine Proteinases and the Pathogenesis of Amebiasis. Clin. Microbiol. Rev. 2000, 13, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Petri, W.A., Jr.; Haque, R.; Mann, B.J. The Bittersweet Interface of Parasite and Host: Lectin-Carbohydrate Interactions during Human Invasion by the Parasite Entamoeba Histolytica. Annu. Rev. Microbiol. 2002, 56, 39. [Google Scholar] [CrossRef]
- Marie, C.; Petri, W.A., Jr. Regulation of Virulence of Entamoeba Histolytica. Annu. Rev. Microbiol. 2014, 68, 493–520. [Google Scholar] [CrossRef] [PubMed]
- Husain, A.; Jeelani, G.; Sato, D.; Nozaki, T. Global Analysis of Gene Expression in Response to L-Cysteine Deprivation in the Anaerobic Protozoan Parasite Entamoeba Histolytica. BMC Genom. 2011, 12, 275. [Google Scholar] [CrossRef] [PubMed]
- Anaya-Velázquez, F.; Padilla-Vaca, F. Virulence of Entamoeba Histolytica: A Challenge for Human Health Research. Future Microbiol. 2011, 6, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Montiel, F.; Mendoza-Macias, C.; Andrade-Guillen, S.; Rangel-Serrano, A.; Paramo-Perez, I.; Rivera-Cuellar, P.E.; España-Sánchez, B.L.; Luna-Barcenas, G.; Anaya-Velazquez, F.; Franco, B.; et al. Plasma Membrane Damage Repair Is Mediated by an Acid Sphingomyelinase in Entamoeba Histolytica. PLoS Pathog. 2019, 15, e1008016. [Google Scholar] [CrossRef]
- Leitsch, D.; Kolarich, D.; Wilson, I.B.H.; Altmann, F.; Duchêne, M. Nitroimidazole Action in Entamoeba Histolytica: A Central Role for Thioredoxin Reductase. PLoS Biol. 2007, 5, e211. [Google Scholar] [CrossRef]
- Ehrenkaufer, G.M.; Eichinger, D.J.; Singh, U. Trichostatin a Effects on Gene Expression in the Protozoan Parasite Entamoeba Histolytica. BMC Genom. 2007, 8, 216. [Google Scholar] [CrossRef]
- Mornico, D.; Hon, C.-C.; Koutero, M.; Weber, C.; Coppée, J.-Y.; Clark, C.G.; Dillies, M.-A.; Guillen, N. RNA Sequencing Reveals Widespread Transcription of Natural Antisense RNAs in Entamoeba Species. Microorganisms 2022, 10, 396. [Google Scholar] [CrossRef]
- Davis, P.H.; Schulze, J.; Stanley, S.L., Jr. Transcriptomic Comparison of Two Entamoeba Histolytica Strains with Defined Virulence Phenotypes Identifies New Virulence Factor Candidates and Key Differences in the Expression Patterns of Cysteine Proteases, Lectin Light Chains, and Calmodulin. Mol. Biochem. Parasitol. 2007, 151, 118–128. [Google Scholar] [CrossRef]
- MacFarlane, R.C.; Singh, U. Identification of Differentially Expressed Genes in Virulent and Nonvirulent Entamoeba Species: Potential Implications for Amebic Pathogenesis. Infect. Immun. 2006, 74, 340–351. [Google Scholar] [CrossRef] [PubMed]
- Davis, P.H.; Zhang, X.; Guo, J.; Townsend, R.R.; Stanley, S.L., Jr. Comparative Proteomic Analysis of Two Entamoeba Histolytica Strains with Different Virulence Phenotypes Identifies Peroxiredoxin as an Important Component of Amoebic Virulence. Mol. Microbiol. 2006, 61, 1523–1532. [Google Scholar] [CrossRef] [PubMed]
- Yanagawa, Y.; Izumiyama, S.; Saito-Nakano, Y.; Nakada-Tsukui, K.; Kobayashi, S.; Yoshida, N.; Kikuchi, Y.; Gatanaga, H.; Oka, S.; Nozaki, T. Gene Expression of Axenically-Isolated Clinical Entamoeba Histolytica Strains and Its Impact on Disease Severity of Amebiasis. PLoS Pathog. 2022, 18, e1010880. [Google Scholar] [CrossRef] [PubMed]
- Kumari, V.; Iyer, L.R.; Roy, R.; Bhargava, V.; Panda, S.; Paul, J.; Verweij, J.J.; Clark, C.G.; Bhattacharya, A.; Bhattacharya, S. Genomic Distribution of SINEs in Entamoeba Histolytica Strains: Implication for Genotyping. BMC Genom. 2013, 14, 432. [Google Scholar] [CrossRef]
- Shah, P.H.; MacFarlane, R.C.; Bhattacharya, D.; Matese, J.C.; Demeter, J.; Stroup, S.E.; Singh, U. Comparative Genomic Hybridizations of Entamoeba Strains Reveal Unique Genetic Fingerprints That Correlate with Virulence. Eukaryot. Cell 2005, 4, 504–515. [Google Scholar] [CrossRef]
- Haghighi, A.; Kobayashi, S.; Takeuchi, T.; Thammapalerd, N.; Nozaki, T. Geographic Diversity among Genotypes of Entamoeba Histolytica Field Isolates. J. Clin. Microbiol. 2003, 41, 3748–3756. [Google Scholar] [CrossRef]
- Weedall, G.D.; Clark, C.G.; Koldkjaer, P.; Kay, S.; Bruchhaus, I.; Tannich, E.; Paterson, S.; Hall, N. Genomic Diversity of the Human Intestinal Parasite Entamoeba Histolytica. Genome Biol. 2012, 13, R38. [Google Scholar] [CrossRef] [PubMed]
- Vicente, J.B.; Ehrenkaufer, G.M.; Saraiva, L.M.; Teixeira, M.; Singh, U. Entamoeba Histolytica Modulates a Complex Repertoire of Novel Genes in Response to Oxidative and Nitrosative Stresses: Implications for Amebic Pathogenesis. Cell. Microbiol. 2009, 11, 51–69. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, C.A.; Houpt, E.; Trapaidze, N.; Fei, Z.; Crasta, O.; Asgharpour, A.; Evans, C.; Martino-Catt, S.; Baba, D.J.; Stroup, S. Impact of Intestinal Colonization and Invasion on the Entamoeba Histolytica Transcriptome. Mol. Biochem. Parasitol. 2006, 147, 163–176. [Google Scholar] [CrossRef]
- Bruchhaus, I.; Roeder, T.; Lotter, H.; Schwerdtfeger, M.; Tannich, E. Differential Gene Expression in Entamoeba Histolytica Isolated from Amoebic Liver Abscess. Mol. Microbiol. 2002, 44, 1063–1072. [Google Scholar] [CrossRef]
- Weber, C.; Koutero, M.; Dillies, M.-A.; Varet, H.; Lopez-Camarillo, C.; Coppée, J.Y.; Hon, C.-C.; Guillén, N. Extensive Transcriptome Analysis Correlates the Plasticity of Entamoeba Histolytica Pathogenesis to Rapid Phenotype Changes Depending on the Environment. Sci. Rep. 2016, 6, 35852. [Google Scholar] [CrossRef]
- Mendoza-Macías, C.L.; Barrios-Ceballos, M.P.; de la Peña, L.P.C.; Rangel-Serrano, A.; Anaya-Velázquez, F.; Mirelman, D.; Padilla-Vaca, F. Entamoeba Histolytica: Effect on Virulence, Growth and Gene Expression in Response to Monoxenic Culture with Escherichia Coli 055. Exp. Parasitol. 2009, 121, 167–174. [Google Scholar] [CrossRef]
- Padilla-Vaca, F.; Ankri, S.; Bracha, R.; Koole, L.A.; Mirelman, D. Down Regulation of Entamoeba Histolytica Virulence by Monoxenic Cultivation with Escherichia Coli O55 Is Related to a Decrease in Expression of the Light (35-Kilodalton) Subunit of the Gal/GalNAc Lectin. Infect. Immun. 1999, 67, 2096–2102. [Google Scholar] [CrossRef]
- Bracha, R.; Mirelman, D. Virulence of Entamoeba Histolytica Trophozoites. Effects of Bacteria, Microaerobic Conditions, and Metronidazole. J. Exp. Med. 1984, 160, 353–368. [Google Scholar] [CrossRef] [PubMed]
- Saito-Nakano, Y.; Makiuchi, T.; Tochikura, M.; Gilchrist, C.A.; Petri, W.A., Jr.; Nozaki, T. ArfX2 GTPase Regulates Trafficking From the Trans-Golgi to Lysosomes and Is Necessary for Liver Abscess Formation in the Protozoan Parasite Entamoeba Histolytica. Front. Cell. Infect. Microbiol. 2021, 11, 1260. [Google Scholar] [CrossRef] [PubMed]
- Biller, L.; Schmidt, H.; Krause, E.; Gelhaus, C.; Matthiesen, J.; Handal, G.; Lotter, H.; Janssen, O.; Tannich, E.; Bruchhaus, I. Comparison of Two Genetically Related Entamoeba Histolytica Cell Lines Derived from the Same Isolate with Different Pathogenic Properties. Proteomics 2009, 9, 4107–4120. [Google Scholar] [CrossRef] [PubMed]
- Biller, L.; Davis, P.H.; Tillack, M.; Matthiesen, J.; Lotter, H.; Stanley, S.L.; Tannich, E.; Bruchhaus, I. Differences in the Transcriptome Signatures of Two Genetically Related Entamoeba Histolytica Cell Lines Derived from the Same Isolate with Different Pathogenic Properties. BMC Genom. 2010, 11, 63. [Google Scholar] [CrossRef]
- Meyer, M.; Fehling, H.; Matthiesen, J.; Lorenzen, S.; Schuldt, K.; Bernin, H.; Zaruba, M.; Lender, C.; Ernst, T.; Ittrich, H. Overexpression of Differentially Expressed Genes Identified in Non-Pathogenic and Pathogenic Entamoeba Histolytica Clones Allow Identification of New Pathogenicity Factors Involved in Amoebic Liver Abscess Formation. PLoS Pathog. 2016, 12, e1005853. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Ghildyal, R.; Bhattacharya, S.; Diamond, L.S. Characterization of a Monoclonal Antibody That Selectively Recognizes a Subset of Entamoeba Histolytica Isolates. Infect. Immun. 1990, 58, 3458–3461. [Google Scholar] [CrossRef]
- Anbar, M.; Bracha, R.; Nuchamowitz, Y.; Li, Y.; Florentin, A.; Mirelman, D. Involvement of a Short Interspersed Element in Epigenetic Transcriptional Silencing of the Amoebapore Gene in Entamoeba Histolytica. Eukaryot. Cell 2005, 4, 1775–1784. [Google Scholar] [CrossRef] [PubMed]
- Diamond, L.S.; Harlow, D.R.; Cunnick, C.C. A New Medium for the Axenic Cultivation of Entamoeba Histolytica and Other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 1978, 72, 431–432. [Google Scholar] [CrossRef] [PubMed]
- Ankri, S.; Padilla-Vaca, F.; Stolarsky, T.; Koole, L.; Katz, U.; Mirelman, D. Antisense Inhibition of Expression of the Light Subunit (35 KDa) of the Gal/GalNac Lectin Complex Inhibits Entamoeba Histolytica Virulence. Mol. Microbiol. 1999, 33, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Nozaki, T.; Asai, T.; Sanchez, L.B.; Kobayashi, S.; Nakazawa, M.; Takeuchi, T. Characterization of the Gene Encoding Serine Acetyltransferase, a Regulated Enzyme of Cysteine Biosynthesis from the Protist Parasites Entamoeba Histolytica and Entamoeba Dispar. J. Biol. Chem. 2002, 274, 32445–32452. [Google Scholar] [CrossRef] [PubMed]
- Vargas, M.A.; Orozco, E. Entamoeba Histolytica: Changes in the Zymodeme of Cloned Nonpathogenic Trophozoites Cultured under Different Conditions. Parasitol. Res. 1993, 79, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Tsutsumi, V.; Mena-Lopez, R.; Anaya-Velazquez, F.; Martinez-Palomo, A. Cellular Bases of Experimental Amebic Liver Abscess Formation. Am. J. Pathol. 1984, 117, 81. [Google Scholar] [PubMed]
- Fotedar, R.; Stark, D.; Beebe, N.; Marriott, D.; Ellis, J.; Harkness, J. Laboratory Diagnostic Techniques for Entamoeba Species. Clin. Microbiol. Rev. 2007, 20, 511–532. [Google Scholar] [CrossRef]
- Calderón, J.; Tovar, R. Loss of Susceptibility to Complement Lysis in Entamoeba Histolytica HM1 by Treatment with Human Serum. Immunology 1986, 58, 467. [Google Scholar]
- Rastew, E.; Morf, L.; Singh, U. Entamoeba Histolytica Rhomboid Protease 1 Has a Role in Migration and Motility as Validated by Two Independent Genetic Approaches. Exp. Parasitol. 2015, 154, 33–42. [Google Scholar] [CrossRef]
- Mora-Galindo, J.; Gutiérrez-Lozano, M.; Anaya-Velázquez, F. Entamoeba Histolytica: Kinetics of Hemolytic Activity, Erythrophagocytosis and Digestion of Erythrocytes. Arch. Med. Res. 1997, 28, 200–201. [Google Scholar] [CrossRef]
- Arias-Negrete, S.; Villagómez-Castro, J.C.; Anaya-Velázquez, F.; Lira-Ortiz, R. Entamoeba Histolytica: A Simplified Method to Quantify Its Cytotoxicity. Int. J. Parasitol. 1991, 21, 373–375. [Google Scholar] [CrossRef]
- Ormerod, M.G.; Sun, X.; Snowden, R.T.; Davies, R.; Fearnhead, H.; Cohen, G.M. Increased Membrane Permeability of Apoptotic Thymocytes: A Flow Cytometric Study. Cytom. J. Int. Soc. Anal. Cytol. 1993, 14, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Trissl, D.; Martínez-Palomo, A.; de la Torre, M.; de la Hoz, R.; Pérez de Suárez, E. Surface Properties of Entamoeba: Increased Rates of Human Erythrocyte Phagocytosis in Pathogenic Strains. J. Exp. Med. 1978, 148, 1137–1143. [Google Scholar] [CrossRef] [PubMed]
- Leippe, M.; Sievertsen, H.J.; Tannich, E.; Horstmann, R.D. Spontaneous Release of Cysteine Proteinases but Not of Pore-Forming Peptides by Viable Entamoeba Histolytica. Parasitology 1995, 111, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Kit, S. GeneChip® Expression Analysis Technical Manual; Affymetrix Inc.: Santa Clara, CA, USA, 2005. [Google Scholar]
- Russell, D.W.; Sambrook, J. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2001; Volume 1. [Google Scholar]
- Saito-Nakano, Y.; Yasuda, T.; Nakada-Tsukui, K.; Leippe, M.; Nozaki, T. Rab5-Associated Vacuoles Play a Unique Role in Phagocytosis of the Enteric Protozoan Parasite Entamoeba Histolytica. J. Biol. Chem. 2004, 279, 49497–49507. [Google Scholar] [CrossRef]
- Debnath, A.; Das, P.; Sajid, M.; McKerrow, J.H. Identification of Genomic Responses to Collagen Binding by Trophozoites of Entamoeba Histolytica. J. Infect. Dis. 2004, 190, 448–457. [Google Scholar] [CrossRef]
- Mirelman, D.; Bracha, R.; Chayen, A.; Aust-Kettis, A.; Diamond, L.S. Entamoeba Histolytica: Effect of Growth Conditions and Bacterial Associates on Isoenzyme Patterns and Virulence. Exp. Parasitol. 1986, 62, 142–148. [Google Scholar] [CrossRef]
- Lujan, H.D.; Diamond, L.S. Cholesterol Requirement and Metabolism in Entamoeba Histolytica. Arch. Med. Res. 1997, 28, 96–97. [Google Scholar]
- Spice, W.M.; Ackers, J.P. The Effect of Axenic versus Xenic Culture Conditions on the Total and Secreted Proteolytic Activity of Entamoeba Histolytica Strains. Arch. Med. Res. 1992, 23, 91–93. [Google Scholar]
- Nitta, T.; Takahama, Y. The Lymphocyte Guard-IANs: Regulation of Lymphocyte Survival by IAN/GIMAP Family Proteins. Trends Immunol. 2007, 28, 58–65. [Google Scholar] [CrossRef]
- König, C.; Honecker, B.; Wilson, I.W.; Weedall, G.D.; Hall, N.; Roeder, T.; Metwally, N.G.; Bruchhaus, I. Taxon-Specific Proteins of the Pathogenic Entamoeba Species E. Histolytica and E. Nuttalli. Front. Cell. Infect. Microbiol. 2021, 11, 641472. [Google Scholar] [CrossRef]
- Tanaka, M.; Makiuchi, T.; Komiyama, T.; Shiina, T.; Osaki, K.; Tachibana, H. Whole Genome Sequencing of Entamoeba Nuttalli Reveals Mammalian Host-Related Molecular Signatures and a Novel Octapeptide-Repeat Surface Protein. PLoS Negl. Trop. Dis. 2019, 13, e0007923. [Google Scholar] [CrossRef] [PubMed]
- Wilson, I.W.; Weedall, G.D.; Lorenzi, H.; Howcroft, T.; Hon, C.-C.; Deloger, M.; Guillén, N.; Paterson, S.; Clark, C.G.; Hall, N. Genetic Diversity and Gene Family Expansions in Members of the Genus Entamoeba. Genome Biol. Evol. 2019, 11, 688–705. [Google Scholar] [CrossRef]
- Naiyer, S.; Kaur, D.; Ahamad, J.; Singh, S.S.; Singh, Y.P.; Thakur, V.; Bhattacharya, A.; Bhattacharya, S. Transcriptomic Analysis Reveals Novel Downstream Regulatory Motifs and Highly Transcribed Virulence Factor Genes of Entamoeba Histolytica. BMC Genom. 2019, 20, 206. [Google Scholar] [CrossRef]
- Ximénez, C.; González, E.; Nieves, M.; Magaña, U.; Morán, P.; Gudiño-Zayas, M.; Partida, O.; Hernández, E.; Rojas-Velázquez, L.; García de León, M.C. Differential Expression of Pathogenic Genes of Entamoeba Histolytica vs E. Dispar in a Model of Infection Using Human Liver Tissue Explants. PLoS ONE 2017, 12, e0181962. [Google Scholar] [CrossRef]
- Nakada-Tsukui, K.; Sekizuka, T.; Sato-Ebine, E.; Escueta-de Cadiz, A.; Ji, D.; Tomii, K.; Kuroda, M.; Nozaki, T. AIG1 Affects in Vitro and in Vivo Virulence in Clinical Isolates of Entamoeba Histolytica. PLoS Pathog. 2018, 14, e1006882. [Google Scholar] [CrossRef]
- Reuber, T.L.; Ausubel, F.M. Isolation of Arabidopsis Genes That Differentiate between Resistance Responses Mediated by the RPS2 and RPM1 Disease Resistance Genes. Plant Cell 1996, 8, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.R.; Verma, A.K.; Paul, J.; Bhattacharya, A. Phagocytosis of Gut Bacteria by Entamoeba Histolytica. Front. Cell. Infect. Microbiol. 2019, 9, 34. [Google Scholar] [CrossRef]
- Varet, H.; Shaulov, Y.; Sismeiro, O.; Trebicz-Geffen, M.; Legendre, R.; Coppée, J.-Y.; Ankri, S.; Guillen, N. Enteric Bacteria Boost Defences against Oxidative Stress in Entamoeba Histolytica. Sci. Rep. 2018, 8, 9042. [Google Scholar] [CrossRef]
- Bär, A.-K.; Phukan, N.; Pinheiro, J.; Simoes-Barbosa, A. The Interplay of Host Microbiota and Parasitic Protozoans at Mucosal Interfaces: Implications for the Outcomes of Infections and Diseases. PLoS Negl. Trop. Dis. 2015, 9, e0004176. [Google Scholar] [CrossRef] [PubMed]
- Leon-Coria, A.; Kumar, M.; Chadee, K. The Delicate Balance between Entamoeba Histolytica, Mucus and Microbiota. Gut Microbes 2020, 11, 118–125. [Google Scholar] [CrossRef]
- Burgess, S.L.; Petri, W.A. The Intestinal Bacterial Microbiome and E. Histolytica Infection. Curr. Trop. Med. Rep. 2016, 3, 71–74. [Google Scholar] [CrossRef]
- Bracha, R.; Kobiler, D.; Mirelman, D. Attachment and Ingestion of Bacteria by Trophozoites of Entamoeba Histolytica. Infect. Immun. 1982, 36, 396–406. [Google Scholar] [CrossRef]
- Bracha, R.; Mirelman, D. Adherence and Ingestion of Escherichia Coli Serotype 055 by Trophozoites of Entamoeba Histolytica. Infect. Immun. 1983, 40, 882–887. [Google Scholar] [CrossRef] [PubMed]
- Fernández-López, L.A.; Gil-Becerril, K.; Galindo-Gómez, S.; Estrada-García, T.; Ximénez, C.; Leon-Coria, A.; Moreau, F.; Chadee, K.; Tsutsumi, V. Entamoeba Histolytica Interaction with Enteropathogenic Escherichia Coli Increases Parasite Virulence and Inflammation in Amebiasis. Infect. Immun. 2019, 87, e00279-19. [Google Scholar] [CrossRef]
- Lu, L.; Loker, E.S.; Zhang, S.-M.; Buddenborg, S.K.; Bu, L. Genome-Wide Discovery, and Computational and Transcriptional Characterization of an AIG Gene Family in the Freshwater Snail Biomphalaria Glabrata, a Vector for Schistosoma Mansoni. BMC Genomics 2020, 21, 190. [Google Scholar] [CrossRef]
- Zhang, S.-M.; Loker, E.S.; Sullivan, J.T. Pathogen-Associated Molecular Patterns Activate Expression of Genes Involved in Cell Proliferation, Immunity and Detoxification in the Amebocyte-Producing Organ of the Snail Biomphalaria Glabrata. Dev. Comp. Immunol. 2016, 56, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Guillen, N. Signals and Signal Transduction Pathways in Entamoeba Histolytica during the Life Cycle and When Interacting with Bacteria or Human Cells. Mol. Microbiol. 2021, 115, 901–915. [Google Scholar] [CrossRef] [PubMed]
- Pascall, J.C.; Rotondo, S.; Mukadam, A.S.; Oxley, D.; Webster, J.; Walker, S.A.; Piron, J.; Carter, C.; Ktistakis, N.T.; Butcher, G.W. The Immune System GTPase GIMAP6 Interacts with the Atg8 Homologue GABARAPL2 and Is Recruited to Autophagosomes. PLoS ONE 2013, 8, e77782. [Google Scholar] [CrossRef] [PubMed]
- Schwefel, D.; Fröhlich, C.; Eichhorst, J.; Wiesner, B.; Behlke, J.; Aravind, L.; Daumke, O. Structural Basis of Oligomerization in Septin-like GTPase of Immunity-Associated Protein 2 (GIMAP2). Proc. Natl. Acad. Sci. USA 2010, 107, 20299–20304. [Google Scholar] [CrossRef]
- Schwefel, D.; Arasu, B.S.; Marino, S.F.; Lamprecht, B.; Köchert, K.; Rosenbaum, E.; Eichhorst, J.; Wiesner, B.; Behlke, J.; Rocks, O. Structural Insights into the Mechanism of GTPase Activation in the GIMAP Family. Structure 2013, 21, 550–559. [Google Scholar] [CrossRef]
- Schwefel, D.; Daumke, O. GTP-Dependent Scaffold Formation in the GTPase of Immunity-Associated Protein Family. Small GTPases 2011, 2, 20299–20304. [Google Scholar] [CrossRef] [PubMed]
- Meng, Z.; Ren, D.; Zhang, K.; Zhao, J.; Jin, X.; Wu, H. Using ESTIMATE Algorithm to Establish an 8-MRNA Signature Prognosis Prediction System and Identify Immunocyte Infiltration-Related Genes in Pancreatic Adenocarcinoma. Aging (Albany NY) 2020, 12, 5048. [Google Scholar] [CrossRef] [PubMed]
- Hanadate, Y.; Saito-Nakano, Y.; Nakada-Tsukui, K.; Nozaki, T. Endoplasmic Reticulum-resident Rab8A GTPase Is Involved in Phagocytosis in the Protozoan Parasite Entamoeba Histolytica. Cell. Microbiol. 2016, 18, 1358–1373. [Google Scholar] [CrossRef] [PubMed]
- Novick, P.; Zerial, M. The Diversity of Rab Proteins in Vesicle Transport. Curr. Opin. Cell Biol. 1997, 9, 496–504. [Google Scholar] [CrossRef]
- Stenmark, H.; Olkkonen, V.M. The Rab Gtpase Family. Genome Biol. 2001, 2, reviews3007.1. [Google Scholar] [CrossRef] [PubMed]
- Stenmark, H. Rab GTPases as Coordinators of Vesicle Traffic. Nat. Rev. Mol. Cell Biol. 2009, 10, 513–525. [Google Scholar] [CrossRef]
- Saito-Nakano, Y.; Loftus, B.J.; Hall, N.; Nozaki, T. The Diversity of Rab GTPases in Entamoeba Histolytica. Exp. Parasitol. 2005, 110, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Nakada-Tsukui, K.; Saito-Nakano, Y.; Husain, A.; Nozaki, T. Conservation and Function of Rab Small GTPases in Entamoeba: Annotation of E. Invadens Rab and Its Use for the Understanding of Entamoeba Biology. Exp. Parasitol. 2010, 126, 337–347. [Google Scholar] [CrossRef]
- Welter, B.H.; Temesvari, L.A. Overexpression of a Mutant Form of EhRabA, a Unique Rab GTPase of Entamoeba Histolytica, Alters Endoplasmic Reticulum Morphology and Localization of the Gal/GalNAc Adherence Lectin. Eukaryot. Cell 2009, 8, 1014–1026. [Google Scholar] [CrossRef]
- Welter, B.H.; Powell, R.R.; Leo, M.; Smith, C.M.; Temesvari, L.A. A Unique Rab GTPase, EhRabA, Is Involved in Motility and Polarization of Entamoeba Histolytica Cells. Mol. Biochem. Parasitol. 2005, 140, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Saito-Nakano, Y.; Mitra, B.N.; Nakada-tsukui, K.; Sato, D.; Nozaki, T. Two Rab7 Isotypes, Eh Rab7A and Eh Rab7B, Play Distinct Roles in Biogenesis of Lysosomes and Phagosomes in the Enteric Protozoan Parasite Entamoeba Histolytica. Cell. Microbiol. 2007, 9, 1796–1808. [Google Scholar] [CrossRef] [PubMed]
- Nakada-Tsukui, K.; Saito-Nakano, Y.; Ali, V.; Nozaki, T. A Retromerlike Complex Is a Novel Rab7 Effector That Is Involved in the Transport of the Virulence Factor Cysteine Protease in the Enteric Protozoan Parasite Entamoeba Histolytica. Mol. Biol. Cell 2005, 16, 5294–5303. [Google Scholar] [CrossRef] [PubMed]
- Mitra, B.N.; Saito-Nakano, Y.; Nakada-Tsukui, K.; Sato, D.; Nozaki, T. Rab11B Small GTPase Regulates Secretion of Cysteine Proteases in the Enteric Protozoan Parasite Entamoeba Histolytica. Cell. Microbiol. 2007, 9, 2112–2125. [Google Scholar] [CrossRef]
- Paunola, E.; Mattila, P.K.; Lappalainen, P. WH2 Domain: A Small, Versatile Adapter for Actin Monomers. FEBS Lett. 2002, 513, 92–97. [Google Scholar] [CrossRef]
- Cruz, F.; Ferry, J.G. Interaction of Iron–Sulfur Flavoprotein with Oxygen and Hydrogen Peroxide. Biochim. Et Biophys. Acta (BBA)-Gen. Subj. 2006, 1760, 858–864. [Google Scholar] [CrossRef] [PubMed]
- Loftus, B.; Anderson, I.; Davies, R.; Alsmark, U.C.M.; Samuelson, J.; Amedeo, P.; Roncaglia, P.; Berriman, M.; Hirt, R.P.; Mann, B.J. The Genome of the Protist Parasite Entamoeba Histolytica. Nature 2005, 433, 865–868. [Google Scholar] [CrossRef]
- Eichhorn, E.; van der Ploeg, J.R.; Leisinger, T. Characterization of a Two-Component Alkanesulfonate Monooxygenase from Escherichia Coli. J. Biol. Chem. 1999, 274, 26639–26646. [Google Scholar] [CrossRef]
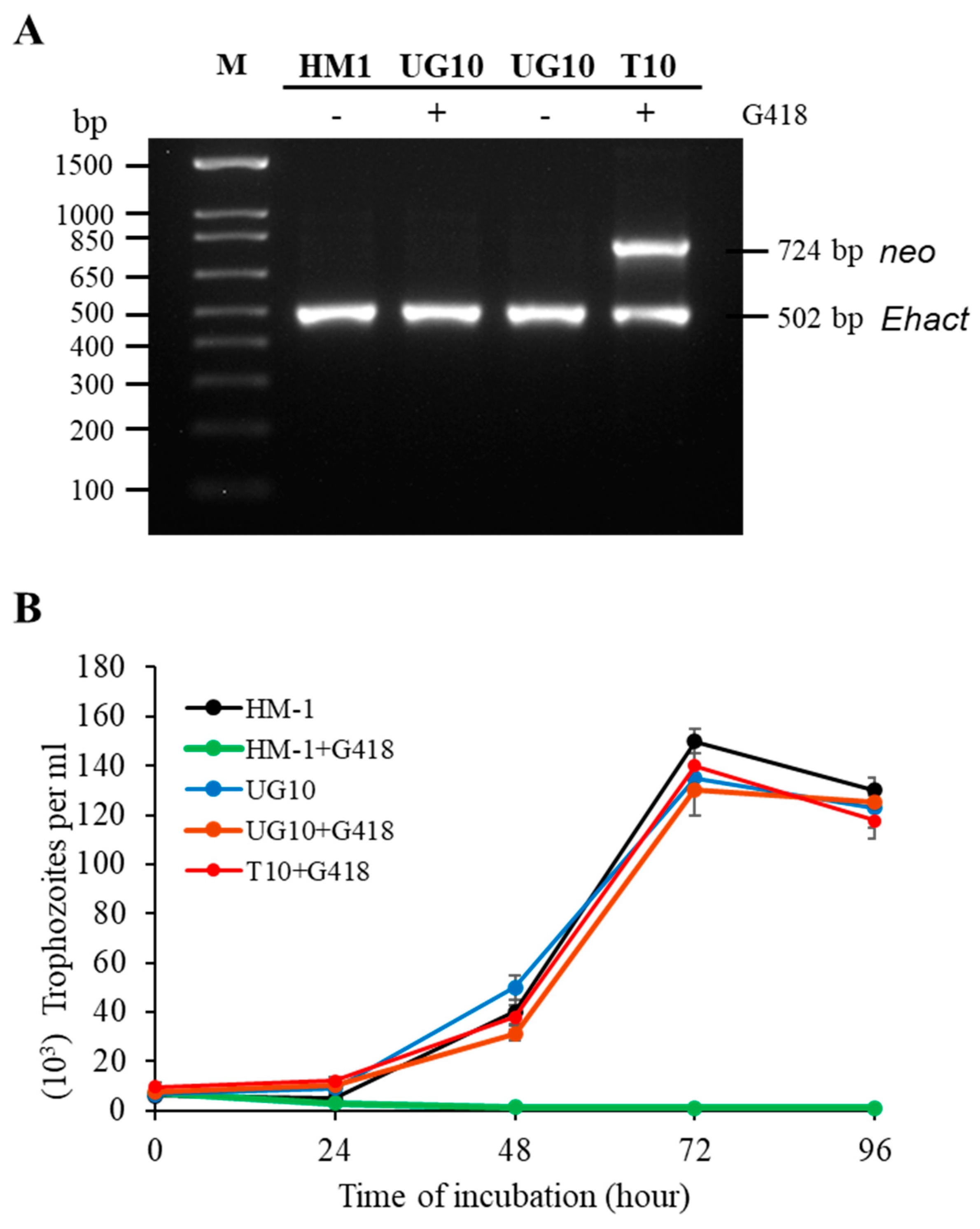

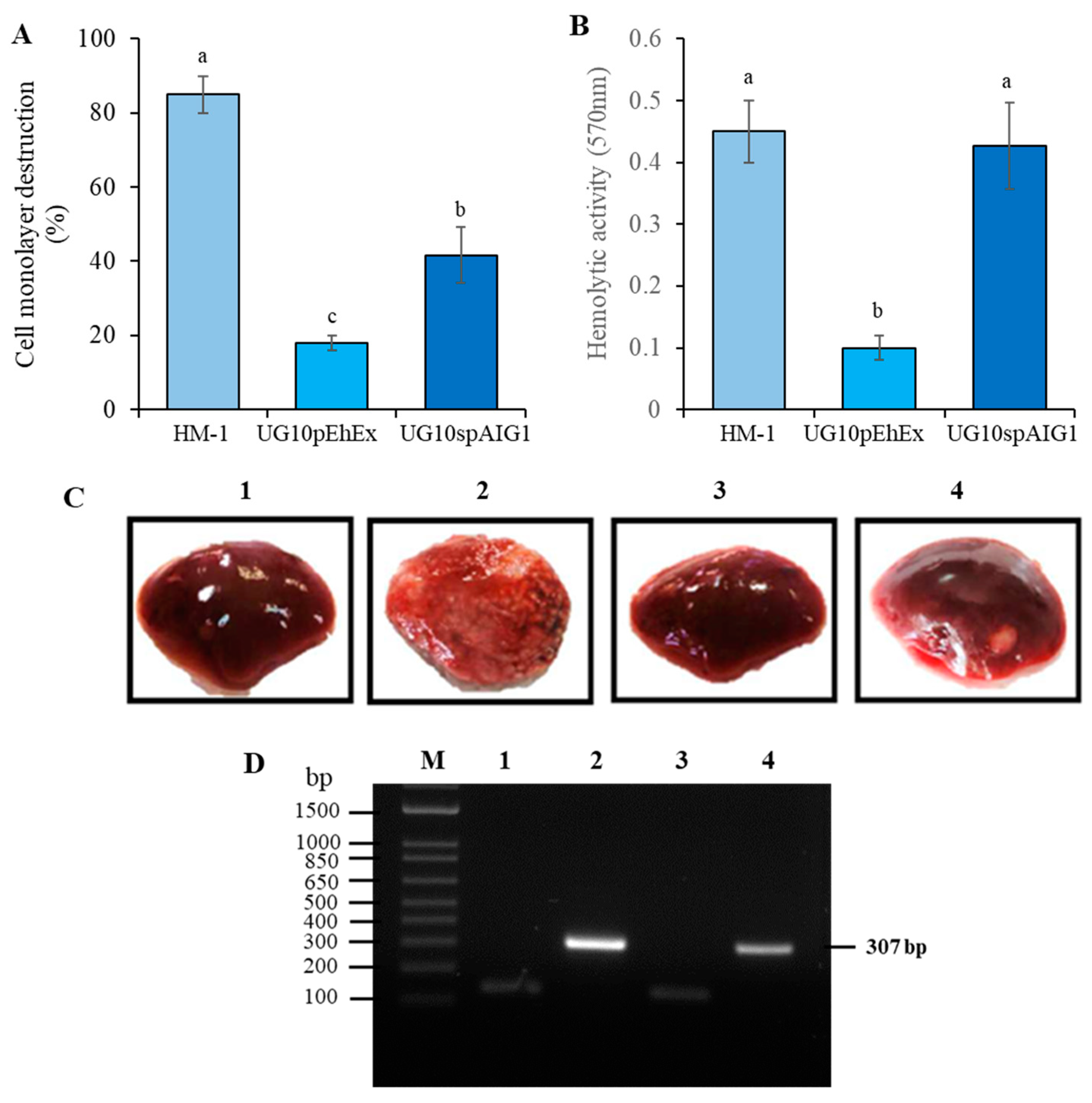
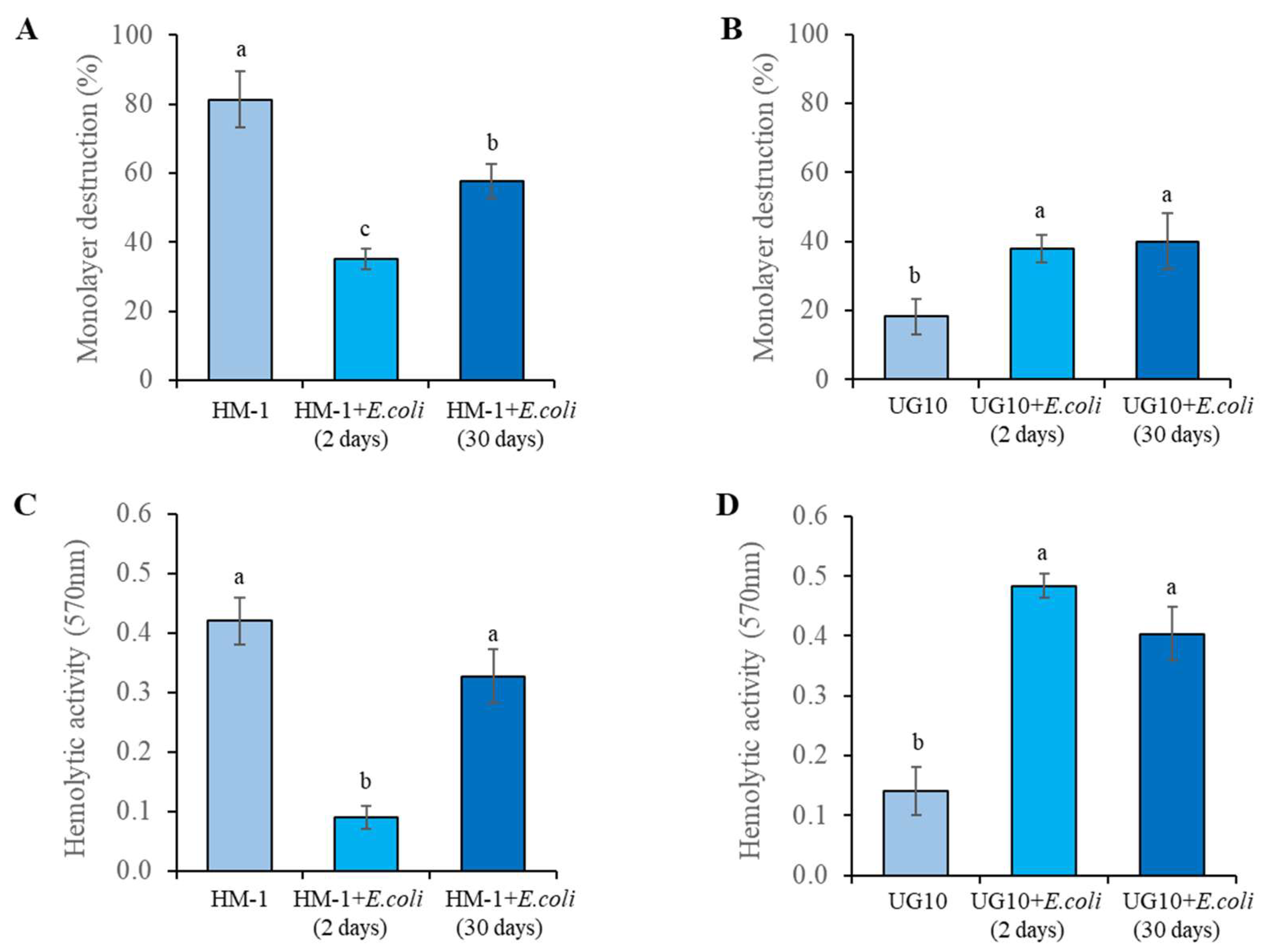

| Biological Activities | HM-1 | G3 | UG10 | Rahman |
|---|---|---|---|---|
| Production of amoebic abscess a | Yes (10/10) | No (0/5) | No (0/10) | No (0/5) |
| Susceptibility to complement b Cytopathic effect c | 47.0 ± 11 87.6 ± 10 | 28.5 ± 7 * 25.0 ± 3 * | 19 ± 6 * 15 ± 4 * | 23 ± 12 * 15 ± 4 * |
| Cytotoxic effect d | 42.0 ± 10 | 32.0 ± 5 * | 7 ± 1 * | 12 ± 2 * |
| Hemolytic activity e | 0.4 ± 0.06 | 0.25 ± 0.02 * | 0.08 ± 0.02 * | 0.10 ± 0.02 * |
| Erythrophagocytosis f 5 min | 5.4 ± 1.2 | 5.7 ± 1.1 | 4.4 ± 0.4 | 2.1 ± 0.3 * |
| 15 min | 7.5 ± 1.1 | 8.0 ± 1.0 | 6.1 ± 0.5 | 2.9 ± 4.5 * |
| 30 min | 10.3 ± 1.0 | 9.6 ± 0.5 | 10.4 ± 1.5 | 4.5 ± 0.5 * |
| Digestion of human erythrocytes g 0 h | 9.00 ± 0.6 | 9.4 ± 0.8 | 8.8 ± 0.5 | 5.4 ± 0.84 * |
| 2 h | 5.00 ± 0.4 | 6.8 ± 0.4 | 6.2 ± 0.3 | 3.2 ± 0.59 * |
| 4 h | 1.32 ± 0.5 | 2.7 ± 0.6 | 2.2 ± 0.3 | 0.6 ± 0.20 * |
| 6 h | 0.45 ± 0.3 | 1.0 ± 0.5 | 0.8 ± 0.2 | 0.3 ± 0.05 |
| Cysteine protease activity (nmol/min/mg) h | 328 ± 46 | 589 ± 75 * | 386 ± 21 | 306 ± 16 |
| Gene Name | Accession Number | Fold Change | Function |
|---|---|---|---|
| > EhRab8B | EHI_127380 | 50.4 | Intracellular trafficking, secretion, and vesicular transport. |
| EhRabF1 | EHI_129740 | 32.2 | Intracellular trafficking and signal transduction mechanisms. |
| AIG1 family protein, putative | EHI_180390 | 31.2 | Bacterial defense mechanisms and stress response. |
| EhaRabX13 | EHI_065790 | 20.9 | Intracellular trafficking and signal transduction mechanisms. |
| EhRab8A | EHI_199820 | 20.4 | Intracellular trafficking, secretion, and vesicular transport. |
| Ras family protein | EHI_118280 | 11.9 | Intracellular trafficking and signal transduction mechanisms. |
| AIG1 family protein putative | EHI_109120 | 11.6 | Bacterial defense mechanisms and stress response. |
| AIG1 family protein | EHI_022500 | 11.5 | Bacterial defense mechanisms and stress response. |
| Lipase, putative | EHI_072130 | 11.0 | Secondary metabolites triglyceride biosynthesis, transport, and catabolism. |
| Ribosomal protein S30, putative | EHI_088600 | 9.8 | Translation, ribosomal structure, and biogenesis. |
| AIG1 family protein | EHI_115160 | 6.4 | Bacterial defense mechanisms and stress response. |
| Hydrolase, carbon-nitrogen family | EHI_035680 | 5.9 | Nitrogen compound metabolic process. |
| Protein kinase putative | EHI_023610 | 4.1 | Protein phosphorylation, inorganic ion transport, energy production, and conversion. |
| Ran GTPase-activation protein, putative | EHI_185290 | 3.0 | Regulates nuclear transport and the union and exchange of GTPs. |
| EhRabX23 | EHI_107140 | 2.9 | Intracellular trafficking and signal transduction mechanisms. |
| Glucosamine-6-phosphate isomerase, putative | EHI_174640 | 2.9 | Carbohydrate transport and metabolism. |
| EhRabC6 | EHI_194280 | 2.8 | Intracellular trafficking, secretion, and vesicular transport. |
| Protein kinase putative | EHI_185230 | 2.7 | Protein phosphorylation, inorganic ion transport, energy production, and conversion. |
| Choline/Ethanolamine kinase, putative | EHI_148580 | 2.5 | Phospholipid metabolism, and catalyzes phosphatidylethanolamine and phosphatidylcholine biosynthesis. |
| EhRabD2 | EHI_164900 | 2.4 | Intracellular trafficking, secretion, and vesicular transport. |
| SKIP/SNW domain protein | EHI_199600 | 2.2 | Bifunctional regulator that works in the nucleus as a splicing factor by integrating into the spliceosome. Transcriptional activator by interacting with the Paf1 complex. |
| Metal-dependent hydrolase, putative | EHI_054690 | 2.2 | Hydrolase activity. |
| WD domain-containing protein | EHI_170080 | 2.1 | Assembly of protein complexes or mediators of transient interplay among other proteins. |
| tRNA (guanine-N(1)-)-methyltransferase | EHI_168400 | 2.1 | Biosynthesis, signal transduction, protein repair, chromatin regulation, and gene silencing. |
| Ubiquitin ligase, putative | EHI_104570 | 2.0 | Ubiquitin mediated proteolysis. |
| Gene Name | Accession Number | Fold Change | Function |
|---|---|---|---|
| WH2 motif domain-containing protein | EHI_050810 | 11.2 | Cell contractility, cell motility, cell trafficking, and cell signaling. |
| EhRab7G | EHI_187090 | 11.1 | Intracellular trafficking, secretion, and vesicular transport. |
| Heat shock protein 70, putative | EHI_123490 | 8.3 | Post-translational modification, protein turnover, chaperones. |
| Iron-sulfur flavoprotein, putative | EHI_022600 | 8.2 | Electron transport and response to oxidative stress. |
| Iron-sulfur flavoprotein, putative | EHI_ 181710 | 6.2 | Electron transport and response to oxidative stress. |
| Iron-sulfur flavoprotein, putative | EHI_067720 | 5.7 | Electron transport and response to oxidative stress. |
| Iron-sulfur flavoprotein, putative | EHI_103260 | 5.4 | Electron transport and response to oxidative stress. |
| Methionine gamma-lyase, putative | EHI_057550 | 5.0 | Amino acid transport and metabolism. |
| SURFACE antigen ariel1-RELATED | EHI_172850 | 4.4 | Surface protein important in adhesion and phagocytosis. |
| Myb-like DNA-binding domain containing protein | EHI_166410 | 4.2 | Encodes for nuclear DNA-binding proteins. |
| U2 snRNP auxiliary factor, putative | EHI_192500 | 3.9 | Interaction with the 3’ splice site dinucleotide AG is essential for regulated splicing. |
| Surface antigen ariel1, putative | EHI_080200 | 3.5 | Surface protein important in adhesion and phagocytosis. |
| Surface antigen ariel1, putative | EHI_101730 | 2.3 | Surface protein important in adhesion and phagocytosis. |
| Vacuolar protein sorting 35, putative | EHI_086580 | 2.1 | Intracellular protein transport, lysosome organization, retrograde transport, endosome to Golgi. |
| Expression Levels (X-Fold) by qRT-PCR | |||||
|---|---|---|---|---|---|
| Protein Name | Gene Accession | HM-1 a | UG10 | G3 | Rahman |
| AIG, putative | EHI_180390 | 1 | −12.7 ± 1.2 (−31.2 b) | −3.4 ± 0.2 | ND |
| EhRab8A | EHI_199820 | 1 | −26.3 ± 1.9 (−20.4 b) | 1 | 1 |
| WH2 motif domain-containing protein | EHI_050810 | 1 | 9.6 ± 0.7 (11.2 b) | 8.3 ± 0.9 | 10.5 ± 0.9 |
| Heat shock protein 70, putative | EHI_123490 | 1 | 12.1 ± 0.9 (8.3 b) | 8.6 ± 0.5 | 1.55 ± 0.1 |
| Strain | ||
|---|---|---|
| Condition | HM-1 | UG10 |
| Axenic | 12.7 ± 1.2 | 1.0 |
| E. coli O55 (2 days) | 6.86 ± 1.1 | 2.35 ± 0.2 |
| E. coli O55 (30 days) | 12.33 ± 1.7 | 4.13 ± 0.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lozano-Mendoza, J.; Ramírez-Montiel, F.; Rangel-Serrano, Á.; Páramo-Pérez, I.; Mendoza-Macías, C.L.; Saavedra-Salazar, F.; Franco, B.; Vargas-Maya, N.; Jeelani, G.; Saito-Nakano, Y.; et al. Attenuation of In Vitro and In Vivo Virulence Is Associated with Repression of Gene Expression of AIG1 Gene in Entamoeba histolytica. Pathogens 2023, 12, 489. https://doi.org/10.3390/pathogens12030489
Lozano-Mendoza J, Ramírez-Montiel F, Rangel-Serrano Á, Páramo-Pérez I, Mendoza-Macías CL, Saavedra-Salazar F, Franco B, Vargas-Maya N, Jeelani G, Saito-Nakano Y, et al. Attenuation of In Vitro and In Vivo Virulence Is Associated with Repression of Gene Expression of AIG1 Gene in Entamoeba histolytica. Pathogens. 2023; 12(3):489. https://doi.org/10.3390/pathogens12030489
Chicago/Turabian StyleLozano-Mendoza, Janeth, Fátima Ramírez-Montiel, Ángeles Rangel-Serrano, Itzel Páramo-Pérez, Claudia Leticia Mendoza-Macías, Faridi Saavedra-Salazar, Bernardo Franco, Naurú Vargas-Maya, Ghulam Jeelani, Yumiko Saito-Nakano, and et al. 2023. "Attenuation of In Vitro and In Vivo Virulence Is Associated with Repression of Gene Expression of AIG1 Gene in Entamoeba histolytica" Pathogens 12, no. 3: 489. https://doi.org/10.3390/pathogens12030489
APA StyleLozano-Mendoza, J., Ramírez-Montiel, F., Rangel-Serrano, Á., Páramo-Pérez, I., Mendoza-Macías, C. L., Saavedra-Salazar, F., Franco, B., Vargas-Maya, N., Jeelani, G., Saito-Nakano, Y., Anaya-Velázquez, F., Nozaki, T., & Padilla-Vaca, F. (2023). Attenuation of In Vitro and In Vivo Virulence Is Associated with Repression of Gene Expression of AIG1 Gene in Entamoeba histolytica. Pathogens, 12(3), 489. https://doi.org/10.3390/pathogens12030489






