First Detection and Molecular Analysis of Leishmania infantum DNA in Sand Flies of Kosovo
Abstract
:1. Introduction
2. Materials and Methods
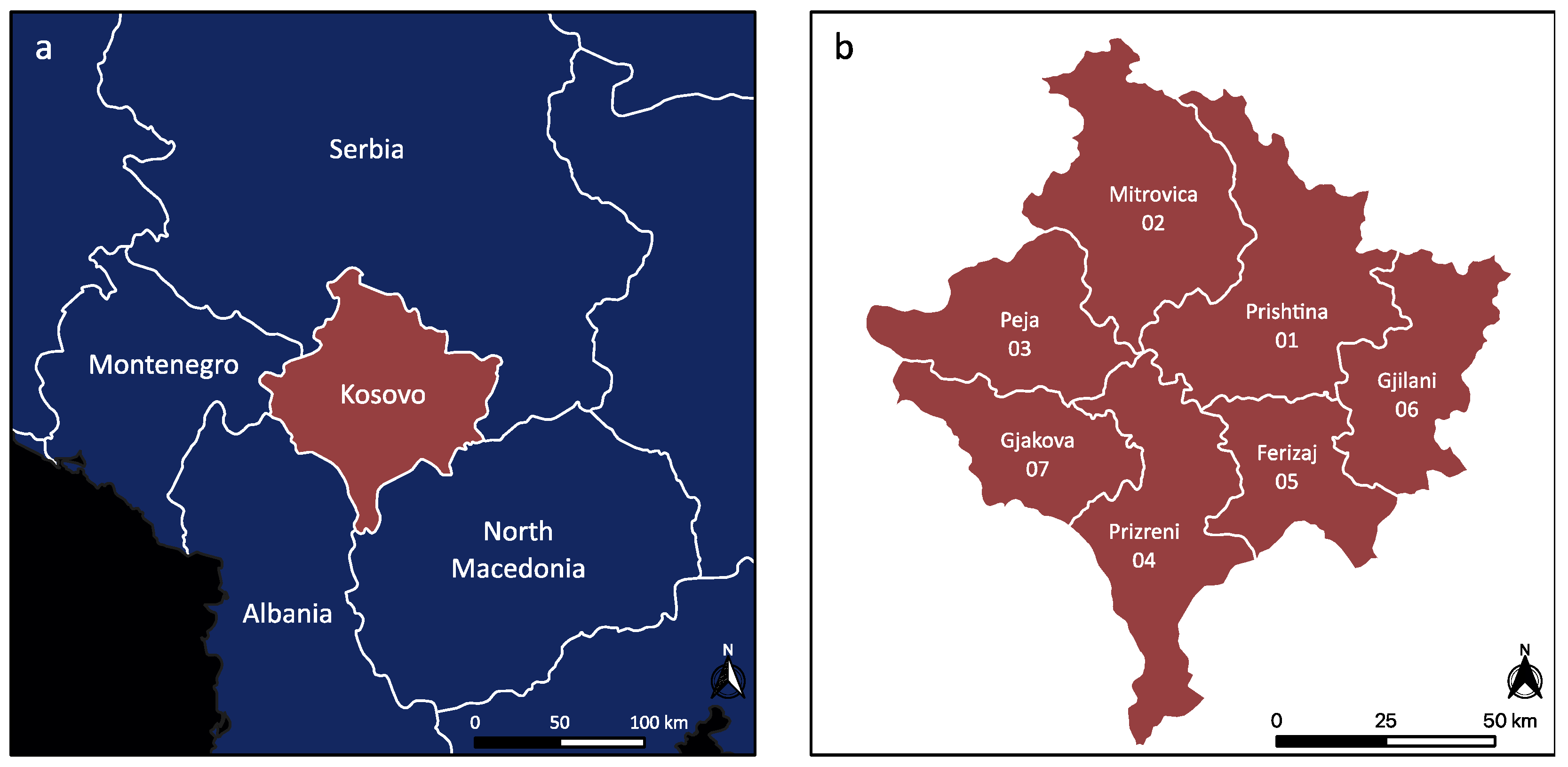
2.1. Study Area
2.2. Entomological Sampling
2.3. Morphological Identification of Sand Flies
2.4. Molecular Analysis
2.5. Detection of Leishmania infantum DNA
2.6. Molecular Identification of Leishmania-Positive Sand Flies
2.7. Molecular Characterization of Detected Leishmania DNA
2.8. Mapping of Sand Fly Occurrence Data
3. Results
3.1. Sand Fly Trapping
3.2. Leishmania DNA Screening
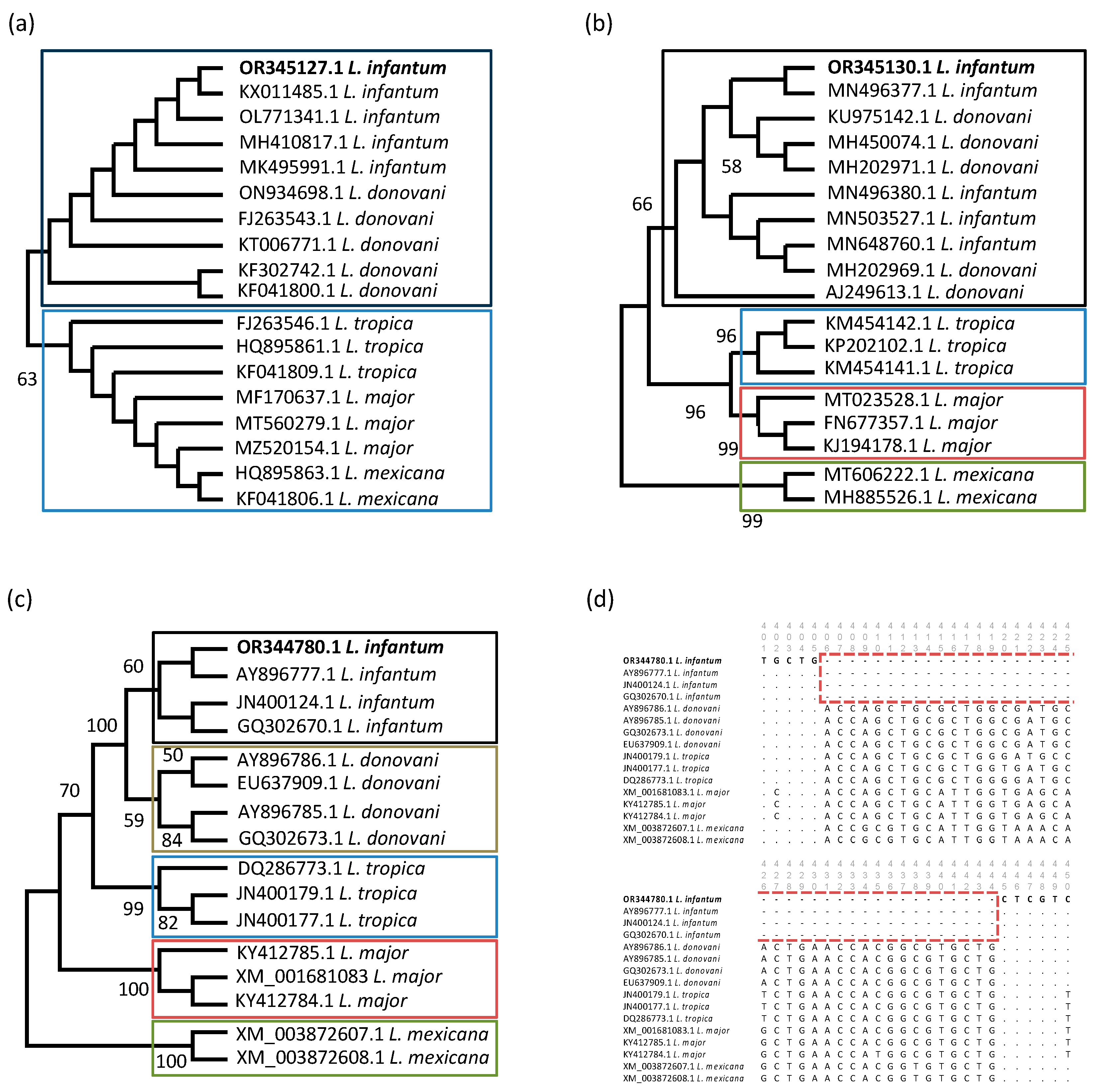
3.3. Molecular Characterization of L. infantum-Positive Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ready, P.D. Biology of Phlebotomine Sand Flies as Vectors of Disease Agents. Annu. Rev. Entomol. 2013, 58, 227–250. [Google Scholar] [CrossRef] [PubMed]
- WHO Fact Sheets: Leishmaniasis. 2021. Available online: https://www.who.int/news-room/fact-sheets/detail/leishmaniasis (accessed on 19 July 2023).
- Inceboz, T. Epidemiology and Ecology of Leishmaniasis. In Current Topics in Neglected Tropical Diseases; IntechOpen: London, UK, 2019. [Google Scholar]
- Ready, P.D. Leishmaniasis emergence in Europe. Eurosurveillance 2010, 15, 19505. [Google Scholar] [CrossRef] [PubMed]
- Dantas-Torres, F.; Miró, G.; Baneth, G.; Bourdeau, P.; Breitschwerdt, E.; Capelli, G.; Cardoso, L.; Day, M.J.; Dobler, G.; Ferrer, L.; et al. Canine Leishmaniasis Control in the Context of One Health. Emerg. Infect. Dis. 2019, 25, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Vaselek, S. Canine leishmaniasis in Balkan—A review of occurrence and epidemiology. Acta Trop. 2021, 224, 106110. [Google Scholar] [CrossRef] [PubMed]
- Lazri, T.; Duscher, G.; Edelhofer, R.; Bytyci, B.; Gjino, P.; Joachim, A. Arthropod-borne parasites of dogs, especially Leishmania, in the Kosovo and Albania. Wien. Klin. Wochenschr. 2008, 120, 54–58. [Google Scholar] [CrossRef] [PubMed]
- Xhekaj, B.; Alishani, M.; Rexhepi, A.; Jakupi, X.; Sherifi, K. Serological survey of canine leishmaniasis in southwestern region of Kosovo. Vet. Ital. 2020, 56, 47–50. [Google Scholar] [CrossRef]
- Xhekaj, B.; Stefanovska, J.; Sherifi, K.; Rexhepi, A.; Bizhga, B.; Rashikj, L.; Nikolovski, M.; Kniha, E.; Cvetkovikj, A. Seroprevalence of canine leishmaniosis in asymptomatic dogs in Kosovo. Parasitol. Res. 2023, 122, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Vaselek, S. Systematic Review: Re-emergence of human leishmaniasis in the Balkans. Trop. Med. Int. Health 2021, 26, 1189–1199. [Google Scholar] [CrossRef]
- Cecílio, P.; Cordeiro-da-Silva, A.; Oliveira, F. Sand flies: Basic information on the vectors of leishmaniasis and their interactions with Leishmania parasites. Commun. Biol. 2022, 5, 305. [Google Scholar] [CrossRef]
- Benallal, K.E.; Garni, R.; Harratfft, Z.; Benallal, K.E.; Volf, P.; Dvořák, V. Phlebotomine sand flies (Diptera: Psychodidae) of the Maghreb region: A systematic review of distribution, morphology, and role in the transmission of the pathogens. PLoS Negl. Trop. Dis. 2022, 16, e0009952. [Google Scholar] [CrossRef]
- Vaselek, S.; Oguz, G.; Ayhan, N.; Ozbel, Y.; Kadriaj, P.; Ćupina, A.I.; Velo, E.; Muja, N.; Baymak, D.; Alishani, M.; et al. Sandfly surveillance and investigation of Leishmania spp. DNA in sandflies in Kosovo. Med. Vet. Entomol. 2020, 34, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Bennai, K.; Tahir, D.; Lafri, I.; Bendjaballah-Laliam, A.; Bitam, I.; Parola, P. Molecular detection of Leishmania infantum DNA and host blood meal identification in Phlebotomus in a hypoendemic focus of human leishmaniasis in northern Algeria. PLoS Negl. Trop. Dis. 2018, 12, e0006513. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D.J. A taxonomic review of the genus Phlebotomus (Diptera: Psychodidae). Bull. Br. Mus. 1982, 45, 121–209. [Google Scholar]
- Dantas-Torres, F.; Tarallo, V.D.; Otranto, D. Morphological keys for the identification of Italian phlebotomine sand flies (Diptera: Psychodidae: Phlebotominae). Parasit. Vectors 2014, 7, 479. [Google Scholar] [CrossRef] [PubMed]
- van Eys, G.J.J.M.; Schoone, G.J.; Kroon, N.C.M.; Ebeling, S.B. Sequence analysis of small subunit ribosomal RNA genes and its use for detection and identification of Leishmania parasites. Mol. Biochem. Parasitol. 1992, 51, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Cruz, I.; Cañavate, C.; Rubio, J.M.; Morales, M.A.; Chicharro, C.; Laguna, F.; Jiménez-Mejías, M.; Sirera, G.; Videla, S.; Alvar, J. A nested polymerase chain reaction (Ln-PCR) for diagnosing and monitoring Leishmania infantum infection in patients co-infected with human immunodeficiency virus. Trans. R. Soc. Trop. Med. Hyg. 2002, 96, 185–189. [Google Scholar] [CrossRef] [PubMed]
- El Tai, N.O.; Osman, F.O.; Far, M.E.; Presber, W.; Schönian, G. Genetic heterogeneity of ribosomal internal transcribed spacer in clinical samples of Leishmania donovani spotted on filter paper as revealed by single-strand conformation polymorphisms and sequencing. Trans. R. Soc. Trop. Med. Hyg. 2000, 94, 575–579. [Google Scholar] [CrossRef]
- Hide, M.; Bañuls, A.L. Species-specific PCR assay for L. infantum/L. donovani discrimination. Acta Trop. 2006, 100, 241–245. [Google Scholar] [CrossRef]
- Mary, C.; Faraut, F.; Lascombe, L.; Dumon, H. Quantification of Leishmania infantum DNA by a real-time PCR assay with high sensitivity. J. Clin. Microbiol. 2004, 42, 5249–5255. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Nicholas, K.B. Genedoc: A Tool for Editing and Annotating Multiple Sequence Alignments. 1997. Available online: http://www.pscedu/biomed/genedoc (accessed on 18 July 2023).
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- QGIS Development Team. QGIS Geographic Information System. Open Source Geospatial Foundation Project. 2019. Available online: http://qgis.osgeo.org (accessed on 18 July 2023).
- Simić, Č.; Živković, V. Prilog poznavanju faune naših flebotomina I deo—Makedonija, južni deo Srbije i Kosmet [Contribution to the knowledge of sandflies in Yugoslavia. Part I—Macedonia, southern Serbia and Kosovo]. Glas. Srp. Akad. Nauk. Med. 1949, 1, 151–181. [Google Scholar]
- Živković, V. Repartition de Phlebotomus chinensis balcanicus Theodor, 1958 (Diptera, Psychodidae) en Yougoslavie. Acta Parasitol. Jugoslavica 1974, 5, 3–9. [Google Scholar]
- Ayhan, N.; Alten, B.; Ivovic, V.; Dvořák, V.; Martinkovic, F.; Omeragic, J.; Stefanovska, J.; Petric, D.; Vaselek, S.; Baymak, D.; et al. Direct evidence for an expanded circulation area of the recently identified Balkan virus (Sandfly fever Naples virus species) in several countries of the Balkan archipelago. Parasit. Vectors 2017, 10, 402. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, V.; Kasap, O.E.; Ivovic, V.; Mikov, O.; Stefanovska, J.; Martinkovic, F.; Omeragic, J.; Pajovic, I.; Baymak, D.; Oguz, G.; et al. Sand flies (Diptera: Psychodidae) in eight Balkan countries: Historical review and region-wide entomological survey. Parasit. Vectors 2020, 13, 573. [Google Scholar] [CrossRef] [PubMed]
- Svobodová, M.; Alten, B.; Zídková, L.; Dvořák, V.; Hlavačková, J.; Myšková, J.; Šeblová, V.; Kasap, O.E.; Belen, A.; Votýpka, J.; et al. Cutaneous leishmaniasis caused by Leishmania infantum transmitted by Phlebotomus tobbi. Int. J. Parasitol. 2009, 39, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Giorgobiani, E.; Lawyer, P.G.; Babuadze, G.; Dolidze, N.; Jochim, R.C.; Tskhvaradze, L.; Kikaleishvili, K.; Kamhawi, S. Incrimination of phlebotomus kandelakii and Phlebotomus balcanicus as vectors of Leishmania infantum in Tbilisi, Georgia. PLoS Negl. Trop. Dis. 2012, 6, e1609. [Google Scholar] [CrossRef]
- Vaselek, S.; Ayhan, N.; Oguz, G.; Erisoz Kasap, O.; Savić, S.; Di Muccio, T.; Gradoni, L.; Ozbel, Y.; Alten, B.; Petrić, D. Sand fly and Leishmania spp. survey in Vojvodina (Serbia): First detection of Leishmania infantum DNA in sand flies and the first record of Phlebotomus (Transphlebotomus) mascittii Grassi, 1908. Parasit. Vectors 2017, 10, 444. [Google Scholar] [CrossRef]
- Velo, E.; Bongiorno, G.; Kadriaj, P.; Myrseli, T.; Crilly, J.; Lika, A.; Mersini, K.; Di Muccio, T.; Bino, S.; Gramiccia, M.; et al. The current status of phlebotomine sand flies in Albania and incrimination of Phlebotomus neglectus (Diptera, Psychodidae) as the main vector of Leishmania infantum. PLoS ONE 2017, 12, e0179118. [Google Scholar] [CrossRef]
- Ayhan, N.; Velo, E.; de Lamballerie, X.; Kota, M.; Kadriaj, P.; Ozbel, Y.; Charrel, R.N.; Bino, S. Detection of Leishmania infantum and a Novel Phlebovirus (Balkan Virus) from Sand Flies in Albania. Vector-Borne Zoonotic Dis. 2016, 16, 802–806. [Google Scholar] [CrossRef]
- Kniha, E.; Walochnik, J.; Poeppl, W.; Mooseder, G.; Obwaller, A.G. Leishmania spp. seropositivity in Austrian soldiers returning from the Kosovo. Wien. Klin. Wochenschr. 2020, 132, 47–49. [Google Scholar] [CrossRef]

| Target | Primer and Probe (5′-3′) | Fragment Size (bp) | Protocol | Reference |
|---|---|---|---|---|
| ssu rRNA (1st round) | R221 (GGTTCCTTTCCTGATTTACG) | ~600 bp | 94 °C/5 min; 15 cycles: 94 °C/30 s, 53 °C/30 s, 72 °C/30 s; 72 °C 10 min | [17] |
| R332 (GGCCGGTAAAGGCCGAATAG) | ||||
| ssu rRNA (2nd round) | R223 (TCCCATCGCAACCTCGGTT) | ~350 bp | 94 °C 5 min; 32 cycles: 94 °C/30 s, 65 °C/30 s, 72 °C/30 s; 72 °C 10 min | [18] |
| R333 (AAAGCGGGCGCGGTGCTG) | ||||
| ITS1 | LITSR (CTGGATCATTTTCCGATG) | ~320 bp | 94 °C/5 min; 34 cycles: 94 °C/20 s, 53 °C/30 s, 72 °C/1 min; 72 °C/10 min | [19] |
| 5.8S (TGATACCACTTATCGCACTT) | ||||
| cpb | cpfF (CGTGACGCCGGTGAAGAAT) | ~740– 780 bp | 94 °C/5 min; 34 cycles: 94 °C/30 s, 62 °C/1min, 72 °C/1 min; 72 °C/10 min | [20] |
| cpbR (CGTGCACTCGGCCGTCTT) | ||||
| kDNA a | F (CTTTTCTGGTCCTCCGGGTAGG) | - | 50 °C/10 min; 95 °C/10 min; 45 cycles: 95 °C/15 s, 60 °C/1 min | [21] |
| R (CCACCCGGCCCTATTTTACACCAA) | ||||
| Probe (FAM-TTTTCGCAGAAC GCCCCTACCCGC-TAMRA) |
| Species | Male | Female (Engorged) | Total | Percentage |
|---|---|---|---|---|
| Ph. perfiliewi | 64 | 651 (110) | 715 | 62.2% |
| Ph. neglectus | 156 | 212 (61) | 368 | 32.0% |
| Ph. tobbi | 5 | 22 (2) | 27 | 2.3% |
| Ph. simici | 3 | 19 (4) | 22 | 1.9% |
| Ph. balcanicus | 4 | 4 (0) | 8 | 0.7% |
| Ph. papatasi | 1 | 1 (0) | 2 | 0.2% |
| S. minuta | 1 | 6 (2) | 7 | 0.6% |
| Total | 234 | 915 (179) | 1149 | 100% |
| Location | Total | Number | Species | Pooled | |
|---|---|---|---|---|---|
| ID | Number | Identified | Ph. perfiliewi | Ph. neglectus | Specimens |
| 02/7 | 133 | 88 | 74 (84.1%) | 14 (15.9%) | 45 (4 pools) |
| 03/7 | 203 | 57 | 56 (98.3%) | 1 (1.7%) | 146 (9 pools) |
| 03/8 | 1002 | 77 | 76 (98.7%) | 1 (1.3%) | 925 (32 pools) |
| 04/9 | 828 | 113 | 87 (77.0%) | 26 (23.0%) | 715 (24 pools) |
| 07/8 | 329 | 37 | 37 (100%) | 0 (0.0%) | 292 (11 pools) |
| total | 2495 | 372 | 330 (88.7%) | 42 (11.3%) | 2123 (80 pools) |
| Species | Prishtina | Mitrovica | Peja | Prizreni | Ferizaj | Gjilani | Gjakova | Total |
|---|---|---|---|---|---|---|---|---|
| Ph. perfiliewi | 4 | 165 | 213 | 92 | 1 | 8 | 232 | 715 |
| Ph. neglectus | 54 | 61 | 12 | 107 | 24 | 53 | 57 | 368 |
| Ph. tobbi | 0 | 0 | 0 | 0 | 1 | 26 | 0 | 27 |
| Ph. simici | 0 | 0 | 0 | 0 | 8 | 13 | 1 | 22 |
| Ph. balcanicus | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 8 |
| Ph. papatasi | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 2 |
| S. minuta | 0 | 0 | 1 | 5 | 0 | 0 | 1 | 7 |
| in mixed pools a | 0 | 45 | 1071 | 715 | 0 | 0 | 292 | 2123 |
| total | 58 | 271 | 1299 | 919 | 34 | 100 | 591 | 3272 |
| Blast Identity Compared to Reference Sequences | ||||
|---|---|---|---|---|
| Gene | L. infantum | L. donovani | L. tropica | L. major |
| ssu rRNA a | 100% (MN757921.1, MK495991.1) | 100% (CP022642.1, ON934698.1) | 99.69% (KF302745.1, GQ332363.1) | 99.69% (MT560279.1) to 99.38% (MZ520154.1) |
| ITS1 a | 100% (MZ362379.1, MN503527.1) | 100% (MH202970.1, OQ184729.1) | 92.39% (KC679052.1) to 90.78% (KY963131.1) | 91.80% (MT423523.1) to 86.51% (ON243845.1) |
| Cpb b | 100% (CP027841.1) to 97.89% (AY896778.1) | 99.75% (EU637909.1) to 98.52% (AY896783.1) | 90.86% (DQ286773.1) to 87.44% (JN400184.1) | 90.15% (JN944175.1) to 88.92% (JN400175.1) |
| Leishmania spp. | L. infantum | L. donovani | L. tropica | L. major | L. mexicana |
|---|---|---|---|---|---|
| L. infantum | - | ||||
| L. donovani | 0.06/0.3/0.8 | - | |||
| L. tropica | 0.3/4.7/9.3 | 0.4/5.0/9.9 | - | ||
| L. major | 0.4/5.5/11.0 | 0.5/5.8/11.6 | 0.1/5.5/8.5 | - | |
| L. mexicana | 0.3/18.2/12.8 | 0.4/17.9/13.4 | 0.0/20.4/12.6 | 0.1/22.0/13.3 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xhekaj, B.; Hoxha, I.; Platzgummer, K.; Kniha, E.; Walochnik, J.; Sherifi, K.; Rexhepi, A.; Behluli, B.; Dvořák, V.; Fuehrer, H.-P.; et al. First Detection and Molecular Analysis of Leishmania infantum DNA in Sand Flies of Kosovo. Pathogens 2023, 12, 1190. https://doi.org/10.3390/pathogens12101190
Xhekaj B, Hoxha I, Platzgummer K, Kniha E, Walochnik J, Sherifi K, Rexhepi A, Behluli B, Dvořák V, Fuehrer H-P, et al. First Detection and Molecular Analysis of Leishmania infantum DNA in Sand Flies of Kosovo. Pathogens. 2023; 12(10):1190. https://doi.org/10.3390/pathogens12101190
Chicago/Turabian StyleXhekaj, Betim, Ina Hoxha, Katharina Platzgummer, Edwin Kniha, Julia Walochnik, Kurtesh Sherifi, Agim Rexhepi, Behlul Behluli, Vit Dvořák, Hans-Peter Fuehrer, and et al. 2023. "First Detection and Molecular Analysis of Leishmania infantum DNA in Sand Flies of Kosovo" Pathogens 12, no. 10: 1190. https://doi.org/10.3390/pathogens12101190
APA StyleXhekaj, B., Hoxha, I., Platzgummer, K., Kniha, E., Walochnik, J., Sherifi, K., Rexhepi, A., Behluli, B., Dvořák, V., Fuehrer, H.-P., Obwaller, A. G., Poeppl, W., Stefanovska, J., & Cvetkovikj, A. (2023). First Detection and Molecular Analysis of Leishmania infantum DNA in Sand Flies of Kosovo. Pathogens, 12(10), 1190. https://doi.org/10.3390/pathogens12101190










