A Putative New Role of Tv-PSP1 Recognizes IRE and ERE Hairpin Structures from Trichomonas vaginalis
Abstract
1. Introduction
2. Materials and Methods
2.1. Transcription in Vitro
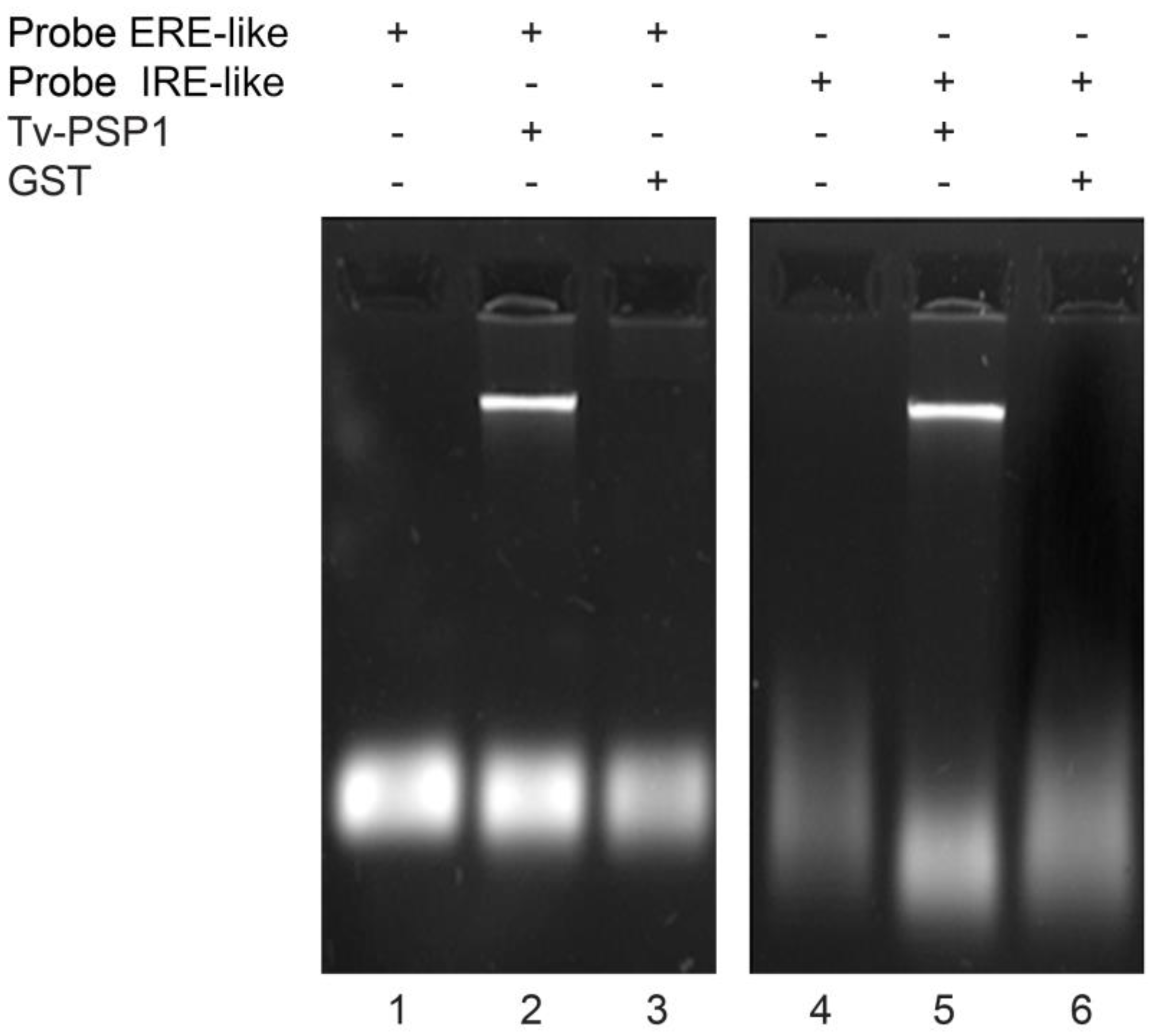
2.2. Purification of Recombinant TV-PSP1 and REMSA Assay
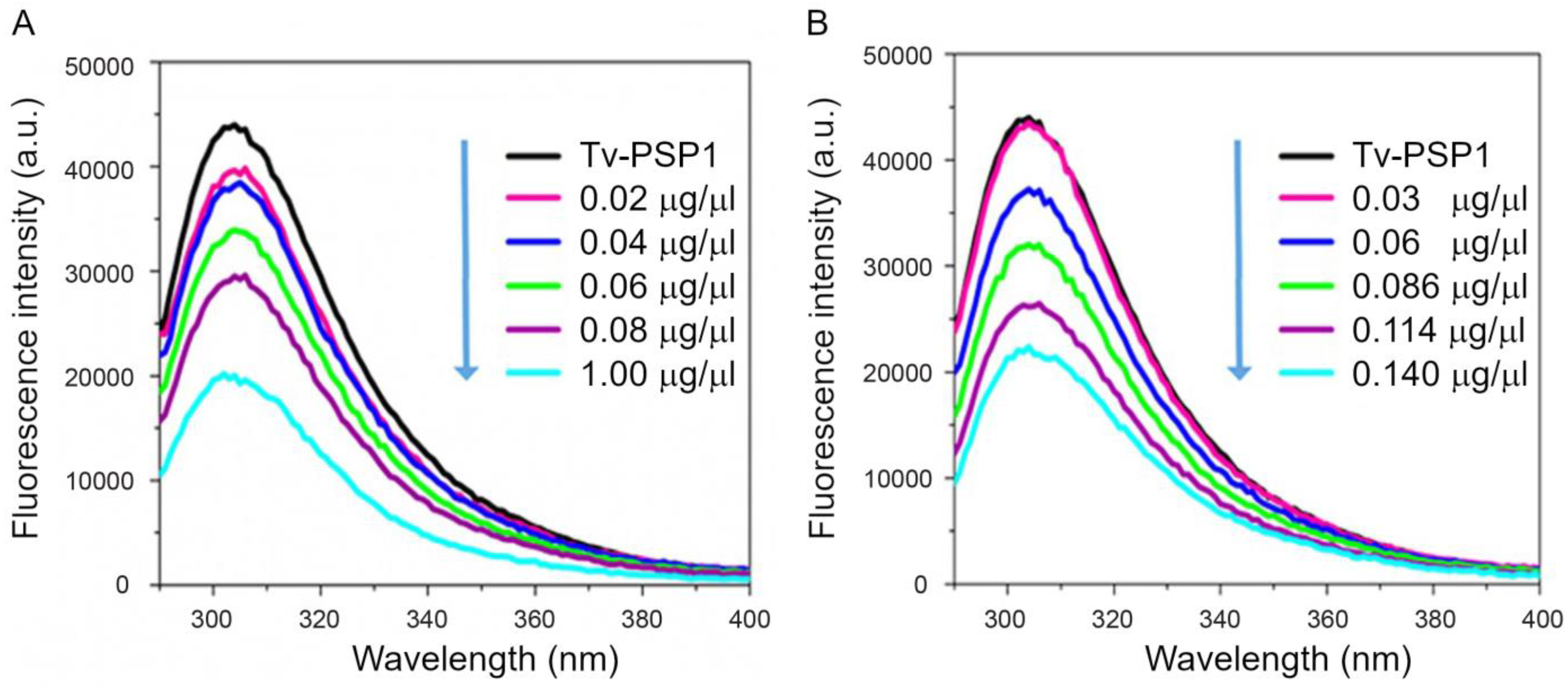
2.3. Intrinsic Fluorescence Studies
2.4. Recombinant Tv-PSP1 Crystallization and Diffraction
2.5. Tv-PSP1 Structure Determination by X-Ray
2.6. Final Crystal Structure Analysis
2.7. Molecular Dynamics Simulations of Tv-PSP1 Trimer Assembles
2.8. Computational Docking: Interaction between Tv-PSP1 Trimer and Stem-Loop Structures
2.9. Binding Free Energy (ΔGb) of Tv-PSP1 Trimer–RNA: Electrostatic and Nonelectrostatic Contributions
2.9.1. Electrostatic Calculations (ΔGelec)
2.9.2. Nonelectrostatic Calculations (ΔGnon-elec)
3. Results
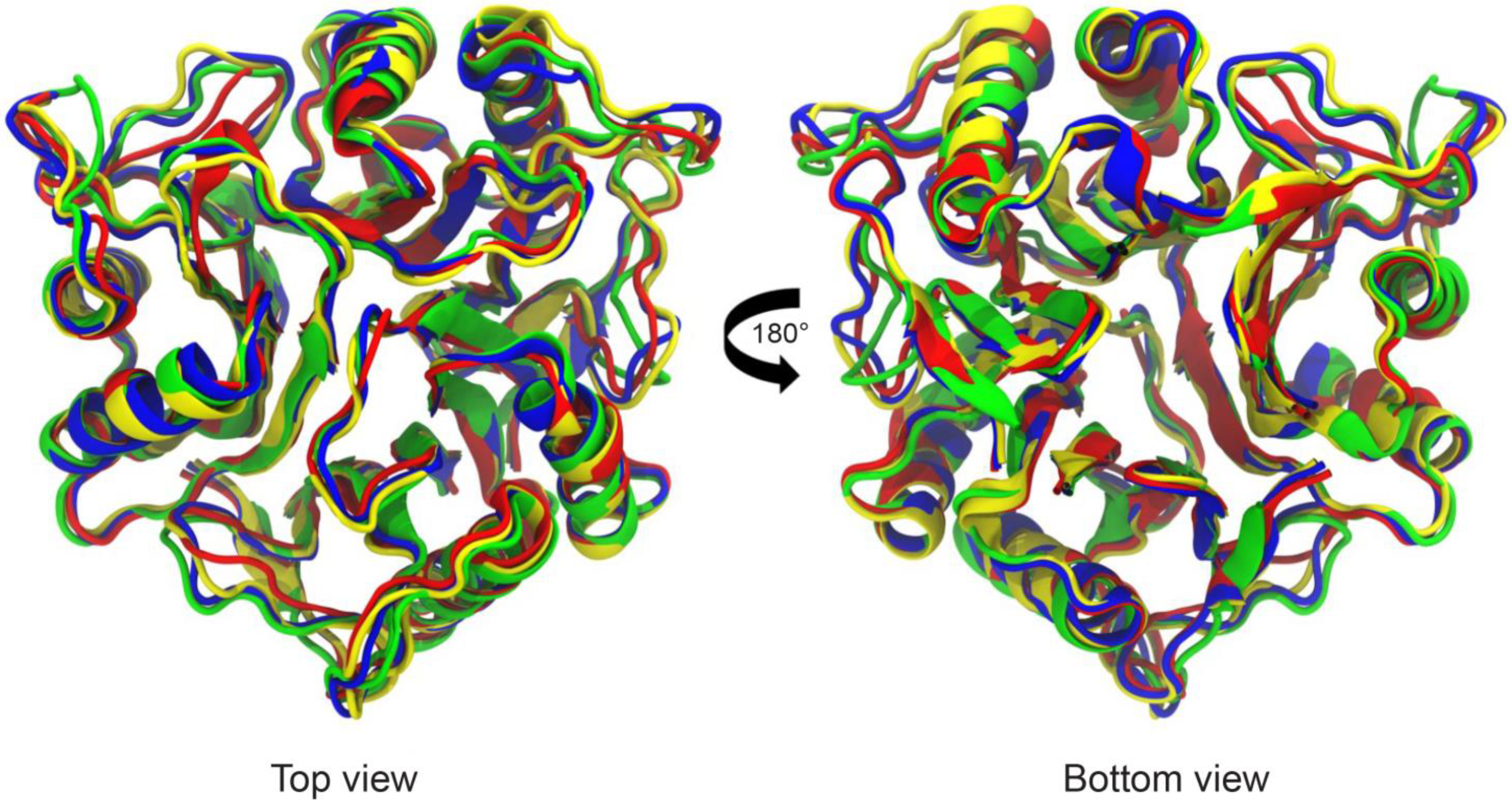
3.1. Crystal Structure and Thermal Stability
3.2. Stem-Loop Structure Interacts with Tv-PSP1
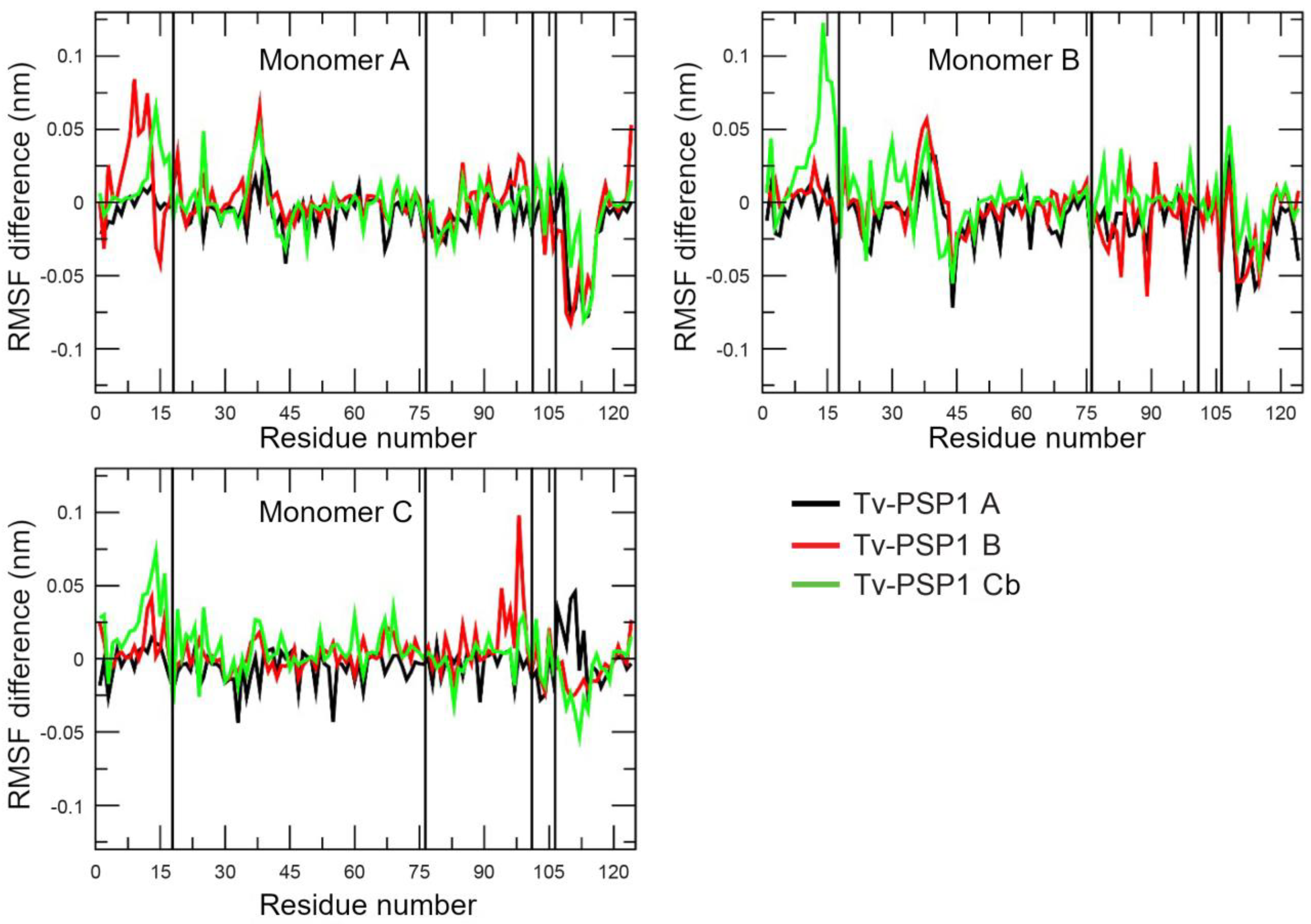
3.3. Tv-PSP1 Trimer Molecular Dynamics and Disulfide Effects on Structure
3.4. Tv-PSP1 Trimer-Element Hairpin-Loop (IRE) Interaction: Docking Studies and Electrostatic Calculations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martin, K.C.; Ephrussi, A. MRNA Localization: Gene Expression in the Spatial Dimension. Cell 2009, 136, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Matoulkova, E.; Michalova, E.; Vojtesek, B.; Hrstka, R. The Role of the 3′ Untranslated Region in Post-Transcriptional Regulation of Protein Expression in Mammalian Cells. RNA Biol. 2012, 9, 563–576. [Google Scholar] [CrossRef] [PubMed]
- Court, D.L.; Gan, J.; Liang, Y.-H.; Shaw, G.X.; Tropea, J.E.; Costantino, N.; Waugh, D.S.; Ji, X. RNase III: Genetics and Function; Structure and Mechanism. Annu. Rev. Genet. 2013, 47, 405–431. [Google Scholar] [CrossRef] [PubMed]
- Marvin, M.C.; Walker, S.C.; Fierke, C.A.; Engelke, D.R. Binding and Cleavage of Unstructured RNA by Nuclear RNase P. RNA 2011, 17, 1429–1440. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Angulo, E.; Calla-Choque, J.; Mancilla-Olea, M.; Arroyo, R. RNA-Binding Proteins in Trichomonas Vaginalis: Atypical Multifunctional Proteins. Biomolecules 2015, 5, 3354–3395. [Google Scholar] [CrossRef]
- Solano-González, E.; Burrola-Barraza, E.; León-Sicairos, C.; Avila-González, L.; Gutiérrez-Escolano, L.; Ortega-López, J.; Arroyo, R. The Trichomonad Cysteine Proteinase TVCP4 Transcript Contains an Iron-Responsive Element. FEBS Lett. 2007, 581, 2919–2928. [Google Scholar] [CrossRef]
- Ramón-Luing, L.d.L.; Rendón-Gandarilla, F.J.; Puente-Rivera, J.; Ávila-González, L.; Arroyo, R. Identification and characterization of the immunogenic cytotoxic TvCP39 proteinase gene of Trichomonas vaginalis. Int. J. Biochem. Cell. Biol. 2011, 43, 1500–1511. [Google Scholar] [CrossRef]
- León-Sicairos, C.R.; León-Felix, J.; Arroyo, R. tvcp12: A novel Trichomonas vaginalis cathepsin L-like cysteine-proteinase encoding gene. Microbiology 2004, 150 Pt 5, 1131–1138. [Google Scholar] [CrossRef][Green Version]
- Carvajal-Gamez, B.I.; Carrillo, L.V.; Torres-Romero, J.C.; Camacho-Nuez, M.; Ponce-Regalado, M.D.; Camarillo, C.L.; Alvarez-Sánchez, M.E. Recombinant Trichomonas Vaginalis EIF-5A Protein Expressed from a Eukaryotic System Binds Specifically to Mammalian and Putative Trichomonal EIF-5A Response Elements (EREs). Parasitol. Int. 2016, 65, 625–631. [Google Scholar] [CrossRef]
- Carvajal-Gamez, B.I.; Quintas-Granados, L.I.; Arroyo, R.; Vázquez-Carrillo, L.I.; Ramón-Luing, L.D.; los, A.; Carrillo-Tapia, E.; Alvarez-Sánchez, M.E. Putrescine-Dependent Re-Localization of TvCP39, a Cysteine Proteinase Involved in Trichomonas Vaginalis Cytotoxicity. PLoS ONE 2014, 9, e107293. [Google Scholar] [CrossRef]
- Enos-Berlage, J.L.; Langendorf, M.J.; Downs, D.M. Complex Metabolic Phenotypes Caused by a Mutation in YjgF, Encoding a Member of the Highly Conserved YER057c/YjgF Family of Proteins. J. Bacteriol. 1998, 180, 6519–6528. [Google Scholar] [CrossRef]
- Kanouchi, H.; Matsumoto, M.; Taga, M.; Yamada, K.; Oka, T.; Toné, S.; Minatogawa, Y. Nuclear Transfer of Perchloric Acid-Soluble Protein by Endoplasmic Reticulum Stressors. Protein Sci. 2005, 14, 2344–2349. [Google Scholar] [CrossRef]
- Kim, J.-M.; Yoshikawa, H.; Shirahige, K. A Member of the YER057c/Yjgf/Uk114 Family Links Isoleucine Biosynthesis and Intact Mitochondria Maintenance in Saccharomyces Cerevisiae: Ile Biosynthesis and Mitochondria Maintenance. Genes Cells 2001, 6, 507–517. [Google Scholar] [CrossRef]
- Morishita, R.; Kawagoshi, A.; Sawasaki, T.; Madin, K.; Ogasawara, T.; Oka, T.; Endo, Y. Ribonuclease Activity of Rat Liver Perchloric Acid-Soluble Protein, a Potent Inhibitor of Protein Synthesis. J. Biol. Chem. 1999, 274, 20688–20692. [Google Scholar] [CrossRef]
- da Fonseca Pires, S.; Fialho, L.C.; Silva, S.O.; Melo, M.N.; de Souza, C.C.; Tafuri, W.L.; Bruna Romero, O.; de Andrade, H.M. Identification of Virulence Factors in Leishmania Infantum Strains by a Proteomic Approach. J. Proteome Res. 2014, 13, 1860–1872. [Google Scholar] [CrossRef]
- López-Rosas, I.; Marchat, L.A.; Olvera, B.G.; Guillen, N.; Weber, C.; Hernández de la Cruz, O.; Ruíz-García, E.; Astudillo-de la Vega, H.; López-Camarillo, C. Proteomic Analysis Identifies Endoribouclease EhL-PSP and EhRRP41 Exosome Protein as Novel Interactors of EhCAF1 Deadenylase. J. Proteom. 2014, 111, 59–73. [Google Scholar] [CrossRef]
- Villalobos-Osnaya, A.; Garza-Ramos, G.; Serratos, I.N.; Millán-Pacheco, C.; González-Robles, A.; Arroyo, R.; Quintas-Granados, L.I.; Alvarez-Sanchez, M.E. Identification of a Perchloric Acid-Soluble Protein (PSP)-like Ribonuclease from Trichomonas Vaginalis. Parasitol. Res. 2018, 117, 3639–3652. [Google Scholar] [CrossRef]
- Kim, H.J.; Kwon, A.-R.; Lee, B.-J. A Novel Chlorination-Induced Ribonuclease YabJ from Staphylococcus Aureus. Biosci. Rep. 2018, 38, BSR20180768. [Google Scholar] [CrossRef]
- Müller, A.; Langklotz, S.; Lupilova, N.; Kuhlmann, K.; Bandow, J.E.; Leichert, L.I.O. Activation of RidA Chaperone Function by N-Chlorination. Nat. Commun. 2014, 5, 5804. [Google Scholar] [CrossRef]
- Park, O.H.; Ha, H.; Lee, Y.; Boo, S.H.; Kwon, D.H.; Song, H.K.; Kim, Y.K. Endoribonucleolytic Cleavage of M6A-Containing RNAs by RNase P/MRP Complex. Mol. Cell 2019, 74, 494–507.e8. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66 Pt 2, 125–132. [Google Scholar] [CrossRef]
- Djinović Carugo, K.; Saraste, M.; Oka, T. Crystallization and Preliminary X-Ray Diffraction Studies of Perchloric Acid Soluble Protein (PSP) from Rat Liver. Acta Crystallogr. D Biol. Crystallogr. 1999, 55 Pt 3, 667–668. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser Crystallographic Software. J. Appl. Crystallogr. 2007, 40 Pt 4, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E.J. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallogr. D Biol. Crystallogr. 1997, 53 Pt 3, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A Comprehensive Python-Based System for Macromolecular Structure Solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66 Pt 2, 213–221. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and Development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66 Pt 4, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-Atom Structure Validation for Macromolecular Crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010, 66 Pt 1, 12–21. [Google Scholar] [CrossRef]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011, 67 Pt 4, 235–242. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Jo, S.; Lim, J.B.; Klauda, J.B.; Im, W. CHARMM-GUI Membrane Builder for Mixed Bilayers and Its Application to Yeast Membranes. Biophys. J. 2009, 97, 50–58. [Google Scholar] [CrossRef]
- Addess, K.J.; Basilion, J.P.; Klausner, R.D.; Rouault, T.A.; Pardi, A. Structure and Dynamics of the Iron Responsive Element RNA: Implications for Binding of the RNA by Iron Regulatory Binding Proteins. J. Mol. Biol. 1997, 274, 72–83. [Google Scholar] [CrossRef] [PubMed]
- McCallum, S.A.; Pardi, A. Refined Solution Structure of the Iron-Responsive Element RNA Using Residual Dipolar Couplings. J. Mol. Biol. 2003, 326, 1037–1050. [Google Scholar] [CrossRef]
- Huang, J.; MacKerell, A.D. CHARMM36 All-Atom Additive Protein Force Field: Validation Based on Comparison to NMR Data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S.-Y. The HDOCK Server for Integrated Protein-Protein Docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, D.; Zhou, P.; Li, B.; Huang, S.-Y. HDOCK: A Web Server for Protein-Protein and Protein-DNA/RNA Docking Based on a Hybrid Strategy. Nucleic Acids Res. 2017, 45, W365–W373. [Google Scholar] [CrossRef]
- Yan, Y.; Wen, Z.; Wang, X.; Huang, S.-Y. Addressing Recent Docking Challenges: A Hybrid Strategy to Integrate Template-Based and Free Protein-Protein Docking. Proteins 2017, 85, 497–512. [Google Scholar] [CrossRef]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An Automated Pipeline for the Setup of Poisson-Boltzmann Electrostatics Calculations. Nucleic Acids Res. 2004, 32, W665–W667. [Google Scholar] [CrossRef]
- MacKerell, A.D.; Bashford, D.; Bellott, M.; Dunbrack, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Levy, R.M.; Zhang, L.Y.; Gallicchio, E.; Felts, A.K. On the Nonpolar Hydration Free Energy of Proteins: Surface Area and Continuum Solvent Models for the Solute–Solvent Interaction Energy. J. Am. Chem. Soc. 2003, 125, 9523–9530. [Google Scholar] [CrossRef]
- Sitkoff, D.; Sharp, K.A.; Honig, B. Accurate Calculation of Hydration Free Energies Using Macroscopic Solvent Models. J. Phys. Chem. 1994, 98, 1978–1988. [Google Scholar] [CrossRef]
- Martínez-Hernández, J.C.; Serratos, I.N.; Millán-Pacheco, C.; Rojo-Domínguez, A.; Padilla-Zúñiga, J. What Comparisons of Natural and Chimeric Contacts Reveal about Inhibition of Human Cathepsins K, L and S by Their Prosegments. J. Mex. Chem. Soc. 2019, 63, 13–24. [Google Scholar] [CrossRef]
- Serratos, I.N.; Olayo, R.; Millán-Pacheco, C.; Morales-Corona, J.; Vicente-Escobar, J.O.; Soto-Estrada, A.M.; Córdoba-Herrera, J.G.; Uribe, O.; Gómez-Quintero, T.; Arroyo-Ornelas, M.Á.; et al. Modeling Integrin and Plasma-Polymerized Pyrrole Interactions: Chemical Diversity Relevance for Cell Regeneration. Sci. Rep. 2019, 9, 7009. [Google Scholar] [CrossRef] [PubMed]
- Sasagawa, T.; Oka, T.; Tokumura, A.; Nishimoto, Y.; Muñoz, S.; Kuwahata, M.; Okita, M.; Tsuji, H.; Natori, Y. Analysis of the Fatty Acid Components in a Perchloric Acid-Soluble Protein. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 1999, 1437, 317–324. [Google Scholar] [CrossRef]
- Manjasetty, B.A.; Delbrück, H.; Pham, D.-T.; Mueller, U.; Fieber-Erdmann, M.; Scheich, C.; Sievert, V.; Büssow, K.; Neisen, F.H.; Weihofen, W.; et al. Crystal Structure of Homo Sapiens Protein Hp14.5: Crystal Structure of Hp14.5. Proteins 2004, 54, 797–800. [Google Scholar] [CrossRef]
- Parsons, L.; Bonander, N.; Eisenstein, E.; Gilson, M.; Kairys, V.; Orban, J. Solution Structure and Functional Ligand Screening of HI0719, a Highly Conserved Protein from Bacteria to Humans in the YjgF/YER057c/UK114 Family. Biochemistry 2003, 42, 80–89. [Google Scholar] [CrossRef]
- Volz, K. A Test Case for Structure-Based Functional Assignment: The 1.2 Å Crystal Structure of the YjgF Gene Product from Escherichia Coli. Protein Sci. 2008, 8, 2428–2437. [Google Scholar] [CrossRef]
- Law, M.J. The Role of Positively Charged Amino Acids and Electrostatic Interactions in the Complex of U1A Protein and U1 Hairpin II RNA. Nucleic Acids Res. 2006, 34, 275–285. [Google Scholar] [CrossRef]
- Blanco, C.; Bayas, M.; Yan, F.; Chen, I.A. Analysis of Evolutionarily Independent Protein-RNA Complexes Yields a Criterion to Evaluate the Relevance of Prebiotic Scenarios. Curr. Biol. 2018, 28, 526–537.e5. [Google Scholar] [CrossRef]
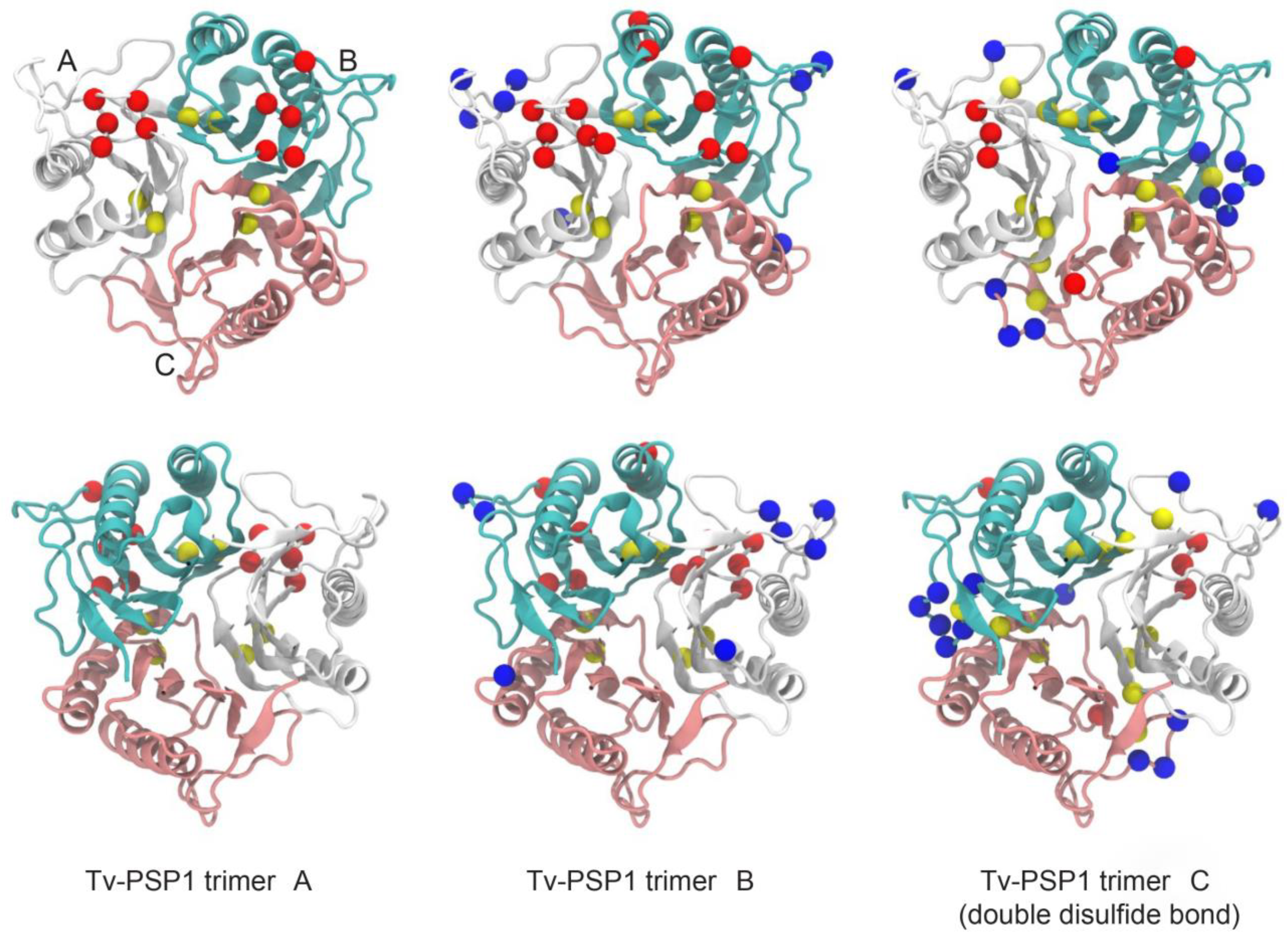

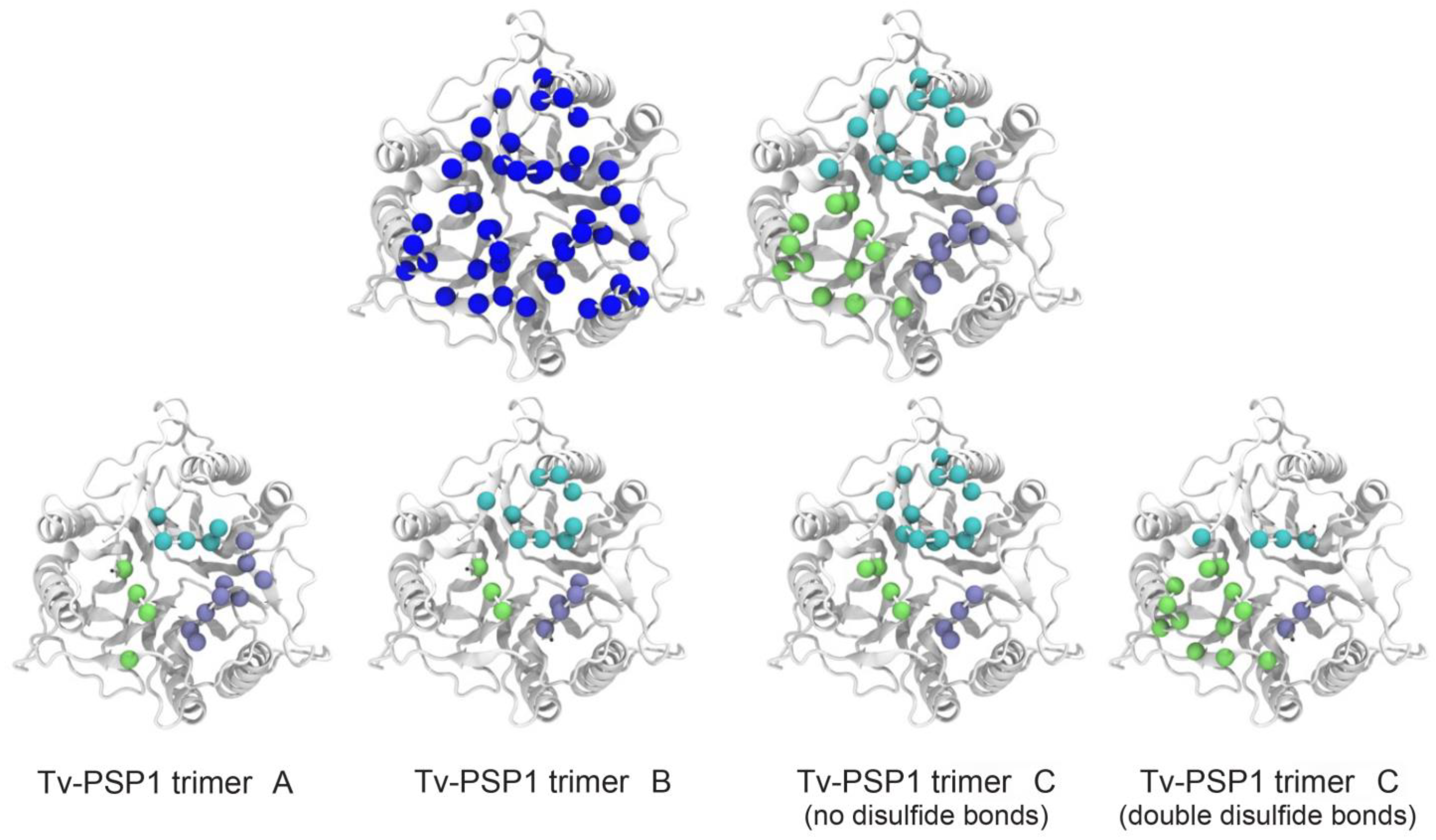
| PDB ID | 7KGC |
|---|---|
| Data collection | |
| Wavelength (Å) | 0.97 |
| Space group | P63 |
| Cell dimensions | |
| a, b, c (Å) | 81.9, 81.9, 129.3 |
| α, β, γ (°) | 90.0, 90.0, 120.0 |
| Resolution (Å) | 25.0–1.95 (2.0–1.95) |
| Unique reflections | 35 775 (2529) |
| I/σI | 14.1 (2.4) |
| Rmerge (%) | 8.2 (44.6) |
| CC1/2 | 0.99 (0.80) |
| Completeness (%) | 99.7 (99.9) |
| Multiplicity | 3.5 (3.5) |
| Mosaicity (°) | 0.3 |
| Refinement | |
| Resolution (Å) | 25.0–1.95 |
| R (%) | 19.5 |
| Rfree (%) | 24.1 |
| Number of atoms | |
| Protein | 3151 |
| TRIS | 24 |
| Water | 295 |
| B factors (Å2) | |
| Protein | 37.7 |
| TRIS | 50.1 |
| Water | 37.9 |
| All atoms | 37.0 |
| Wilson plot | 23.7 |
| RMSD | |
| Bond lengths (Å) | 0.017 |
| Bond angles (°) | 1.474 |
| Ramachandran plot | |
| Most favored regions (%) | 97.78 |
| Additional allowed regions (%) | 1.97 |
| Disallowed regions (%) | 0.25 |
| Protein | Disulfide Bond |
|---|---|
| Tv-PSP1 trimer A | CYS 76-CYS 104 (Chain A) CYS 76-CYS 104 (Chain B) CYS 76-CYS 104 (Chain C) |
| Tv-PSP1 trimer B | CYS 76-CYS 104 (Chain A) CYS 76-CYS 104 (Chain B) CYS 76-CYS 104 (Chain C) |
| Tv-PSP1 trimer C | Without disulfide bridge |
| Tv-PSP1 trimer Cb | CYS 76-CYS 104 (Chain A) CYS 76-CYS 104 (Chain B) CYS 76-CYS 104 (Chain C) CYS 18 (Chain A)-CYS 101 (Chain B) CYS 18 (Chain C)-CYS 101 (Chain A) CYS 18 (Chain B)-CYS 101 (Chain C) |
| B Factor Value Average by Residue of Main Chain Atoms | ASA of Interfaces and Surfaces on the Crystal ** | ||||||
|---|---|---|---|---|---|---|---|
| AMC | Minimum | Maximum | Interfaces | A | B | C | |
| A * | 19.70 | 12.60 | 49.74 | ASAbio | 1904.80 | 2079.10 | 2015.7 |
| B * | 24.05 | 14.67 | 52.98 | ASAcrys | 1275.91 | 1132.26 | 788.26 |
| C | 55.85 | 30.73 | 88.48 | Surface | A | B | C |
| D | 73.41 | 38.99 | 102.85 | ASAm | 6756.50 | 6910.50 | 6790.90 |
| ASAtrimer | 14,555.4 | 14,494.2 | 14,325.5 | ||||
| RMSD between Monomers | RMSD between Trimers | |||||
|---|---|---|---|---|---|---|
| B | C | TB | TC | |||
| A | RMSD (atoms) | 0.227 (105) | 0.310 (107) | TA | 0.241 (315) | 0.361 (318) |
| A | RMSD (all atoms) | 0.750 (124) | 0.683 (124) | TA | 0.760 (372) | 0.676 (372) |
| B | RMSD (atoms) | 0.00 | 0.352 (93) | TB | 0.00 | 0.402 (267) |
| B | Atoms (all atoms) | 0.00 | 0.951 (124) | TB | 0.00 | 0.980 (372) |
| Cut off 0.6 Å. | Cut off 0.6 Å. | |||||
| Tv-PSP1 A | Tv-PSP1 B | Tv-PSP1 C | Tv-PSP1 Cb | |
|---|---|---|---|---|
| Tv-PSP1 A | 0.0 | 1.08 | 0.97 | 1.13 |
| Tv-PSP1 B | 0.00 | 1.22 | 1.31 | |
| Tv-PSP1 C | 0.00 | 1.19 | ||
| Tv-PSP1 Cb | 0.00 |
| A | B | C | ||
|---|---|---|---|---|
| Tv-PSP1 A | No | No | No | RMSF (+) |
| Tv-PSP1 A | L7 | L7 | No | RMSF (−) |
| Tv-PSP1 B | L1 | No | No | RMSF (+) |
| Tv-PSP1 B | L7 | L7 | No | RMSF (−) |
| Tv-PSP1 C | Ns | L1 | L1 | RMSF (+) |
| Tv-PSP1 Cb | L7 | Ns | Ns | RMSF (−) |
| + | − | Differences |
| Complexes | ΔGsolv (kJ/mol) | ΔGcoul (kJ/mol) | ΔGnon-elec (kJ/mol) | ΔGb * (kJ/mol) |
|---|---|---|---|---|
| Tv-PSP1 trimer A IRE | 142 | −849 | −37 | −744 |
| Tv-PSP1 trimer B IRE | 178 185 212 214 189 129 | −823 −698 −726 −686 −614 −252 | −37 −35 −33 −35 −34 −26 | −682 −548 −547 −507 −459 −149 |
| Tv-PSP1 trimer C IRE | 215 198 | −664 −639 | −34 −35 | −483 −477 |
| Tv-PSP1 trimer Cb IRE | 233 218 273 340 320 | −788 −479 −479 −409 −381 | −44 −31 −40 −43 −39 | −599 −292 −246 −112 −100 |
| Complex | ΔGsolv (kJ/mol) | ΔGcoul (kJ/mol) | ΔGnon-elec (kJ/mol) | ΔGb * (kJ/mol) |
|---|---|---|---|---|
| Tv-PSP1 trimer A–IRE | 136 269 | −832 −631 | −31 −37 | −727 −399 |
| Tv-PSP1 trimer B–IRE | 154 201 188 152 | −836 −639 −482 −343 | −41 −35 −32 −28 | −723 −473 −326 −219 |
| Tv-PSP1 trimer C–IRE | 296 | −46 | −37 | −213 |
| Tv-PSP1 trimer Cb–IRE | 177 | 12 | −32 | 157 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Millán-Pacheco, C.; Arreola, R.; Villalobos-Osnaya, A.; Garza-Ramos, G.; Serratos, I.N.; Díaz-Vilchis, A.; Rudiño-Piñera, E.; Alvarez-Sanchez, M.E. A Putative New Role of Tv-PSP1 Recognizes IRE and ERE Hairpin Structures from Trichomonas vaginalis. Pathogens 2023, 12, 79. https://doi.org/10.3390/pathogens12010079
Millán-Pacheco C, Arreola R, Villalobos-Osnaya A, Garza-Ramos G, Serratos IN, Díaz-Vilchis A, Rudiño-Piñera E, Alvarez-Sanchez ME. A Putative New Role of Tv-PSP1 Recognizes IRE and ERE Hairpin Structures from Trichomonas vaginalis. Pathogens. 2023; 12(1):79. https://doi.org/10.3390/pathogens12010079
Chicago/Turabian StyleMillán-Pacheco, César, Rodrigo Arreola, Alma Villalobos-Osnaya, Georgina Garza-Ramos, Iris N. Serratos, Adelaida Díaz-Vilchis, Enrique Rudiño-Piñera, and María Elizbeth Alvarez-Sanchez. 2023. "A Putative New Role of Tv-PSP1 Recognizes IRE and ERE Hairpin Structures from Trichomonas vaginalis" Pathogens 12, no. 1: 79. https://doi.org/10.3390/pathogens12010079
APA StyleMillán-Pacheco, C., Arreola, R., Villalobos-Osnaya, A., Garza-Ramos, G., Serratos, I. N., Díaz-Vilchis, A., Rudiño-Piñera, E., & Alvarez-Sanchez, M. E. (2023). A Putative New Role of Tv-PSP1 Recognizes IRE and ERE Hairpin Structures from Trichomonas vaginalis. Pathogens, 12(1), 79. https://doi.org/10.3390/pathogens12010079






