Janthinobacterium sp. Strain SLB01 as Pathogenic Bacteria for Sponge Lubomirskia baikalensis
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling of Sponges and Cell Culture of Primmorphs
2.2. Bacteria Isolation
2.3. Microscopy
2.4. Genome Sequencing
2.5. Genome Assembly, Annotation and Phylogenetic Relationship
2.6. Statistical Analysis
3. Results
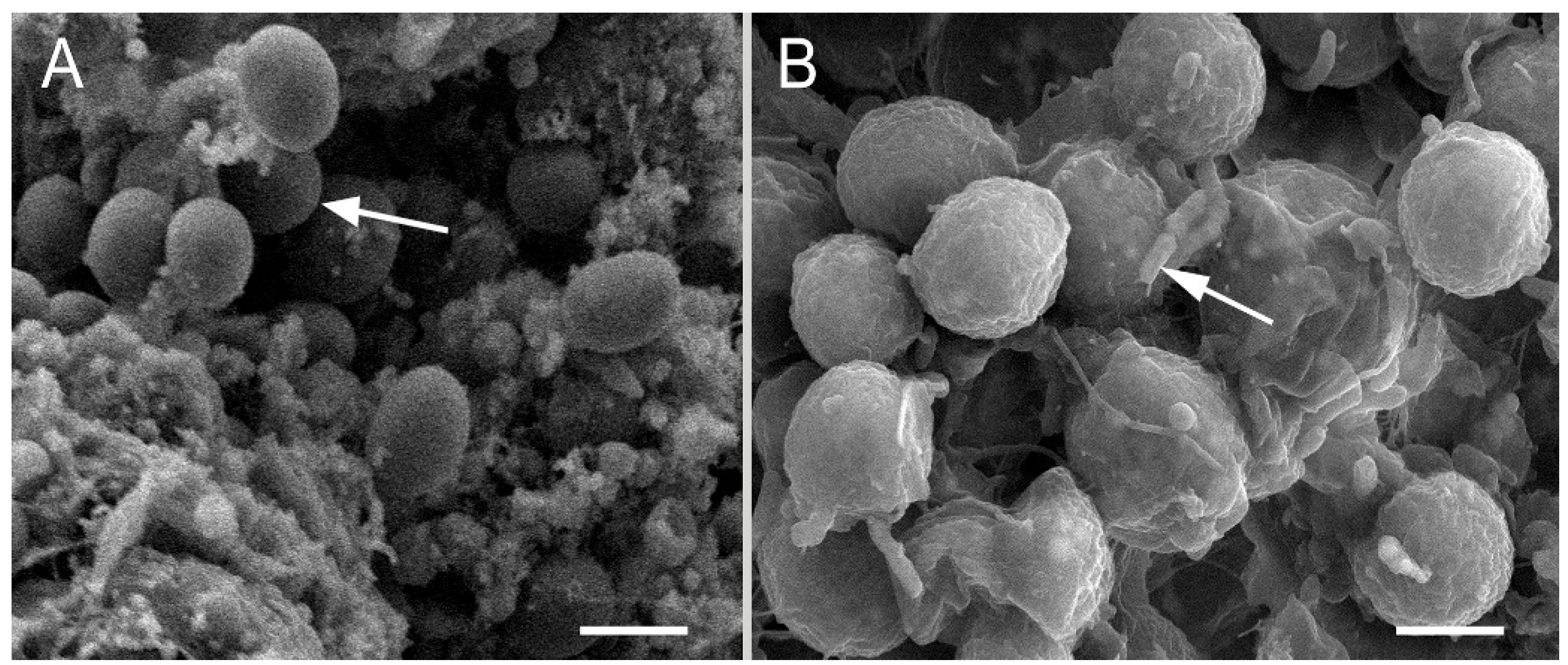
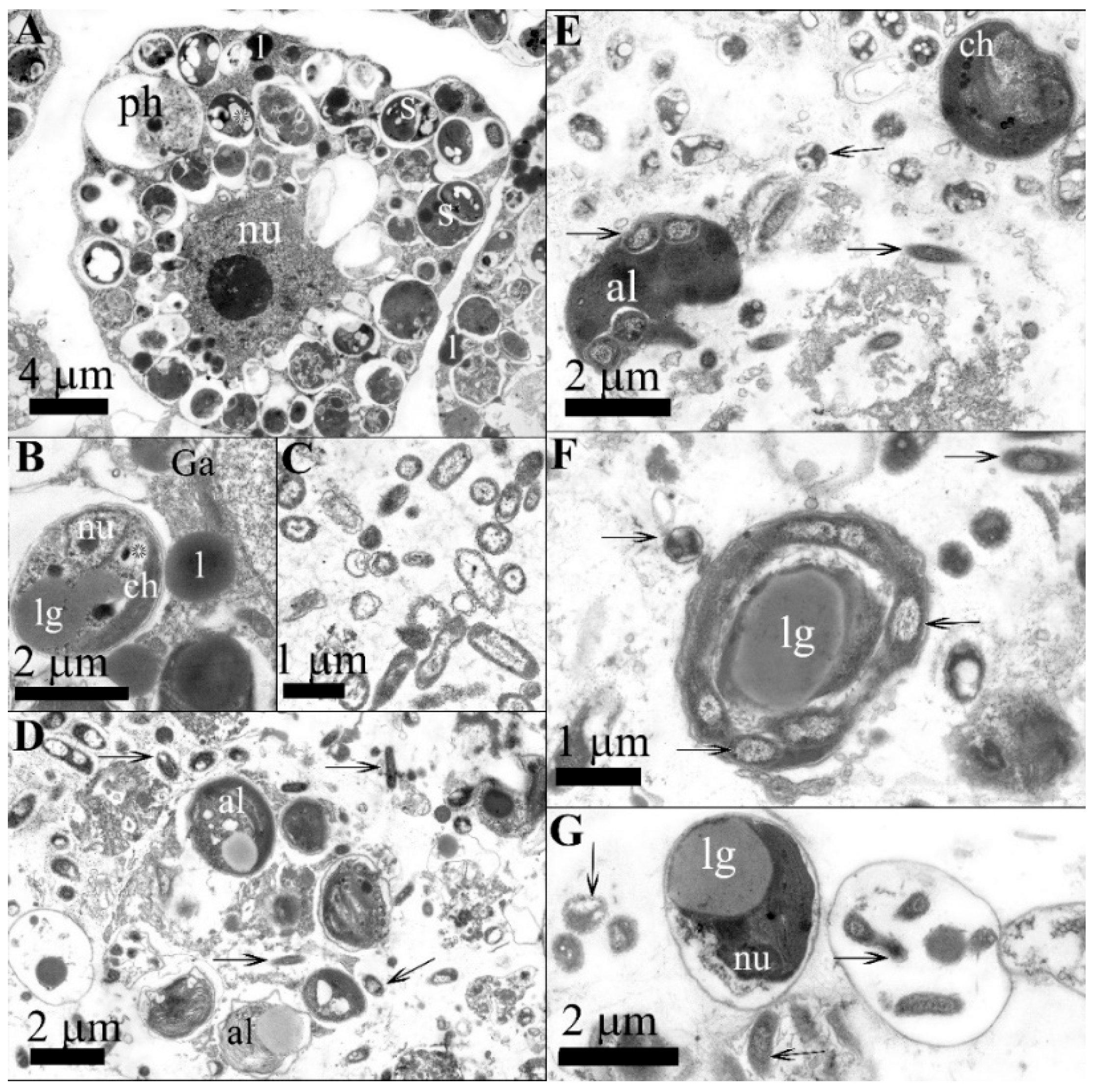
3.1. Bacterial Isolation and Microscopy
3.2. Comparison of Genomes of Janthinobacterium spp. Strains
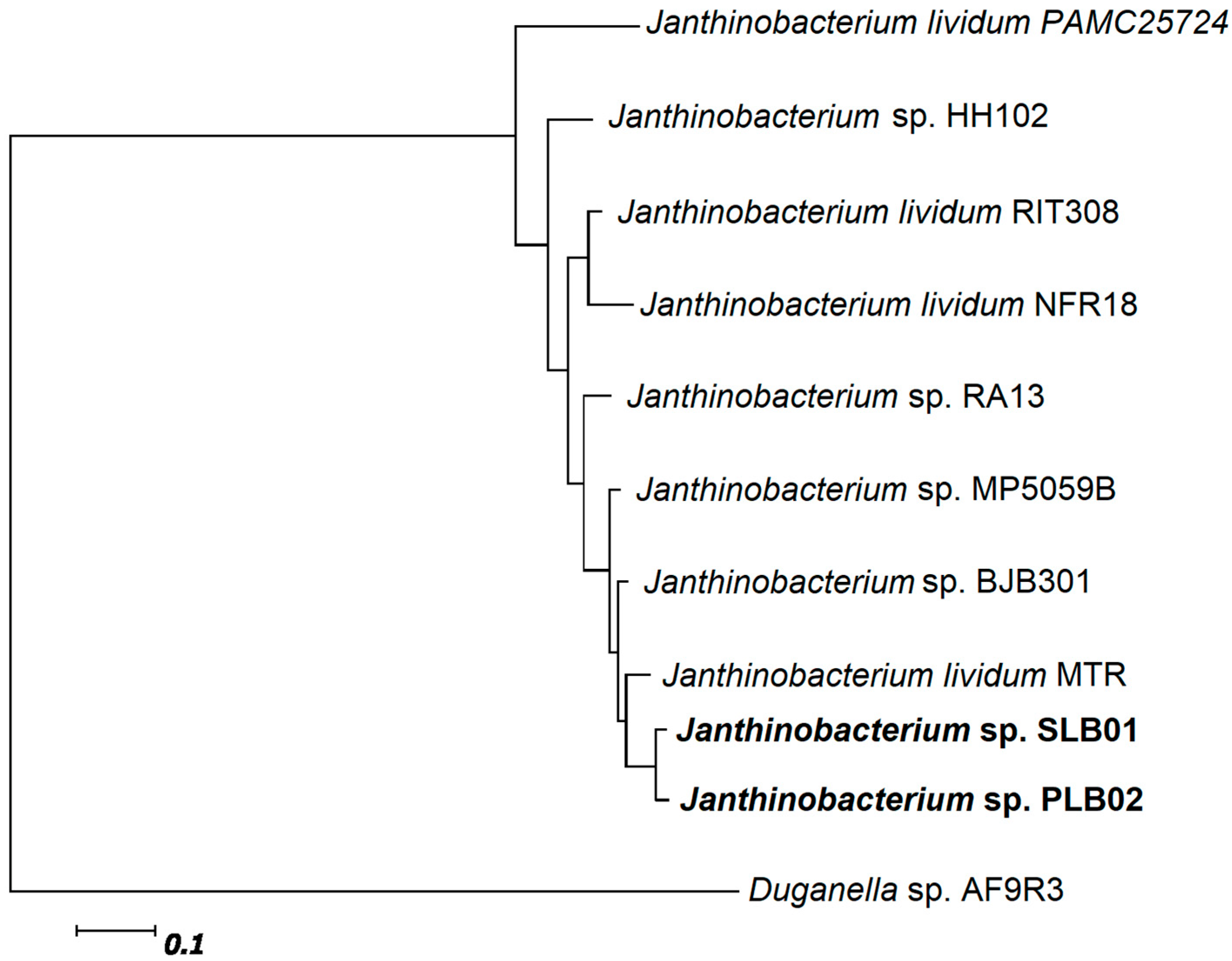
3.3. Phylogenetic Relationship
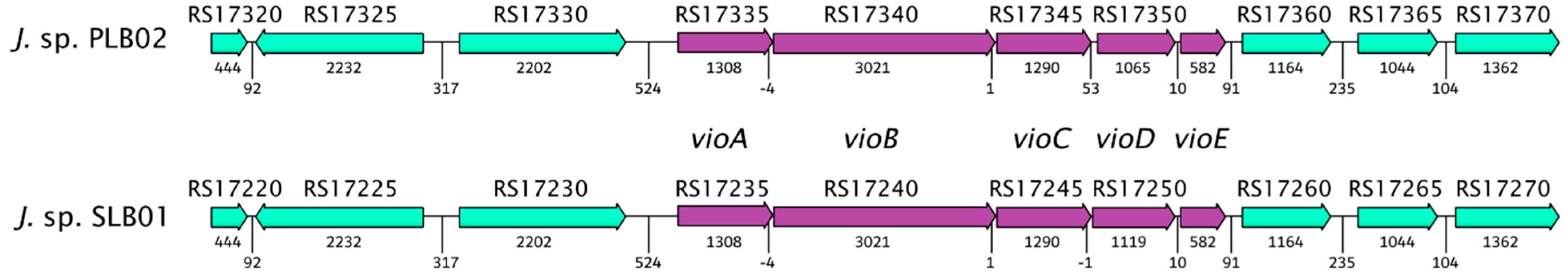
3.4. Analysis of the Virulence Genes
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hedges, S.I.B.; Blair, J.E.; Venturi, M.L.; Shoe, J.L. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol. Biol. 2004, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Osinga, R.; Armstrong, E.; Burgess, J.G.; Hoffmann, F.; Reitner, J.; Schumann-Kindel, G. Sponge–microbe associations and their importance for sponge bioprocess engineering. Hydrobiologia 2001, 461, 55–62. [Google Scholar] [CrossRef]
- Hentschel, U.; Fieseler, L.; Wehrl, M.; Gernert, C.; Steinert, M.; Hacker, J.; Horn, M. Microbial Diversity of Marine Sponges. Prog. Mol. Subcell. Biol. 2003, 37, 59–88. [Google Scholar] [CrossRef] [PubMed]
- Fieseler, L.; Horn, M.; Wagner, M.; Hentschel, U. Discovery of the Novel Candidate Phylum “Poribacteria” in Marine Sponges. Appl. Environ. Microbiol. 2004, 70, 3724–3732. [Google Scholar] [CrossRef]
- Taylor, M.W.; Radax, R.; Steger, D.; Wagner, M. Sponge-Associated Microorganisms: Evolution, Ecology, and Biotechnological Potential. Microbiol. Mol. Biol. Rev. 2007, 71, 295–347. [Google Scholar] [CrossRef]
- Weisz, J.B.; Lindquist, N.; Martens, C.S. Do associated microbial abundances impact marine demosponge pumping rates and tissue densities? Oecologia 2007, 155, 367–376. [Google Scholar] [CrossRef]
- Webster, N.; Taylor, M.W. Marine sponges and their microbial symbionts: Love and other relationships. Environ. Microbiol. 2011, 14, 335–346. [Google Scholar] [CrossRef]
- Thomas, T.; Moitinho-Silva, L.; Lurgi, M.; Björk, J.; Easson, C.; Garcia, M.D.C.A.; Olson, J.B.; Erwin, P.M.; Lopez-Legentil, S.; Luter, H.; et al. Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun. 2016, 7, 11870. [Google Scholar] [CrossRef]
- Laffy, P.W.; Wood-Charlson, E.M.; Turaev, D.; Jutz, S.; Pascelli, C.; Botté, E.S.; Bell, S.C.; Peirce, T.E.; Weynberg, K.D.; van Oppen, M.J.H.; et al. Reef invertebrate viromics: Diversity, host specificity and functional capacity. Environ. Microbiol. 2018, 20, 2125–2141. [Google Scholar] [CrossRef]
- Efremova, S.M. New genus and new sponge species of the family Lubomirskiidae Rezvoy, 1936. In Index of Animal Species InHabiting Lake Baikal and Its Catchment Area; Book 1; Timoshkin, O.A., Ed.; Nauka: Novosibirsk, Russia, 2004; Volume 1, pp. 1261–1278. (In Russian) [Google Scholar]
- Manconi, R.; Pronzato, R. Global diversity of sponges (Porifera: Spongillina) in freshwater. In Freshwater Animal Diversity Assessment. Developments in Hydrobiology; Balian, E.V., Lévêque, C., Segers, H., Martens, K., Eds.; Springer: Dordrecht, The Netherlands, 2007; Volume 198, pp. 27–33. [Google Scholar] [CrossRef]
- Latyshev, N.; Zhukova, N.; Efremova, S.; Imbs, A.; Glysina, O. Effect of habitat on participation of symbionts in formation of the fatty acid pool of fresh-water sponges of Lake Baikal. Comp. Biochem. Physiol. Part B Comp. Biochem. 1992, 102, 961–965. [Google Scholar] [CrossRef]
- Bil, K.; Titlyanov, E.; Berner, T.; Fomina, I.; Muscatine, L. Some aspects of the physiology and biochemistry of Lubomirska bai-calensis, a sponge from Lake Baikal containing symbiotic algae. Symbiosis 1999, 26, 179–191. [Google Scholar]
- Chernogor, L.; Denikina, N.; Kondratov, I.; Solovarov, I.; Khanaev, I.; Belikov, S.; Ehrlich, H. Isolation and identification of the microalgal symbiont from primmorphs of the endemic freshwater sponge Lubomirskia baicalensis (Lubomirskiidae, Porifera). Eur. J. Phycol. 2013, 48, 497–508. [Google Scholar] [CrossRef]
- Kravtsova, L.S.; Izhboldina, L.A.; Khanaev, I.V.; Pomazkina, G.V.; Rodionova, E.V.; Domysheva, V.M.; Sakirko, M.V.; Tomberg, I.V.; Kostornova, T.Y.; Kravchenko, O.S.; et al. Nearshore benthic blooms of filamentous green algae in Lake Baikal. J. Great Lakes Res. 2014, 40, 441–448. [Google Scholar] [CrossRef]
- Khanaev, I.V.; Kravtsova, L.S.; Maikova, O.O.; Bukshuk, N.A.; Sakirko, M.V.; Kulakova, N.V.; Butina, T.V.; Nebesnykh, I.A.; Belikov, S.I. Current state of the sponge fauna (Porifera: Lubomirskiidae) of Lake Baikal: Sponge disease and the problem of conservation of diversity. J. Great Lakes Res. 2018, 44, 77–85. [Google Scholar] [CrossRef]
- Belikov, S.; Belkova, N.; Butina, T.; Chernogor, L.; Kley, A.M.-V.; Nalian, A.; Rorex, C.; Khanaev, I.; Maikova, O.; Feranchuk, S. Diversity and shifts of the bacterial community associated with Baikal sponge mass mortalities. PLoS ONE 2019, 14, e0213926. [Google Scholar] [CrossRef]
- Chernogor, L.; Klimenko, E.; Khanaev, I.; Belikov, S. Microbiome analysis of healthy and diseased sponges Lubomirskia baicalensis by using cell cultures of primmorphs. PeerJ 2020, 8, e9080. [Google Scholar] [CrossRef]
- Chernogor, L.; Bakhvalova, K.; Belikova, A.; Belikov, S. Isolation and Properties of the Bacterial Strain Janthinobacterium sp. SLB01. Microorganisms 2022, 10, 1071. [Google Scholar] [CrossRef]
- Petrushin, I.S.; Belikov, S.I.; Chernogor, L.I. Draft genome sequence of Janthinobacterium sp. strain SLB01, isolated from the diseased sponge Lubomirskia baicalensis. Microbiol. Resour. Announc. 2019, 8, e01108. [Google Scholar] [CrossRef]
- Belikov, S.I.; Petrushin, I.S.; Chernogor, L.I. Genome Analysis of the Janthinobacterium sp. Strain SLB01 from the Diseased Sponge of the Lubomirskia baicalensis. Curr. Issues Mol. Biol. 2021, 43, 2220–2237. [Google Scholar] [CrossRef]
- Petrushin, I.; Belikov, S.; Chernogor, L. Cooperative Interaction of Janthinobacterium sp. SLB01 and Flavobacterium sp. SLB02 in the Diseased Sponge Lubomirskia baicalensis. Int. J. Mol. Sci. 2020, 21, 8128. [Google Scholar] [CrossRef]
- Justo, G.Z.; Durán, N.; Ferreira, C.V.; Melon, P.S.; Cordi, L.; Martins, D. Violacein: Properties and biological activities. Biotechnol. Appl. Biochem. 2007, 48, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, A.L.; Göcke, Y.; Bolten, C.; Brock, N.L.; Dickschat, J.S.; Wittmann, C. Microbial production of the drugs violacein and deoxyviolacein: Analytical development and strain comparison. Biotechnol. Lett. 2011, 34, 717–720. [Google Scholar] [CrossRef] [PubMed]
- Garrity, G.M.; Bell, J.A.; Lilburn, T.E. Family II Oxalobacteraceae fam. nov. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Garrity, G.M., Brenner, D.J., Krieg, N.R., Staley, J., Eds.; Springer: New York, NY, USA, 2005; Volume 2, p. 623. [Google Scholar] [CrossRef]
- Gillis, M.; De Ley, J. The Genera Chromobacterium and Janthinobacterium. In The Prokaryotes: Handbook on the Biology of Bacteria; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; Volume 7, p. 737. [Google Scholar] [CrossRef]
- Gillis, M.; Logan, N.A. Janthinobacterium. In Bergey’s Manual of Systematics of Archaea and Bacteria; Trujillo, M.E., Dedysh, S., DeVos, P., Hedlund, B., Kämpfer, P., Rainey, F.A., Whitman, W.B., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA; Bergey’s Manual Trust: Athen, GA, USA, 2015; pp. 1–12. [Google Scholar] [CrossRef]
- Bingle, L.E.; Bailey, C.M.; Pallen, M.J. Type VI secretion: A beginner’s guide. Curr. Opin. Microbiol. 2008, 11, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Gao, N.; Xia, M.; Dai, J.; Yu, D.; An, W.; Li, S.; Liu, S.; He, P.; Zhang, L.; Wu, Z.; et al. Both widespread PEP-CTERM proteins and exopolysaccharides are required for floc formation of Zoogloea resiniphila and other activated sludge bacteria. Environ. Microbiol. 2018, 20, 1677–1692. [Google Scholar] [CrossRef] [PubMed]
- Chernogor, L.I.; Denikina, N.N.; Belikov, S.I.; Ereskovsky, A.V. Long-Term Cultivation of Primmorphs from Freshwater Baikal Sponges Lubomirskia baikalensis. Mar. Biotechnol. 2011, 13, 782–792. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.A.; Korobeynikov, A.; Lapidus, A.; Prjibelski, A.D.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling Single-Cell Genomes and Mini-Metagenomes From Chimeric MDA Products. J. Comput. Biol. 2013, 20, 714–737. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Armstrong, J.; Raney, B.J.; Streeter, I.; Dunn, M.; Yang, F.; Odom, D.; Flicek, P.; Keane, T.M.; Thybert, D.; et al. Chromosome assembly of large and complex genomes using multiple references. Genome Res. 2018, 28, 1720–1732. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Waterhouse, R.M.; Seppey, M.; Simão, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO Applications from Quality Assessments to Gene Prediction and Phylogenomics. Mol. Biol. Evol. 2018, 35, 543–548. [Google Scholar] [CrossRef]
- Asnicar, F.; Thomas, A.M.; Beghini, F.; Mengoni, C.; Manara, S.; Manghi, P.; Zhu, Q.; Bolzan, M.; Cumbo, F.; May, U.; et al. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat. Commun. 2020, 11, 2500. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Börnigen, D.; Morgan, X.; Huttenhower, C. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat. Commun. 2013, 4, 2304. [Google Scholar] [CrossRef] [PubMed]
- Haack, F.S.; Poehlein, A.; Kröger, C.; Voigt, C.A.; Piepenbring, M.; Bode, H.B.; Daniel, R.; Schäfer, W.; Streit, W.R. Molecular Keys to the Janthinobacterium and Duganella spp. Interaction with the Plant Pathogen Fusarium graminearum. Front. Microbiol. 2016, 7, 1668. [Google Scholar] [CrossRef] [PubMed]
- Van Teeseling, M.C.F.; de Pedro, M.A.; Cava, F. Determinants of Bacterial Morphology: From Fundamentals to Possibilities for Antimicrobial Targeting. Front. Microbiol. 2017, 8, 1264. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.I.; Salama, N.R. The gram-negative bacterial periplasm: Size matters. PLoS Biol. 2018, 16, e2004935. [Google Scholar] [CrossRef] [PubMed]
- Matz, C.; Webb, J.; Schupp, P.J.; Phang, S.Y.; Penesyan, A.; Egan, S.; Steinberg, P.; Kjelleberg, S. Marine Biofilm Bacteria Evade Eukaryotic Predation by Targeted Chemical Defense. PLoS ONE 2008, 3, e2744. [Google Scholar] [CrossRef]
- Silverman, J.M.; Brunet, Y.R.; Cascales, E.; Mougous, J.D. Structure and Regulation of the Type VI Secretion System. Annu. Rev. Microbiol. 2012, 66, 453–472. [Google Scholar] [CrossRef]
- Wang, J.; Brodmann, M.; Basler, M. Assembly and Subcellular Localization of Bacterial Type VI Secretion Systems. Annu. Rev. Microbiol. 2019, 73, 621–638. [Google Scholar] [CrossRef]
- Valdes, N.; Soto, P.; Cottet, L.; Alarcon, P.; Gonzalez, A.; Castillo, A.; Corsini, G.; Tello, M. Draft genome sequence of Janthinobacterium lividum strain MTR reveals its mechanism of capnophilic behavior. Stand. Genom. Sci. 2015, 10, 110. [Google Scholar] [CrossRef]
- Alexander, B.E.; Mueller, B.; Vermeij, M.J.; van der Geest, H.H.; de Goeij, J.M. Biofouling of inlet pipes affects water quality in running seawater aquaria and compromises sponge cell proliferation. PeerJ 2015, 3, e1430. [Google Scholar] [CrossRef]
- Pantanella, F.; Berlutti, F.; Passariello, C.; Sarli, S.; Morea, C.; Schippa, S. Violacein and biofilm production in Janthinobacterium lividum. J. Appl. Microbiol. 2006, 102, 992–999. [Google Scholar] [CrossRef] [PubMed]
- Coenye, T. The family Burkholderiaceae. In The Prokaryotes: Alphaproteobacteria and Betaproteobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: New York, NY, USA, 2014; pp. 759–776. [Google Scholar] [CrossRef]
- Webster, N.S.; Smith, L.D.; Heyward, A.J.; Watts, J.; Webb, R.I.; Blackall, L.L.; Negri, A.P. Metamorphosis of a Scleractinian Coral in Response to Microbial Biofilms. Appl. Environ. Microbiol. 2004, 70, 1213–1221. [Google Scholar] [CrossRef] [PubMed]
- Olson, J.; Gochfeld, D.; Slattery, M. Aplysina red band syndrome: A new threat to Caribbean sponges. Dis. Aquat. Org. 2006, 71, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Hoegh-Guldberg, O.; Mumby, P.J.; Hooten, A.J.; Steneck, R.S.; Greenfield, P.; Gomez, E.; Harvell, C.D.; Sale, P.F.; Edwards, A.J.; Caldeira, K.; et al. Coral Reefs Under Rapid Climate Change and Ocean Acidification. Science 2007, 318, 1737–1742. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.W.; Richardson, L.L. A meta-analysis of 16S rRNA gene clone libraries from the polymicrobial black band disease of corals. FEMS Microbiol. Ecol. 2010, 75, 231–241. [Google Scholar] [CrossRef]
- Pita, L.; Rix, L.; Slaby, B.M.; Franke, A.; Hentschel, U. The sponge holobiont in a changing ocean: From microbes to ecosystems. Microbiome 2018, 6, 46. [Google Scholar] [CrossRef]
- Webster, N.S. Sponge disease: A global threat? Environ. Microbiol. 2007, 9, 1363–1375. [Google Scholar] [CrossRef]
- Stabili, L.; Cardone, F.; Alifano, P.; Tredici, S.M.; Piraino, S.; Corriero, G.; Gaino, E. Epidemic Mortality of the Sponge Ircinia variabilis (Schmidt, 1862) Associated to Proliferation of a Vibrio Bacterium. Microb. Ecol. 2012, 64, 802–813. [Google Scholar] [CrossRef]
- Di Camillo, C.G.; Bartolucci, I.; Cerrano, C.; Bavestrello, G. Sponge disease in the Adriatic Sea. Mar. Ecol. 2012, 34, 62–71. [Google Scholar] [CrossRef]
- Blanquer, A.; Uriz, M.J.; Cebrian, E.; Galand, P.E. Snapshot of a Bacterial Microbiome Shift during the Early Symptoms of a Massive Sponge Die-Off in the Western Mediterranean. Front. Microbiol. 2016, 7, 752. [Google Scholar] [CrossRef]
- Webster, N.S.; Xavier, J.R.; Freckelton, M.; Motti, C.A.; Cobb, R. Shifts in microbial and chemical patterns within the marine sponge Aplysina aerophoba during a disease outbreak. Environ. Microbiol. 2008, 10, 3366–3376. [Google Scholar] [CrossRef] [PubMed]
- Luter, H.M.; Whalan, S.; Webster, N.S. Exploring the Role of Microorganisms in the Disease-Like Syndrome Affecting the Sponge Ianthella basta. Appl. Environ. Microbiol. 2010, 76, 5736–5744. [Google Scholar] [CrossRef] [PubMed]
- Erwin, P.M.; Pita, L.; López-Legentil, S.; Turon, X. Stability of Sponge-Associated Bacteria over Large Seasonal Shifts in Temperature and Irradiance. Appl. Environ. Microbiol. 2012, 78, 7358–7368. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Liu, M.; Simister, R.; Webster, N.; Thomas, T. Marine microbial symbiosis heats up: The phylogenetic and functional response of a sponge holobiont to thermal stress. ISME J. 2013, 7, 991–1002. [Google Scholar] [CrossRef]
- Fujise, L.; Yamashita, H.; Suzuki, G.; Sasaki, K.; Liao, L.M.; Koike, K. Moderate Thermal Stress Causes Active and Immediate Expulsion of Photosynthetically Damaged Zooxanthellae (Symbiodinium) from Corals. PLoS ONE 2014, 9, e114321. [Google Scholar] [CrossRef] [PubMed]
- Sweet, M.; Bulling, M.; Cerrano, C. A novel sponge disease caused by a consortium of micro-organisms. Coral Reefs 2015, 34, 871–883. [Google Scholar] [CrossRef]
- Webster, N.; Negri, A.; Webb, R.; Hill, R. A spongin-boring a-proteobacterium is the etiological agent of disease in the Great Barrier Reef sponge Rhopaloeides odorabile. Mar. Ecol. Prog. Ser. 2002, 232, 305–309. [Google Scholar] [CrossRef]
- Choudhury, J.D.; Pramanik, A.; Webster, N.; Llewellyn, L.E.; Gachhui, R.; Mukherjee, J. The Pathogen of the Great Barrier Reef Sponge Rhopaloeides odorabile Is a New Strain of Pseudoalteromonas agarivorans Containing Abundant and Diverse Virulence-Related Genes. Mar. Biotechnol. 2015, 17, 463–478. [Google Scholar] [CrossRef]

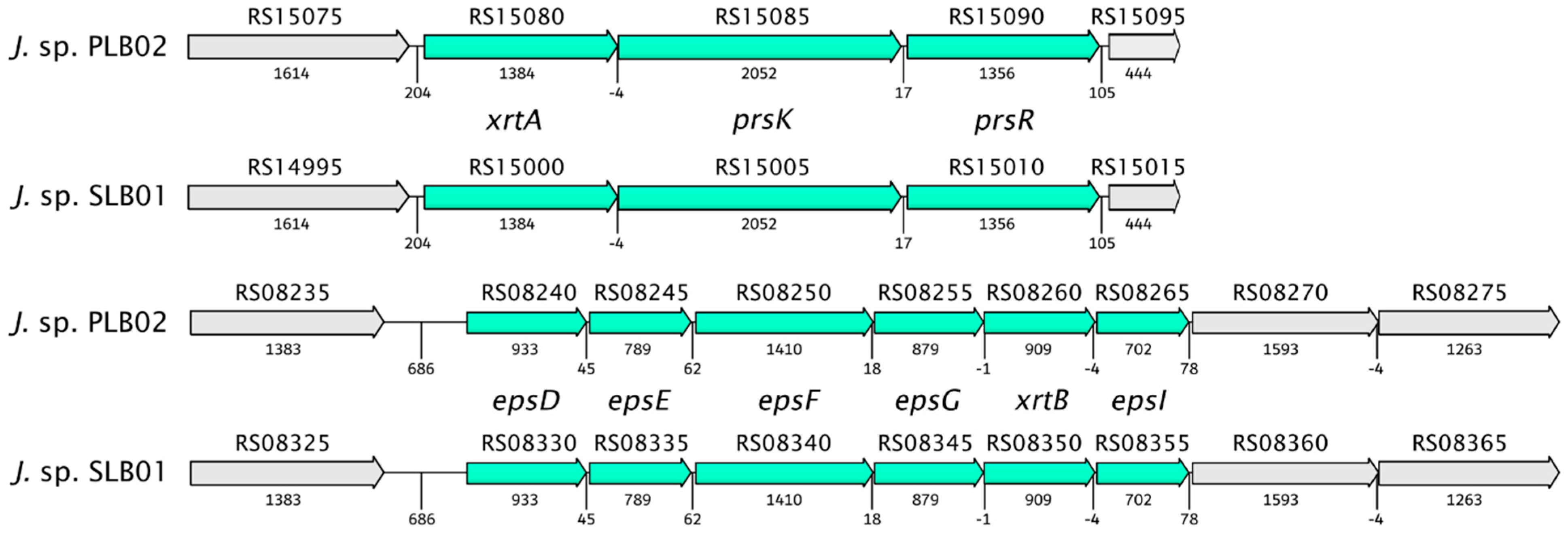
| Property | Janthinobacterium sp. Strain PLB02 | Janthinobacterium sp. Strain SLB01 |
|---|---|---|
| Classification | Proteobacteria, Betaproteobacteria, Burkholderiales, Oxalobacteraceae, Janthinobacterium | |
| Gram stain | negative | |
| Cell shape | rod shape | |
| Motility | twitching | |
| Temperature | 15–22 °C | |
| Pigmentation | purple | |
| Host | primmorphs L. baikalensis sponge L. baikalensis | |
| Sequencing method | Illumina MiSeq (2 × 250) | |
| Raw reads | 7,471,704 | 12,099,942 * |
| Genome coverage | 180x | 180x |
| GenBank accession number | NZ_CP071520.1 | VZAB00000000.1 |
| Genome size, bp | 6,417,505 | 6,467,981 |
| Assembly method | SPAdes version 3.11.0 | |
| Annotation method | NCBI Prokaryotic Genome Annotation Pipeline | |
| Number of contigs | 1 | 2 |
| GC content | 62.65% | 62.63% |
| Number of genes | 5651 | 5643 |
| Protein-coding sequences | 5510 | 5502 |
| tRNAs | 76 | 76 |
| Noncoding RNAs | 4 | 4 |
| Pseudogenes | 57 | 56 |
| Gene Name | Locus Tag | Begin | End | Homology % | Length, bp | ||||
|---|---|---|---|---|---|---|---|---|---|
| Strain | SLB01 | PLB02 | SLB01 | PLB02 | SLB01 | PLB02 | SLB01 | PLB02 | |
| vioA | F3B38_RS17235 | J3P46_17335 | 1353472 | 3909260 | 1354779 | 3910567 | 100 | 1308 | 1308 |
| vioB | F3B38_RS17240 | J3P46_17340 | 1354776 | 3910564 | 1357796 | 3913584 | 100 | 3021 | 3021 |
| vioC | F3B38_RS17245 | J3P46_17345 | 1357798 | 3913586 | 1359087 | 3914875 | 100 | 1290 | 1290 |
| vioD | F3B38_RS17250 | J3P46_17350 | 1359087 | 3914929 | 1360205 | 3915993 | 100 | 1119 | 1065 |
| vioE | F3B38_RS17255 | J3P46_17355 | 1360216 | 3916004 | 1360216 | 3916585 | 100 | 582 | 582 |
| Component | Locus Tag (PLB02) | Length, aa (PLB02) | Locus Tag (SLB01) | Length, aa (SLB01) | Identity % | Annotation |
|---|---|---|---|---|---|---|
| RpoN | J3P46_RS03515 | 1479 | F3B38_RS03495 | 1479 | 100 | RNA polymerase factor sigma-54 (RpoN) |
| PepA | J3P46_RS08815 | 1494 | F3B38_RS08900 | 1494 | 100 | leucyl aminopeptidase (PepA Zooglea sp.) Over expression could circumvent the requirement of rpoN, prsK and prsR for the floc-forming phenotype by fixing the exopolysaccharides to bacterial cells. |
| XrtA | J3P46_RS15080 | 1389 | F3B38_RS15000 | 1389 | 100 | TIGR03013 family PEP-CTERM/XrtA system glycosyltransferase putative exosortase XrtA (previously called EpsH) |
| PrsK | J3P46_RS15085 | 2052 | F3B38_RS15005 | 2052 | 100 | PEP-CTERM system histidine kinase PrsK Sensor histidine kinase of a two-component system |
| PrsR | J3P46_RS15090 | 1356 | F3B38_RS15010 | 1356 | 100 | PEP-CTERM-box response regulator transcription factor PrsR Sensor histidine kinase of a two-component system |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chernogor, L.; Eliseikina, M.; Petrushin, I.; Chernogor, E.; Khanaev, I.; Belikov, S.I. Janthinobacterium sp. Strain SLB01 as Pathogenic Bacteria for Sponge Lubomirskia baikalensis. Pathogens 2023, 12, 8. https://doi.org/10.3390/pathogens12010008
Chernogor L, Eliseikina M, Petrushin I, Chernogor E, Khanaev I, Belikov SI. Janthinobacterium sp. Strain SLB01 as Pathogenic Bacteria for Sponge Lubomirskia baikalensis. Pathogens. 2023; 12(1):8. https://doi.org/10.3390/pathogens12010008
Chicago/Turabian StyleChernogor, Lubov, Marina Eliseikina, Ivan Petrushin, Ekaterina Chernogor, Igor Khanaev, and Sergei I. Belikov. 2023. "Janthinobacterium sp. Strain SLB01 as Pathogenic Bacteria for Sponge Lubomirskia baikalensis" Pathogens 12, no. 1: 8. https://doi.org/10.3390/pathogens12010008
APA StyleChernogor, L., Eliseikina, M., Petrushin, I., Chernogor, E., Khanaev, I., & Belikov, S. I. (2023). Janthinobacterium sp. Strain SLB01 as Pathogenic Bacteria for Sponge Lubomirskia baikalensis. Pathogens, 12(1), 8. https://doi.org/10.3390/pathogens12010008






