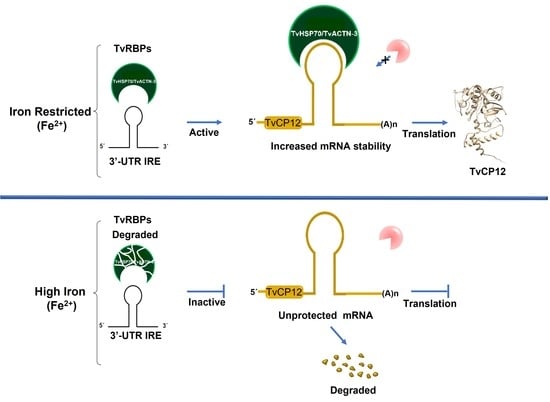
The Non-Canonical Iron-Responsive Element of IRE-tvcp12 Hairpin Structure at the 3′-UTR of Trichomonas vaginalis TvCP12 mRNA That Binds TvHSP70 and TvACTN-3 Can Regulate mRNA Stability and Amount of Protein
Abstract
1. Introduction
2. Materials and Methods
2.1. Parasites and Culture Conditions
2.2. RT-PCR Assays
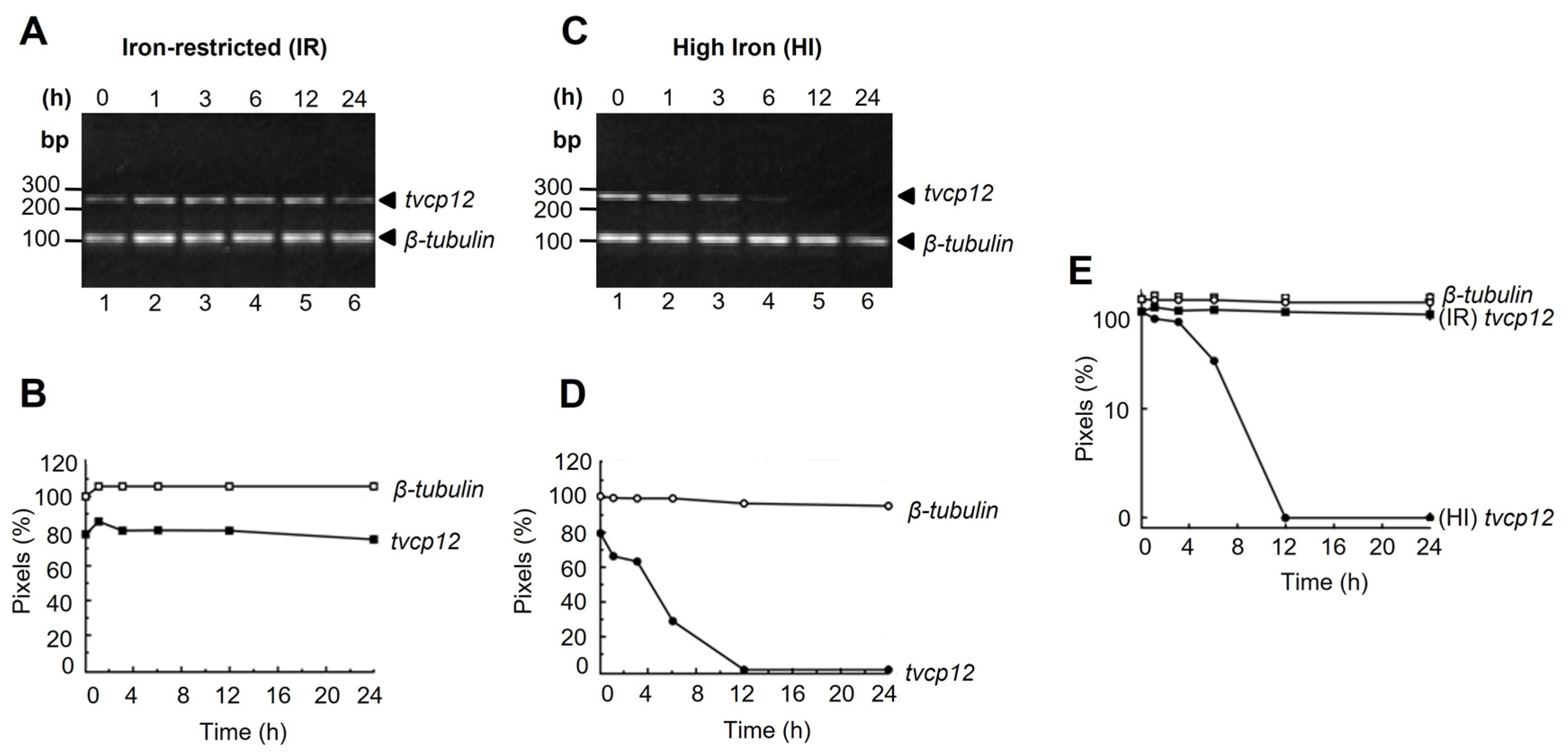
2.3. Tvcp12 mRNA Stability Assays
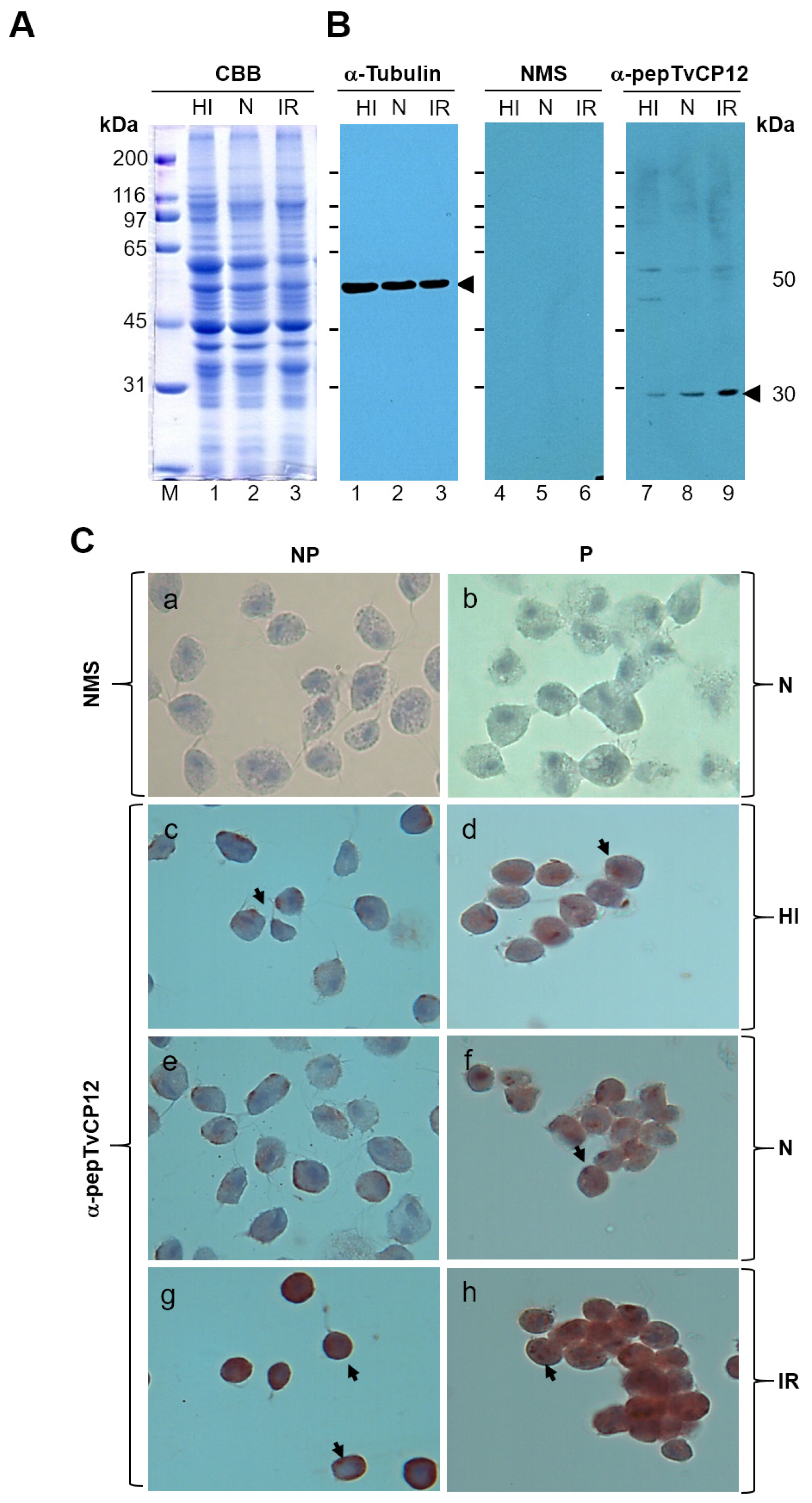
2.4. Western Blotting Assays
2.5. Immunocytochemical Assay
2.6. Purification of Recombinant Proteins
2.7. Cytoplasmic Extracts from HeLa Cells and Trichomonad Parasites
2.8. In Vitro Transcription of IRE-like Sequences
2.9. RNA Electrophoretic Mobility Shift Assay (REMSA)
2.10. UV Cross-Linking Assays
2.11. Northwestern Blotting (NWB) Assay
3. Results
3.1. The tvcp12 mRNA Stability Is Higher in Iron-Restricted Trichomonads
3.2. The tvcp12 Gene Expression Is Also Upregulated by IR Conditions at the Protein Level
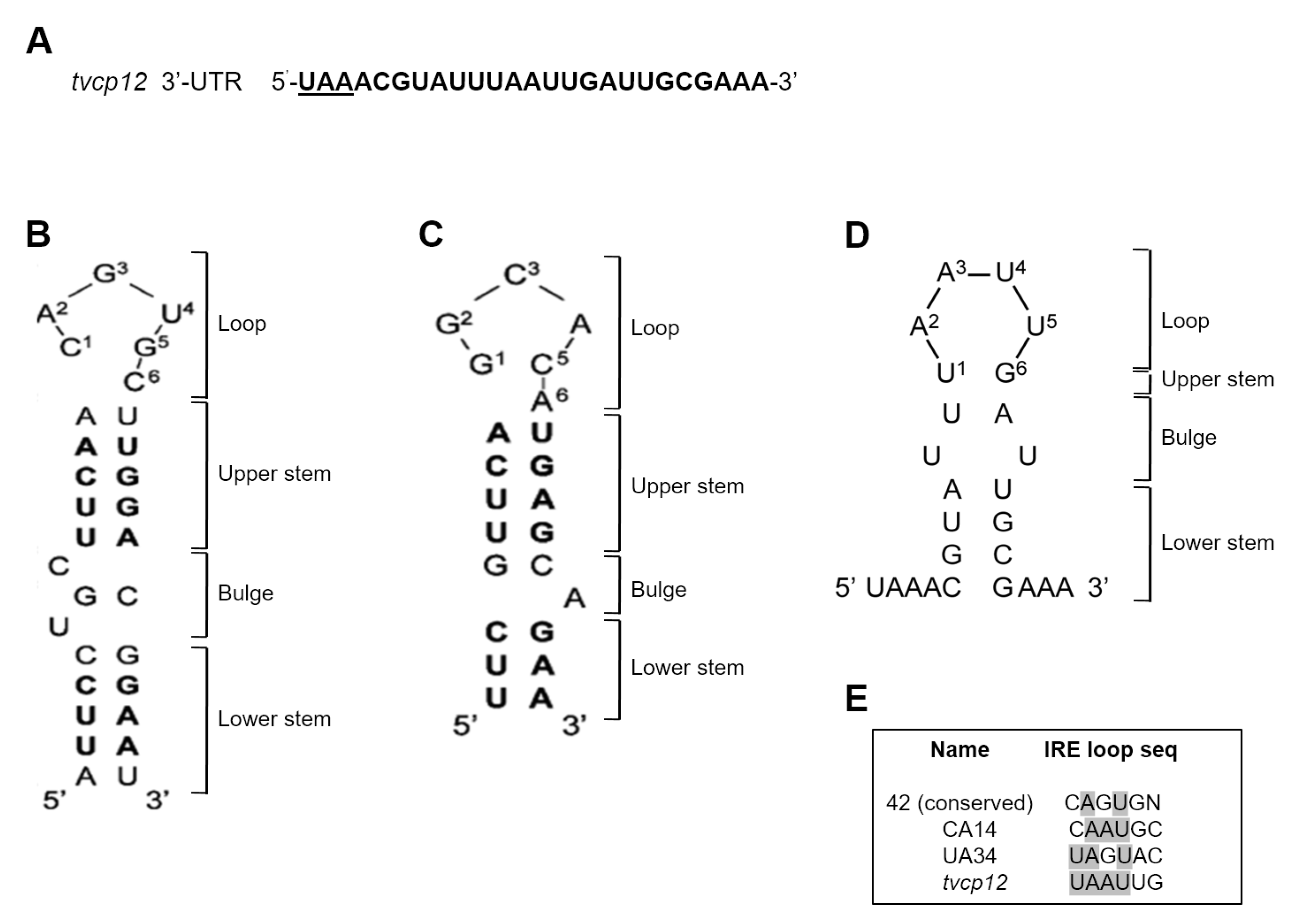
3.3. An IRE-like Structure Was Found at the 3′-UTR of the Tvcp12 mRNA
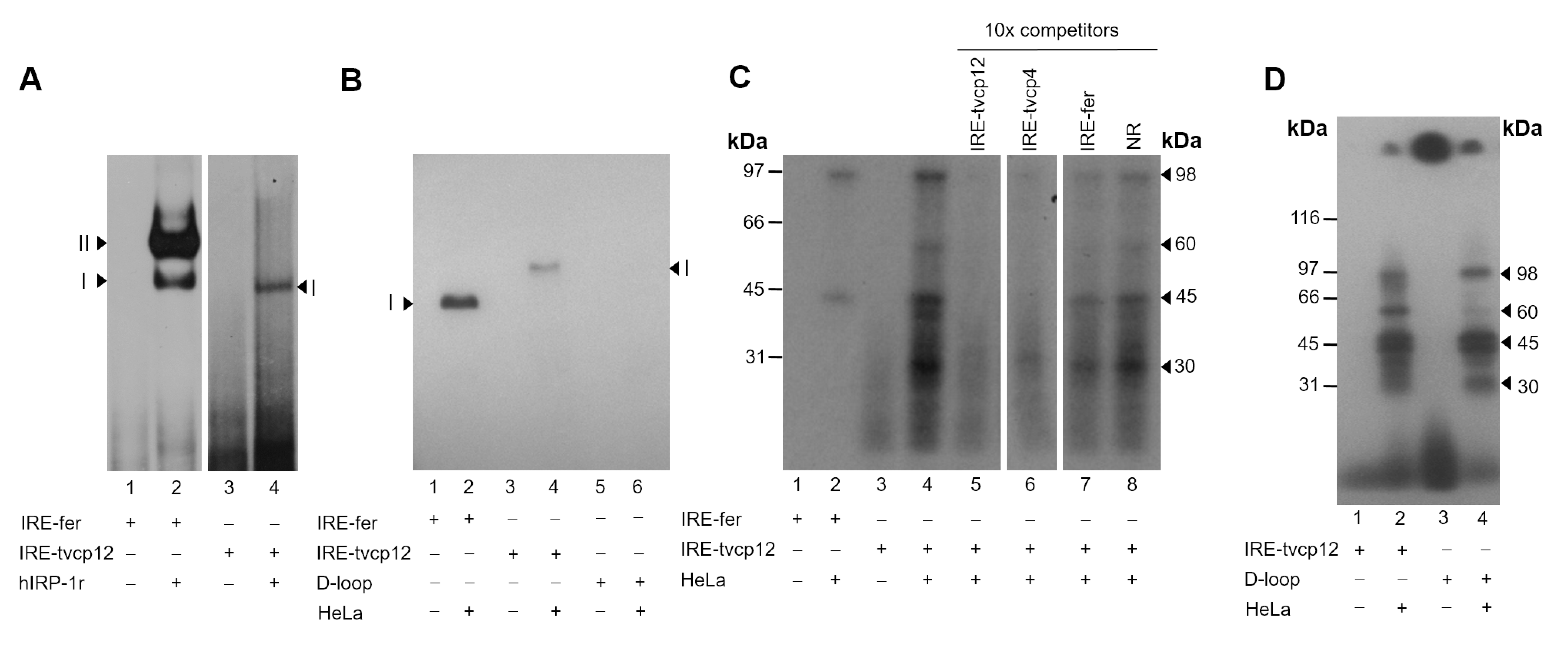
3.4. The IRE-tvcp12 Binds to Human IRPs
3.5. The IRE-tvcp12 Hairpin Specifically Interacts with IRP Proteins from HeLa Cytoplasmic Extracts
3.6. The Loop Sequence of the IRE tvcp12 Is Necessary for Binding to HeLa IRP Proteins
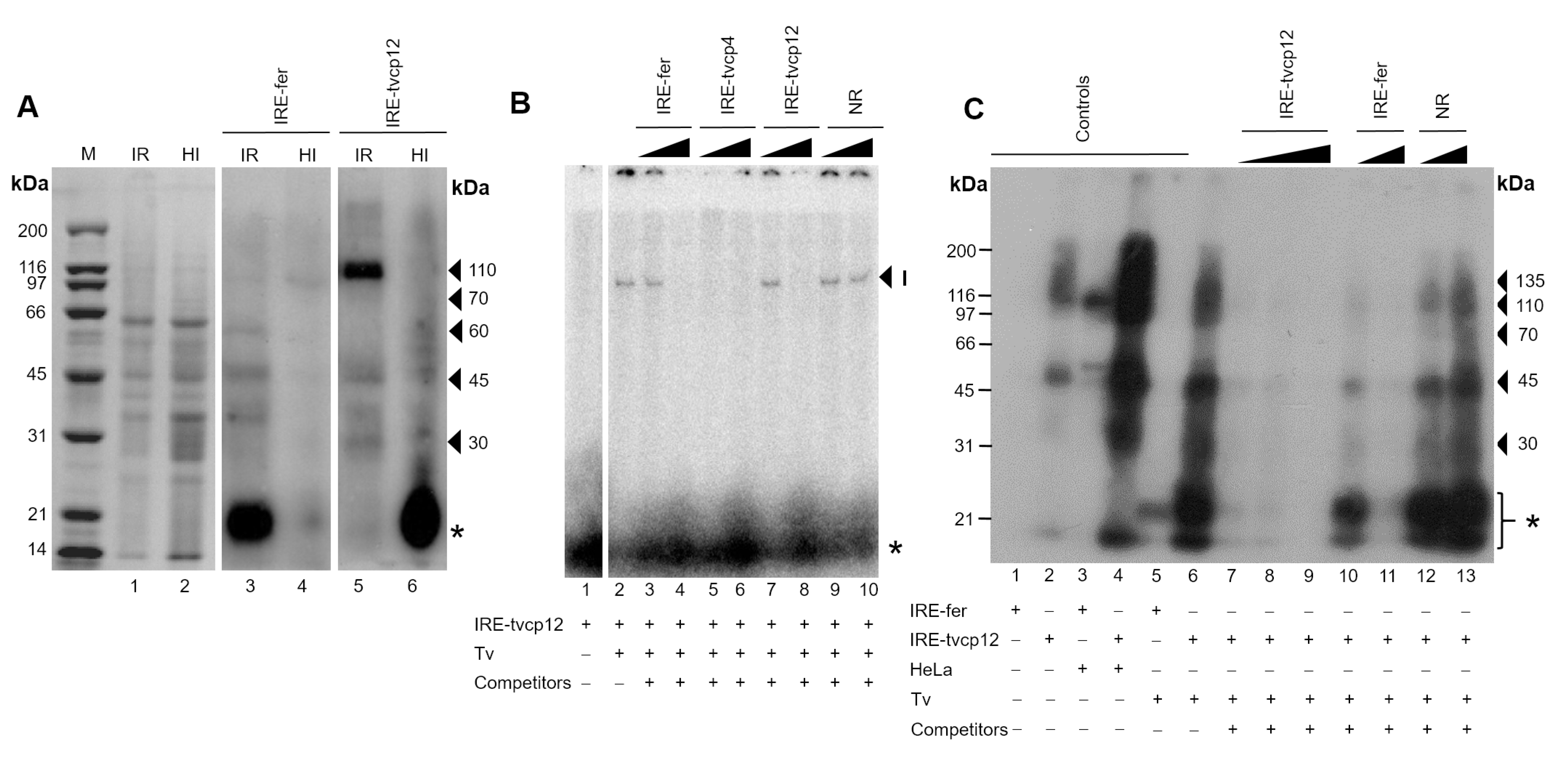
3.7. Effect of Iron on RNA-Protein Complex Formation between the IRE-tvcp12 Hairpin and Trichomonad Cytoplasmic Proteins
3.8. The Non-Canonical IRE-tvcp12 Hairpin Specifically Interacts with Atypical Cytoplasmic RNA-Binding Proteins from Trichomonad Cytoplasmic Extracts
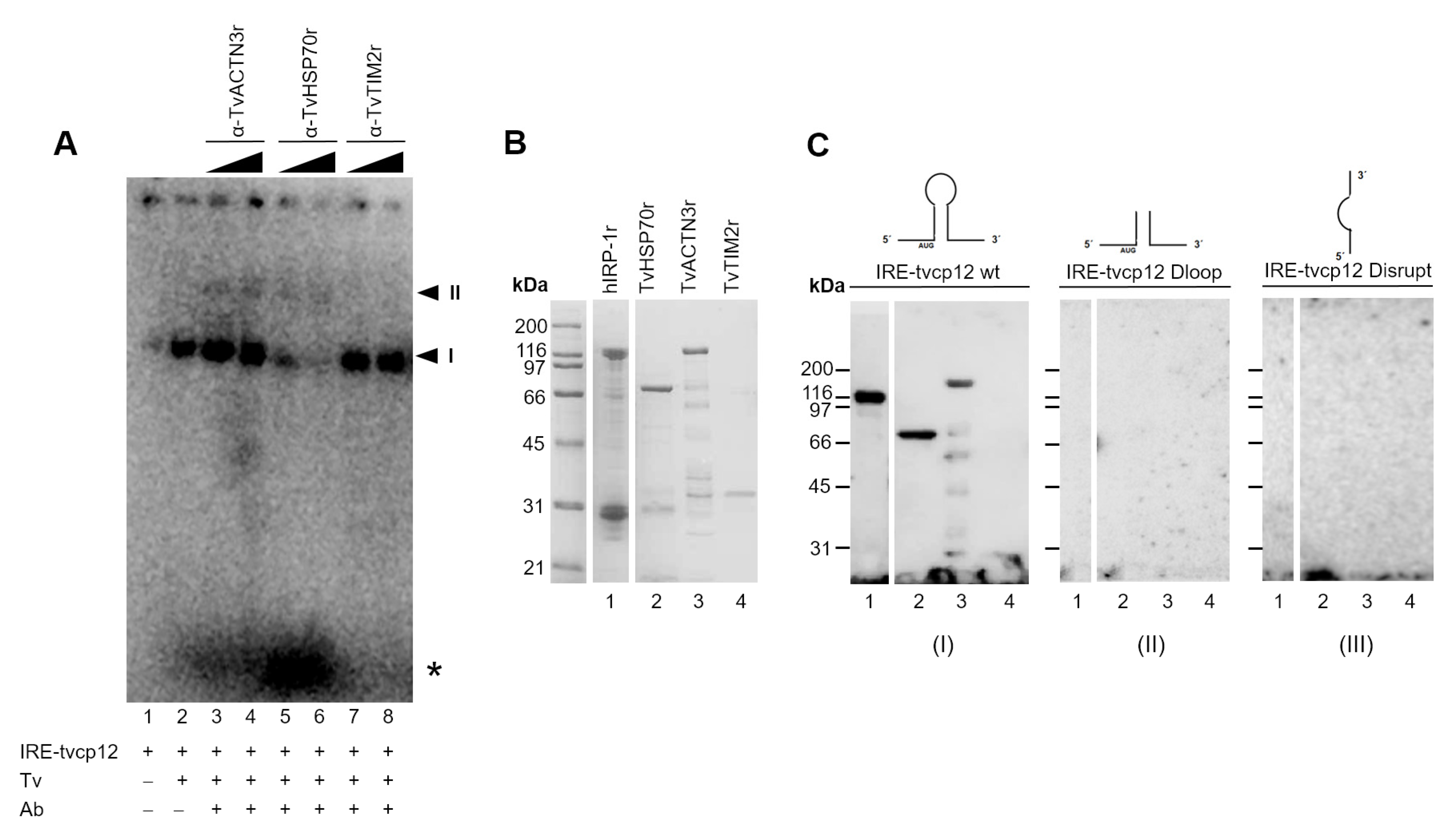
3.9. The Polyclonal Anti-TvACTN3r and Anti-TvHSP70r Antibodies Supershifted RNA-Protein Complex I
3.10. The IRE-tvcp12 Binds to TvACTN3 and TvHSP70, Two Atypical RNA-Binding Proteins in Trichomonads
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kühn, L.C. How iron controls iron. Cell. Metab. 2009, 10, 439–441. [Google Scholar] [CrossRef] [PubMed]
- Volz, K. The functional duality of iron regulatory protein 1. Curr. Opin. Struct. Biol. 2008, 18, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Hentze, M.W.; Muckenthaler, M.U.; Andrews, N.C. Balancing acts: Molecular control of mammalian iron metabolism. Cell 2004, 117, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Pantopoulos, K. Iron metabolism and the IRE/IRP regulatory system: An update. Ann. N. Y. Acad. Sci. 2004, 1012, 1–13. [Google Scholar] [CrossRef]
- Rouault, T.A. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat. Chem. Biol. 2006, 2, 406–414. [Google Scholar] [CrossRef]
- Testa, U. Recent developments in the understanding of iron metabolism. Hematol. J. 2002, 3, 63–89. [Google Scholar] [CrossRef]
- Henderson, B.R.; Kühn, L.C. Differential modulation of the RNA-binding proteins IRP-1 and IRP-2 in response to iron. IRP-2 inactivation requires translation of another protein. J. Biol. Chem. 1995, 270, 20509–20515. [Google Scholar] [CrossRef]
- Salahudeen, A.A.; Thompson, J.W.; Ruiz, J.C.; Ma, H.W.; Kinch, L.N.; Li, Q.M.; Grishin, N.V.; Bruick, R.K. An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science 2009, 326, 722–726. [Google Scholar] [CrossRef]
- Vashisht, A.A.; Zumbrennen, K.B.; Huang, X.; Powers, D.N.; Durazo, A.; Sun, D.; Bhaskaran, N.; Persson, A.; Uhlen, M.; Sangfelt, O.; et al. Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 2009, 326, 718–721. [Google Scholar] [CrossRef]
- World Health Organization. Global Sector Strategy of Health against Infections Sexual Transmission 2016–2021; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Carlton, J.M.; Hirt, R.P.; Silva, J.C.; Delcher, A.L.; Schatz, M.; Zhao, Q.; Wortman, J.R.; Bidwell, S.L.; Alsmark, U.C.M.; Besteiro, S.; et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 2007, 315, 207–212. [Google Scholar] [CrossRef]
- Arroyo, R.; Cárdenas-Guerra, R.E.; Figueroa-Angulo, E.E.; Puente-Rivera, J.; Zamudio-Prieto, O.; Ortega-López, J. Trichomonas vaginalis cysteine proteinases: Iron response in gene expression and proteolytic activity. BioMed Res. Int. Spec. Issue Iron Parasites 2015, 2015, 946787. [Google Scholar]
- Alvarez-Sánchez, M.E.; Avila-González, L.; Becerril-García, C.; Fattel-Facenda, L.; Arroyo, R. A novel cysteine proteinase (CP65) of Trichomonas vaginalis involved in cytotoxicity. Microb. Pathog. 2000, 28, 193–202. [Google Scholar] [CrossRef]
- Arroyo, R.; Alderete, J.F. Trichomonas vaginalis surface proteinase activity is necessary for parasite adherence to epithelial cells. Infect. Immun. 1989, 57, 2991–2997. [Google Scholar] [CrossRef]
- Arroyo, R.; Alderete, J.F. Two Trichomonas vaginalis surface proteinases bind to host epithelial cells and are related to levels of cytoadherence and cytotoxicity. Arch. Med. Res. 1995, 26, 279–285. [Google Scholar]
- Cárdenas-Guerra, R.E.; Arroyo, R.; de Andrade, I.R.; Benchimol, M.; Ortega-López, J. Iron-induced cysteine proteinase TvCP4 plays a key role in Trichomonas vaginalis haemolysis. Microbes Infect. 2013, 15, 959–968. [Google Scholar] [CrossRef]
- Hernández-Gutiérrez, R.; Ortega-López, J.; Arroyo, R. A 39-kDa cysteine proteinase CP39 from Trichomonas vaginalis, which is negatively affected by iron, may be involved in trichomonal cytotoxicity. J. Eukaryot. Microbiol. 2003, 50, 696–698. [Google Scholar] [CrossRef]
- Hernández-Gutiérrez, R.; Ávila-González, L.; Ortega-López, J.; Cruz-Talonia, F.; Gómez-Gutiérrez, G.; Arroyo, R. Trichomonas vaginalis: Characterization of a 39-kDa cysteine proteinase found in patient vaginal secretions. Exp. Parasitol. 2004, 107, 125–135. [Google Scholar] [CrossRef]
- Figueroa-Angulo, E.E.; Rendón-Gandarilla, F.J.; Puente-Rivera, J.; Calla-Choque, J.S.; Cárdenas-Guerra, R.E.; Ortega-López, J.; Quintas-Granados, L.I.; Alvarez-Sánchez, M.E.; Arroyo, R. The effects of environmental factors on the virulence of Trichomonas vaginalis. Microbes Infect. 2012, 14, 1411–1427. [Google Scholar] [CrossRef]
- Sommer, U.; Costello, C.E.; Hayes, G.R.; Beach, D.H.; Gilbert, R.O.; Lucas, J.J.; Singh, B.N. Identification of Trichomonas vaginalis cysteine proteases that induce apoptosis in human vaginal epithelial cells. J. Biol. Chem. 2005, 280, 23853–23860. [Google Scholar] [CrossRef]
- Mendoza-López, M.R.; Becerril-García, C.; Fattel-Facenda, L.V.; Avila-González, L.; Ruíz-Tachiquín, M.E.; Ortega-López, J.; Arroyo, R. CP30, a cysteine proteinase involved in Trichomonas vaginalis cytoadherence. Infect. Immun. 2000, 68, 4907–4912. [Google Scholar] [CrossRef]
- Provenzano, D.; Alderete, J.F. Analysis of human immunoglobulin-degrading cysteine proteinases of Trichomonas vaginalis. Infect. Immun. 1995, 63, 3388–3395. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Rivas, L.A.; Lorenzo-Benito, S.; Sánchez-Rodríguez, D.B.; Miranda-Ozuna, J.F.T.; Euceda-Padilla, E.A.; Ortega-López, J.; Chávez-Munguía, B.; Lagunes-Guillén, A.; Velazquez-Valassi, B.; Jasso-Villazul, L.; et al. The effect of iron on Trichomonas vaginalis TvCP2, a cysteine proteinase found in vaginal secretions of trichomoniasis patients. Parasitology 2020, 147, 760–774. [Google Scholar] [CrossRef] [PubMed]
- Hernández, H.M.; Marcet, R.; Sarracent, J. Biological roles of cysteine proteinases in the pathogenesis of Trichomonas vaginalis. Parasite 2014, 21, 54. [Google Scholar] [CrossRef] [PubMed]
- Klemba, M.; Goldberg, D.E. Biological roles of proteases in parasitic protozoa. Annu. Rev. Biochem. 2002, 71, 275–305. [Google Scholar] [CrossRef]
- Alvarez-Sánchez, M.E.; Solano-González, E.; Yánez-Gómez, C.; Arroyo, R. Negative iron regulation of the CP65 cysteine proteinase cytotoxicity in Trichomonas vaginalis. Microbes Infect. 2007, 9, 1597–1605. [Google Scholar] [CrossRef]
- Kummer, S.; Hayes, G.R.; Gilbert, R.O.; Beach, D.H.; Lucas, J.J.; Singh, B.N. Induction of human host cell apoptosis by Trichomonas vaginalis cysteine proteinases is modulated by parasite exposure to iron. Microb. Pathog. 2008, 44, 197–203. [Google Scholar] [CrossRef]
- León-Sicairos, C.R.; León-Felix, J.; Arroyo, R. Tvcp12: A novel Trichomonas vaginalis cathepsin L-like cysteine proteinase-encoding gene. Microbiology 2004, 150, 1131–1138. [Google Scholar] [CrossRef]
- Solano-González, E.; Burrola-Barraza, V.; León-Sicairos, C.R.; Ávila-González, L.; Gutiérrez-Escolano, L.; Ortega-López, J.; Arroyo, R. The trichomonad cysteine proteinase TVCP4 transcript contains an iron-responsive element. FEBS Lett. 2007, 581, 2919–2928. [Google Scholar] [CrossRef]
- Gorrell, T.E. Effect of culture medium iron content on the biochemical composition and metabolism of Trichomonas vaginalis. J. Bacteriol. 1985, 161, 1228–1230. [Google Scholar] [CrossRef]
- Lehker, M.; Arroyo, R.; Alderete, J.F. The regulation by iron of the synthesis of adhesins and cytoadherence levels in the protozoan Trichomonas vaginalis. J. Exp. Med. 1991, 174, 311–318. [Google Scholar] [CrossRef]
- Wilson, M.E.; Britigan, B.E. Iron acquisition by parasitic protozoa. Parasitol. Today 1998, 14, 348–353. [Google Scholar] [CrossRef]
- Tsai, C.D.; Liu, H.W.; Tai, J.H. Characterization of an iron-responsive promoter in the protozoan pathogen Trichomonas vaginalis. J. Biol. Chem. 2002, 277, 5153–5162. [Google Scholar] [CrossRef]
- Ong, S.-J.; Hsu, H.-M.; Liu, H.-W.; Chu, C.-H.; Tai, J.-H. Multifarious transcriptional regulation of adhesion protein gene ap65-1 by a novel Myb1 protein in the protozoan parasite Trichomonas vaginalis. Eukaryot. Cell. 2006, 5, 391–399. [Google Scholar] [CrossRef]
- Ong, S.-J.; Hsu, H.-M.; Liu, H.-W.; Chu, C.-H.; Tai, J.-H. Activation of multifarious transcription of an adhesion protein ap65-1 gene by a novel Myb2 protein in the protozoan parasite Trichomonas vaginalis. J. Biol. Chem. 2007, 282, 6716–6725. [Google Scholar] [CrossRef]
- Hsu, H.M.; Ong, S.J.; Lee, M.C.; Tai, J.H. Transcriptional regulation of an iron-inducible gene by differential and alternate promoter entries of multiple Myb proteins in the protozoan parasite Trichomonas vaginalis. Eukaryot. Cell. 2009, 8, 362–372. [Google Scholar] [CrossRef]
- Hsu, H.M.; Lee, Y.; Indra, D.; Wei, S.Y.; Liu, H.W.; Chang, L.C.; Chen, C.; Ong, S.J.; Tai, J.H. Iron-inducible nuclear translocation of a Myb3 transcription factor in the protozoan parasite Trichomonas vaginalis. Eukaryot. Cell. 2012, 11, 1441–1450. [Google Scholar] [CrossRef]
- Smith, A.J.; Chudnovsky, L.; Simoes-Barbosa, A.; Delgadillo-Correa, M.G.; Jonsson, Z.O.; Wohlschlegel, J.A.; Johnson, P.J. Novel core promoter elements and a cognate transcription factor in the divergent unicellular eukaryote Trichomonas vaginalis. Mol. Cell. Biol. 2011, 31, 1444–1458. [Google Scholar] [CrossRef]
- Calla-Choque, J.S.; Figueroa-Angulo, E.E.; Avila-González, L.; Arroyo, R. α-Actinin TvACTN3 of Trichomonas vaginalis is an RNA-binding protein that may participate in its post-transcriptional iron regulatory mechanism. BioMed Res. Int. 2014, 2014, 424767. [Google Scholar] [CrossRef]
- Figueroa-Angulo, E.E.; Calla-Choque, J.S.; Mancilla-Olea, M.I.; Arroyo, R. RNA-binding proteins in Trichomonas vaginalis: Atypical multifunctional proteins. Biomolecules special issue in “RNA-Binding Proteins—Structure, Function, Networks and Disease”. Biomolecules 2015, 5, 3354–3395. [Google Scholar] [CrossRef]
- Diamond, L.S. The establishment of various trichomonads of animals and man in axenic cultures. J. Parasitol. 1957, 43, 488–490. [Google Scholar] [CrossRef]
- Madico, G.; Quinn, T.; Rompalo, A.; Mckee, K.T.; Gaydos, C.A. Diagnosis of Trichomonas vaginalis infection by PCR using vaginal swab samples. J. Clin. Microbiol. 1998, 36, 3205–3210. [Google Scholar] [CrossRef] [PubMed]
- Higgins, C.F.; Causton, H.C.; Dance, G.S.G.; Mudd, E.A. The role of the 3′ end in mRNA stability and decay. In Control of Messenger RNA Stability; Belasco, J.G., Brawerman, G., Eds.; Academic Press: San Diego, CA, USA, 1993; pp. 13–30. [Google Scholar]
- Campbell, K.S.; Bedzyk, W.D.; Cambier, J.C. Manipulation of B-cell antigen receptor tyrosine phosphorylation using aluminum fluoride and sodium orthovanadate. Mol. Immunol. 1995, 32, 1283–1294. [Google Scholar] [CrossRef] [PubMed]
- Henderson, B.R.; Menotti, E.; Bonnard, C.; Kühn, L.C. Optimal sequence and structure of iron-responsive elements. J. Biol. Chem. 1994, 269, 17481–17489. [Google Scholar] [CrossRef] [PubMed]
- Henderson, B.R.; Menotti, E.; Kühn, L.C. Iron regulatory proteins 1 and 2 distinct sets of RNA target sequences. J. Biol. Chem. 1996, 271, 4900–4908. [Google Scholar] [CrossRef]
- Miranda-Ozuna, J.F.T.; Hernández-García, M.S.; Brieba, L.G.; Benítez-Cardoza, C.G.; Ortega-López, J.; González-Robles, A.; Arroyo, R. The glycolytic enzyme triosephosphate isomerase of Trichomonas vaginalis is a surface-associated protein induced by glucose that functions as a laminin- and fibronectin-binding protein. Infect. Immun. 2016, 84, 2878–2894. [Google Scholar] [CrossRef]
- Popovic, Z.; Templeton, D.M. A Northwestern blotting approach for studying iron regulatory element-binding proteins. Mol. Cell. Biochem. 2005, 268, 67–74. [Google Scholar] [CrossRef]
- Zhao, S.L.; Liang, C.Y.; Zhang, W.J.; Tang, X.C.; Peng, H.Y. Mapping the RNA-binding domain on the DpCPV VP4. Arch. Virol. 2006, 151, 273–283. [Google Scholar] [CrossRef]
- Torres-Romero, J.C.; Arroyo, R. Responsiveness of Trichomonas vaginalis to iron concentrations: Evidence for a posttranscriptional iron regulation by an IRE/IRP-like system. Infect. Genet. Evol. 2009, 9, 1065–1074. [Google Scholar] [CrossRef]
- Espinosa, N.; Hernandez, R.; López-Griego, L.; López-Villasenor, I. Separable putative polyadenylation and cleavage motifs in Trichomonas vaginalis mRNAs. Gene 2002, 289, 81–86. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The Vienna RNA Websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef]
- Addes, K.J.; Basilion, J.P.; Klausner, R.D.; Rouault, T.A.; Pardi, A.J. Structure and dynamics of the iron-responsive element RNA: Implications for binding of the RNA by iron regulatory binding proteins. J. Mol. Biol. 1997, 274, 72–83. [Google Scholar] [CrossRef]
- Gou, B.; Phillips, J.D.; Yu, Y.; Leibold, E.A. Iron regulates the intracellular degradation of iron regulatory protein 2 by the proteasome. J. Biol. Chem. 1995, 270, 21645–21651. [Google Scholar]
- Dycke, C.; Bougault, C.; Gaillard, J.; Andrieu, J.P.; Pantopoulos, K.; Moulis, J.M. Human iron regulatory protein 2 is easily cleaved in its specific domain: Consequences for the haem-binding properties of the protein. Biochem. J. 2007, 408, 429–439. [Google Scholar] [CrossRef]
- Binder, R.; Horowitz, J.A.; Basilion, J.P.; Koeller, D.M.; Klausner, R.D.; Harford, J.B. Evidence that the pathway of transferrin receptor mRNA degradation involves an endonucleolytic cleavage within the 3′ UTR and does not involve poly(A) tail shortening. EMBO J. 1994, 13, 1969–1980. [Google Scholar] [CrossRef]
- Leipuviene, R.; Theil, C. The family of iron-responsive RNA structures regulated by changes in cellular iron and oxygen. Cell. Mol. Life Sci. 2007, 64, 2945–2955. [Google Scholar] [CrossRef]
- Liston, D.R.; Johnson, P. Gene transcription in Trichomonas vaginalis. Parasitol. Today 1998, 14, 261–265. [Google Scholar] [CrossRef]
- Malquori, L.; Carsetti, L.; Ruberti, G. The 3′ UTR of the human CTLA4 mRNA can regulate mRNA stability and translational efficiency. Biochim. Biophys. Acta 2008, 1779, 60–65. [Google Scholar] [CrossRef]
- Hentze, M.; Caughman, S.; Casey, J.; Koeller, D.; Rouault, T.; Harford, J.; Klausner, R. A model for the structure and function of iron-responsive elements. Gene 1988, 72, 201–208. [Google Scholar] [CrossRef]
- Piccenelli, P.; Samuelsson, T. Evolution of the iron-responsive element. RNA 2007, 13, 952–966. [Google Scholar] [CrossRef]
- Dos Santos, C.O.; Dore, L.C.; Valentine, E.; Shelat, S.G.; Hardison, R.C.; Ghosh, M.; Wang, W.; Eisenstein, R.S.; Costa, F.F.; Weiss, M.J.; et al. An iron-responsive element-like stem-loop regulates (alpha)-hemoglobin-stabilizing protein mRNA. J. Biol. Chem. 2008, 283, 26956–26964. [Google Scholar] [CrossRef] [PubMed]
- Soto-Castro, L.; Plata-Guzmán, L.Y.; Figueroa-Angulo, E.E.; Calla-Choque, J.S.; Reyes-López, M.; De la Garza, M.; León-Sicairos, N.; Garzón-Tiznado, J.A.; Arroyo, R.; León-Sicairos, C. Iron Responsive-like Elements in the parasite Entamoeba histolytica. Microbiology 2019, 165, 366. [Google Scholar] [CrossRef] [PubMed]
- Plata-Guzmán, L.Y.; Arroyo, R.; León-Sicairos, N.; Canizález-Román, A.; López-Moreno, H.S.; Chávez-Ontiveros, J.; Garzón-Tiznado, J.A.; León-Sicairos, C. Stem–loop structures in iron-regulated mRNAs of Giardia duodenalis. Int. J. Environ. Res. Public. Health 2023, 20, 3556. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
León-Sicairos, C.R.; Figueroa-Angulo, E.E.; Calla-Choque, J.S.; Arroyo, R. The Non-Canonical Iron-Responsive Element of IRE-tvcp12 Hairpin Structure at the 3′-UTR of Trichomonas vaginalis TvCP12 mRNA That Binds TvHSP70 and TvACTN-3 Can Regulate mRNA Stability and Amount of Protein. Pathogens 2023, 12, 586. https://doi.org/10.3390/pathogens12040586
León-Sicairos CR, Figueroa-Angulo EE, Calla-Choque JS, Arroyo R. The Non-Canonical Iron-Responsive Element of IRE-tvcp12 Hairpin Structure at the 3′-UTR of Trichomonas vaginalis TvCP12 mRNA That Binds TvHSP70 and TvACTN-3 Can Regulate mRNA Stability and Amount of Protein. Pathogens. 2023; 12(4):586. https://doi.org/10.3390/pathogens12040586
Chicago/Turabian StyleLeón-Sicairos, Claudia R., Elisa E. Figueroa-Angulo, Jaeson S. Calla-Choque, and Rossana Arroyo. 2023. "The Non-Canonical Iron-Responsive Element of IRE-tvcp12 Hairpin Structure at the 3′-UTR of Trichomonas vaginalis TvCP12 mRNA That Binds TvHSP70 and TvACTN-3 Can Regulate mRNA Stability and Amount of Protein" Pathogens 12, no. 4: 586. https://doi.org/10.3390/pathogens12040586
APA StyleLeón-Sicairos, C. R., Figueroa-Angulo, E. E., Calla-Choque, J. S., & Arroyo, R. (2023). The Non-Canonical Iron-Responsive Element of IRE-tvcp12 Hairpin Structure at the 3′-UTR of Trichomonas vaginalis TvCP12 mRNA That Binds TvHSP70 and TvACTN-3 Can Regulate mRNA Stability and Amount of Protein. Pathogens, 12(4), 586. https://doi.org/10.3390/pathogens12040586