Independent Circulation of Leishmania major and Leishmania tropica in Their Respective Sandfly Vectors for Transmission of Zoonotic and Chronic Cutaneous Leishmaniasis Co-Existing in a Mixed Focus of Central Tunisia
Abstract
1. Introduction
2. Material and Methods
2.1. Study Site
2.2. Sandfly Trapping and Identification
2.3. Detection of Leishmania DNA in Female Sandflies
2.4. Detection of Leishmania DNA, DNA Sequencing and Phylogenetic Analysis
2.5. Data Analysis
Sandfly Species Diversity
- pi = the proportional abundance or percent abundance of a species present (pi = ni/N).
- ni = the number of individuals counted for the species.
- N = the total number of individuals counted, all species combined.
- S = the total or cardinal number of the list of present species.
3. Results
3.1. Sandfly Fauna
3.2. Leishmania Detection
3.3. Leishmania DNA Sequencing and Phylogenetic Analysis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alvar, J.; Vélez, I.D.; Bern, C.; Herrero, M.; Desjeux, P.; Cano, J.; Jannin, J.; de Boer, M. Leishmaniasis worldwide and global estimates of its incidence. PLoS ONE 2012, 7, e35671. [Google Scholar] [CrossRef] [PubMed]
- Kallel, K.; Pratlong, F.; Belhadj, S.; Cherif, F.; Hammami, M.; Dedet, J.P.; Chaker, E. Cutaneous leishmaniasis in Tunisia: Results of the iso-enzymatic characterization of 71 Strains. Ann. Trop. Med. Parasitol. 2005, 99, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Aoun, K.; Amri, F.; Chouihi, E.; Haouas, N.; Bedoui, K.; Benikhlef, R.; Ghrab, J.; Babba, H.; Chahed, M.K.; Harrat, Z.; et al. Épidémiologie de Leishmania (L.) infantum, L. major et L. killicki en Tunisie: Résultats et analyse de l’identification de 226 isolats humains et canins et revue de la littérature. Bull. Soc. Pathol. Exot. 2008, 101, 323–328. [Google Scholar] [CrossRef]
- Haouas, N.; Chaker, E.; Chargui, N.; Gorcii, M.; Belhadj, S.; Kallel, K.; Aoun, K.; Akrout, F.M.; Ben Said, M.; Pratlong, F.; et al. Geographical distribution updating of Tunisian leishmaniasis foci: About the isoenzymatic analysis of 694 Strains. Acta Trop. 2012, 124, 221–228. [Google Scholar] [CrossRef]
- Aoun, K.; Bouratbine, A. Cutaneous leishmaniasis in North Africa: A Review. Parasite 2014, 21, 11820. [Google Scholar] [CrossRef]
- Derbali, M.; Chelbi, I.; Ben Hadj Ahmed, S.; Zhioua, E. Leishmania major Yakimoff et Schokhor, 1914 (Kinetoplastida: Trypanosomatidae) chez Meriones shawi Duvernoy, 1842 (Rodentia: Gerbillidae): Persistance de l’infection du mérion et de son infectivité pour le phlébotome vecteur Phlebotomus (Phlebotomus) papatasi. Bull. Soc. Pathol. Exot. 2012, 105, 399–402. [Google Scholar] [CrossRef] [PubMed]
- Ismail, R.B.; Gramiccia, M.; Gradoni, L.; Helal, H.; Rachid, M.S.B.; Ben Ismail, R.; Gramiccia, M.; Gradoni, L.; Helal, H.; Ben Rachid, M.S. Isolation of Leishmania major from Phlebotomus papatasi in Tunisia. Trans. R. Soc. Trop. Med. Hyg. 1987, 81, 749. [Google Scholar] [CrossRef]
- Rioux, J.A.; Petter, F.; Zahaf, A.; Lanotte, G.; Houin, R.; Jarry, D.; Perieres, J.; Martini, A.; Sarhani, S. Isolation of Leishmania major Yakimoff and Shokhor, 1914 (Kinetoplastida-Trypanosomatidae) in Meriones shawi-shawi (Duvernoy, 1842) (Rodentia-Gerbillidae) in Tunisia. Ann. Parasitol. Hum. Comp. 1986, 61, 139–145. [Google Scholar] [CrossRef]
- Ben-Ismail, R.; Ben Rachid, M.S.; Gradoni, L.; Gramiccia, M.; Helal, H.; Bach-Hamba, D. Zoonotic cutaneous leishmaniasis in Tunisia: Study of the disease reservoir in the Douara area. Ann. Soc. Belg. Med. Trop. 1987, 67, 335–343. [Google Scholar]
- Fichet-Calvet, E.; Jomâa, I.; Ben Ismail, R.; Ashford, R.W. Leishmania major infection in the fat sand rat Psammomys obesus in Tunisia: Interaction of host and parasite populations. Ann. Trop. Med. Parasitol. 2003, 97, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Ghawar, W.; Toumi, A.; Snoussi, M.A.; Chlif, S.; Zâatour, A.; Boukthir, A.; Bel Haj Hamida, N.; Chemkhi, J.; Diouani, M.F.; Ben Salah, A. Leishmania major infection among Psammomys obesus and Meriones shawi: Reservoirs of zoonotic cutaneous leishmaniasis in Sidi Bouzid (Central Tunisia). Vector Borne Zoonotic. Dis. 2011, 11, 1561–1568. [Google Scholar] [CrossRef] [PubMed]
- Derbali, M.; Polyakova, L.; Boujaama, A.; Burruss, D.; Cherni, S.; Barhoumi, W.; Chelbi, I.; Poché, R.; Zhioua, E. Laboratory and field evaluation of rodent bait treated with fipronil for feed through and systemic control of Phlebotomus papatasi. Acta Trop. 2014, 135, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Ghawar, W.; Snoussi, M.A.; Bel Hadj Hamida, N.; Boukthir, A.; Yazidi, R.; Chaâbane, S.; Chemkhi, J.; Zâatour, A.; Ben Salah, A. First report of natural infection of Least Weasel (Mustela Nivalis Linnaeus, 1776) with Leishmania major in Tunisia. Vector Borne Zoonotic. Dis. 2011, 11, 1507–1509. [Google Scholar] [CrossRef]
- Chemkhi, J.; Souguir, H.; Ali, I.B.; Driss, M.; Guizani, I.; Guerbouj, S. Natural infection of Algerian hedgehog, Atelerix Algirus (Lereboullet 1842) with Leishmania parasites in Tunisia. Acta Trop. 2015, 150, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Ghawar, W.; Bettaieb, J.; Salem, S.; Snoussi, M.A.; Jaouadi, K.; Yazidi, R.; Ben-Salah, A. Natural infection of Ctenodactylus gundi by Leishmania major in Tunisia. Acta Trop. 2018, 177, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Ben Othman, S.; Ghawar, W.; Chaouch, M.; Ayari, C.; Chemkhi, J.; Cancino-Faure, B.; Tomás-Pérez, M.; Alcover, M.M.; Riera, C.; Ben Salah, A.; et al. First detection of Leishmania DNA in Psammomys obesus and Psammomys vexillaris: Their potential involvement in the epidemiology of leishmaniasis in Tunisia. Infect. Genet. Evol. 2018, 59, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Chelbi, I.; Kaabi, B.; Béjaoui, M.; Derbali, M.; Zhioua, E.; BéJaoui, M.; Zhioua, E. Spatial correlation between Phlebotomus papatasi Scopoli (Diptera: Psychodidae) and incidence of zoonotic cutaneous leishmaniasis in Tunisia. J. Med. Entomol. 2009, 46, 400–402. [Google Scholar] [CrossRef] [PubMed]
- Chelbi, I.; Derbali, M.; Al-Ahmadi, Z.; Zaafouri, B.; El Fahem, A.; Zhioua, E. Phenology of Phlebotomus papatasi (Diptera: Psychodidae) relative to the seasonal prevalence of zoonotic cutaneous leishmaniasis in Central Tunisia. J. Med. Entomol. 2007, 44, 385–388. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ben Salah, A.; Kamarianakis, Y.; Chlif, S.; Ben Alaya, N.; Prastacos, P. Zoonotic cutaneous leishmaniasis in Central Tunisia: Spatio-temporal dynamics. Int. J. Epidemiol. 2007, 36, 991–1000. [Google Scholar] [CrossRef] [PubMed]
- Chalghaf, B.; Chlif, S.; Mayala, B.; Ghawar, W.; Bettaieb, J.; Harrabi, M.; Benie, G.B.; Michael, E.; Ben Salah, A. Ecological niche modeling for the prediction of the geographic distribution of lutaneous leishmaniasis in Tunisia. Am. J. Trop. Med. Hyg. 2016, 94, 844–851. [Google Scholar] [CrossRef] [PubMed]
- Rioux, J.-A.; Lanotte, G.; Pratlong, F. Leishmania killicki n. sp. (Kinetoplasitda, Trypanosomatidae). In Leishmania: Taxonomie et Phylogenèse: Applications Eco-Epidémiologiques; IMEEE: Montpellier, France, 1986; pp. 139–142. [Google Scholar]
- Bousslimi, N.; Aoun, K.; Ben-Abda, I.; Ben-Alaya-Bouafif, N.; Raouane, M.; Bouratbine, A. Epidemiologic and clinical features of cutaneous leishmaniasis in Southeastern Tunisia. Am. J. Trop. Med. Hyg. 2010, 83, 1034–1039. [Google Scholar] [CrossRef] [PubMed]
- Tabbabi, A.; Bousslimi, N.; Rhim, A.; Aoun, K.; Bouratbine, A. Short Report: First report on natural infection of Phlebotomus sergenti with Leishmania promastigotes in the cutaneous leishmaniasis focus in Southeastern Tunisia. Am. J. Trop. Med. Hyg. 2011, 85, 646–647. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bousslimi, N.; Ben-Ayed, S.; Ben-Abda, I.; Aoun, K.; Bouratbine, A. Natural infection of North African gundi (Ctenodactylus gundi) by Leishmania tropica in the focus of cutaneous leishmaniasis, Southeast Tunisia. Am. J. Trop. Med. Hyg. 2012, 86, 962–965. [Google Scholar] [CrossRef]
- Bouratbine, A.; Aoun, K.; Ghrab, J.; Harrat, Z.; Ezzedini, M.S.; Etlijani, S. Spread of Leishmania killicki to Central and South-West Tunisia. Parasite 2005, 12, 59–63. [Google Scholar] [CrossRef]
- Haouas, N.; Chargui, N.; Chaker, E.; Ben Said, M.; Babba, H.; Belhadj, S.; Kallel, K.; Pratlong, F.; Dedet, J.P.; Mezhoud, H.; et al. Anthroponotic cutaneous leishmaniasis in Tunisia: Presence of Leishmania killicki outside its original focus of Tataouine. Trans. R. Soc. Trop. Med. Hyg. 2005, 99, 499–501. [Google Scholar] [CrossRef] [PubMed]
- Haouas, N.; Gorcii, M.; Chargui, N.; Aoun, K.; Bouratbine, A.; Messaadi Akrout, F.; Masmoudi, A.; Zili, J.; Ben Said, M.; Pratlong, F.; et al. Leishmaniasis in Central and Southern Tunisia: Current geographical distribution of zymodemes. Parasite 2007, 14, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Gounot, M.; Le Houerou, H.N. 1967. Essai de synthèse sur la végétation et la phyto-écologie tunisiennes. In Élément de Botanique et de Phyto-Ecologie Tunisiennes; Tome, I., Nabli, M.A., Eds.; Faculty of Sciences of Tunis and MAB: Tunis, Tunisia, 1989; 387p. [Google Scholar]
- Bellali, H.; Hchaichi, A.; Harizi, C.; Mrabet, A.; Chahed, M.K. Comparison between active surveillance and passive detection of zoonotic cutaneous leishmaniasis in endemic rural areas in Central Tunisia, 2009 to 2014. Asian Pacific J. Trop. Dis. 2015, 5, 515–519. [Google Scholar] [CrossRef]
- Ghrab, J.; Rhim, A.; Bach-Hamba, D.; Chahed, M.K.; Aoun, K.; Nouira, S.; Bouratbine, A. Phlebotominae (Diptera: Psychodidae) of human leishmaniosis sites in Tunisia. Parasite 2006, 13, 23–33. [Google Scholar] [CrossRef]
- Barhoumi, W.; Chelbi, I.; Fares, W.; Zhioua, S.; Abbas, M.; Derbali, M.; Ramalho-Ortigao, M.; Zhioua, E. Risk Assessment of the role of the ecotones in the transmission of zoonotic cutaneous leishmaniasis in Central Tunisia. Int. J. Environ. Res. Public Health 2021, 18, 9274. [Google Scholar] [CrossRef]
- Croset, H.; Rioux, J.; Maistre, M.; Bayar, N. Les phlébotomes de la Tunisie (Diptera, Phlebotominae). Mise au point systématique, chorologique et éthologique. Ann. Parasitol. Hum. Comp. 1978, 53, 711–749. [Google Scholar] [CrossRef]
- Pesson, B.; Ready, J.S.; Benabdennbi, I.; Martín-Sánchez, J.; Esseghir, S.; Cadi-Soussi, M.; Morillas-Marquez, F.; Ready, P.D. Sandflies of the Phlebotomus perniciosus complex: Mitochondrial introgression and a new sibling species of P. longicuspis in the Moroccan Rif. Med. Vet. Entomol. 2004, 18, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Boussaa, S.; Boumezzough, A.; Remy, P.E.; Glasser, N.; Pesson, B. Morphological and isoenzymatic differentiation of Phlebotomus perniciosus and Phlebotomus longicuspis (Diptera: Psychodidae) in Southern Morocco. Acta Trop. 2008, 106, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Noyes, H.A.; Reyburn, H.; Bailey, J.W.; Smith, D. A nested-PCR-based schizodeme method for identifying Leishmania kinetoplast minicircle classes directly from clinical samples and its application to the study of the epidemiology of Leishmania tropica in Pakistan. J. Clin. Microbiol. 1998, 36, 2877–2881. [Google Scholar] [CrossRef] [PubMed]
- Parvizi, P.; Ready, P.D. Nested PCRs and sequencing of nuclear ITS-rDNA fragments detect three Leishmania species of gerbils in sandflies from Iranian foci of zoonotic cutaneous leishmaniasis. Trop. Med. Int. Health 2008, 13, 1159–1171. [Google Scholar] [CrossRef] [PubMed]
- Parvizi, P.; Mauricio, I.; Aransay, A.M.; Miles, M.A.; Ready, P.D. First detection of Leishmania major in peridomestic Phlebotomus papatasi from Isfahan Province, Iran: Comparison of nested PCR of nuclear ITS ribosomal DNA and semi-Nested PCR of minicircle kinetoplast DNA. Acta Trop. 2005, 93, 75–83. [Google Scholar] [CrossRef]
- Parvizi, P.; Moradi, G.; Akbari, G.; Farahmand, M.; Ready, P.D.; Piazak, N.; Assmar, M.; Amirkhani, A. PCR detection and sequencing of parasite ITS-RDNA gene from reservoirs host of zoonotic cutaneous leishmaniasis in Central Iran. Parasitol. Res. 2008, 103, 1273–1278. [Google Scholar] [CrossRef] [PubMed]
- Barhoumi, W.; Fares, W.; Cherni, S.; Derbali, M.; Dachraoui, K.; Chelbi, I.; Ramalho-Ortigao, M.; Beier, J.C.; Zhioua, E. Changes of sand fly populations and Leishmania infantum infection rates in an irrigated village located in arid Central Tunisia. Int. J. Environ. Res. Public Health 2016, 13, 329. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Spellerberg, I.A.N.F.; Fedor, P.J. Tribute to Claude Shannon (1916-2001) and a plea for more rigorous use of species diversity and the “Shannon-Wiener” Index. Glob. Ecol. Biogeogr. 2003, 12, 177–179. [Google Scholar] [CrossRef]
- Chelbi, I.; Zhioua, E. Confirmation de la présence en Tunisie de Sergentomyia (Sintonius) clydei (Sinton, 1928). Bull. Soc. Pathol. Exot. 2012, 105, 396–398. [Google Scholar] [CrossRef]
- Chemkhi, J.; Guerbouj, S.; Saadawi, W.; Shaibi, T.; Faris, S.; Ghawar, W.; Boukthir, A.; Guizani, I.; Annajar, B.B.; Ben Salah, A. Presence of Sergentomyia (Parrotomyia) lewisi (Diptera: Psychodidae) in Tunisia. J. Med. Entomol. 2019, 56, 560–564. [Google Scholar] [CrossRef]
- Zaatour, W.; Marilleau, N.; Giraudoux, P.; Martiny, N.; Amara, A.B.H.; Ben Miled, S. An agent-based model of a cutaneous leishmaniasis reservoir host, Meriones shawi. Ecol. Modell. 2021, 443, 109455. [Google Scholar] [CrossRef]
- Orshan, L.; Elbaz, S.; Ben-Ari, Y.; Akad, F.; Afik, O.; Ben-Avi, I.; Dias, D.; Ish-Shalom, D.; Studentsky, L.; Zonstein, I. Distribution and dispersal of Phlebotomus papatasi (Diptera: Psychodidae) in a zoonotic cutaneous leishmaniasis focus, the Northern Negev, Israel. PLoS Negl. Trop. Dis. 2016, 10, e0004819. [Google Scholar] [CrossRef] [PubMed]
- Chelbi, I.; Mathlouthi, O.; Zhioua, S.; Fares, W.; Boujaama, A.; Cherni, S.; Barhoumi, W.; Dachraoui, K.; Derbali, M.; Abbass, M.; et al. The Impact of illegal waste sites on the transmission of zoonotic cutaneous leishmaniasis in Central Tunisia. Int. J. Environ. Res. Public Health 2021, 18, 66. [Google Scholar] [CrossRef]
- Jaouadi, K.; Bettaieb, J.; Bennour, A.; Salem, S.; Rjeibi, M.R.; Chaabane, S.; Yazidi, R.; Khabouchi, N.; Gharbi, A.; Ben Salah, A. First report on natural infection of Phlebotomus sergenti with Leishmania tropica in a classical focus of Leishmania major in Tunisia. Am. J. Trop. Med. Hyg. 2017, 97, 291–294. [Google Scholar] [CrossRef]
- Tabbabi, A.; Ghrab, J.; Aoun, K.; Ready, P.D.; Bouratbine, A. Habitats of the sandfly vectors of Leishmania tropica and L. major in a mixed focus of cutaneous leishmaniasis in Southeast Tunisia. Acta Trop. 2011, 119, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Jaouadi, K.; Haouas, N.; Chaara, D.; Gorcii, M.; Chargui, N.; Augot, D.; Pratlong, F.; Dedet, J.P.; Ettlijani, S.; Mezhoud, H.; et al. First Detection of Leishmania killicki (Kinetoplastida, Trypanosomatidae) in Ctenodactylus gundi (Rodentia, Ctenodactylidae), a possible reservoir of human cutaneous leishmaniasis in Tunisia. Parasite Vectors 2011, 4, 4–6. [Google Scholar] [CrossRef] [PubMed]
- Kamhawi, S.; Modi, G.B.; Pimenta, P.F.P.; Rowton, E.; Sacks, D.L. The vectorial competence of Phlebotomus sergenti is specific for Leishmania tropica and is controlled by species-specific, lipophosphoglycan-mediated midgut attachment. Parasitology 2000, 121, 25–33. [Google Scholar] [CrossRef]
- Volf, P.; Myskova, J. Sand flies and Leishmania: Specific versus permissive vectors. Trends Parasitol. 2007, 23, 91–92. [Google Scholar] [CrossRef]
- Moin-Vaziri, V.; Depaquit, J.; Yaghoobi-Ershadi, M.R.; Oshaghi, M.A.; Derakhshandeh-Peykar, P.; Ferté, H.; Kaltenbach, M.; Bargues, M.D.; Léger, N.; Nadim, A. Intraspecific variation within Phlebotomus sergenti Parrot (1917) (Diptera: Psychodidae) based on MtDNA sequences in Islamic Republic of Iran. Acta Trop. 2007, 102, 29–37. [Google Scholar] [CrossRef]
- Ait Kbaich, M.; Mhaidi, I.; Ezzahidi, A.; Dersi, N.; El Hamouchi, A.; Riyad, M.; Akarid, K.; Lemrani, M. New epidemiological pattern of cutaneous leishmaniasis in two pre-Saharan arid provinces, Southern Morocco. Acta Trop. 2017, 173, 11–16. [Google Scholar] [CrossRef]
- Hmamouch, A.; El Alem, M.M.; Hakkour, M.; Amarir, F.; Daghbach, H.; Habbari, K.; Fellah, H.; Bekhti, K.; Sebti, F. Circulating species of Leishmania at microclimate area of Boulemane Province, Morocco: Impact of environmental and human factors. Parasite Vectors 2017, 10, 100. [Google Scholar] [CrossRef] [PubMed]
- Mohamed Mahmoud, E.A.; Faiza, S.; Lemine, M.; Smaine, C.; Adlaoui, E.B.; Khalid, H.; Abderrahim, S.; Hajiba, F. Geographical distribution and new Situation of Leishmania species after the control of cutaneous Leishmaniasis foci in Errachidia Province, Morocco, in 2014. Biomed. Res. Int. 2016, 2016, 8642373. [Google Scholar] [CrossRef] [PubMed]
- Reithinger, R.; Dujardin, J.-C.; Louzir, H.; Pirmez, C.; Alexander, B.; Brooker, S. Cutaneous leishmaniasis. Lancet. Infect. Dis. 2007, 7, 581–596. [Google Scholar] [CrossRef]
- Rhajaoui, M.; Nasereddin, A.; Fellah, H.; Azmi, K.; Amarir, F.; Al-Jawabreh, A.; Ereqat, S.; Planer, J.; Abdeen, Z. New Clinico-epidemiologic profile of cutaneous leishmaniasis, Morocco. Emerg. Infect. Dis. 2007, 13, 1358–1360. [Google Scholar] [CrossRef] [PubMed]
- Baghad, B.; Razanapinaritra, R.; Maksouri, H.; El Bouri, H.; Outlioua, A.; Fellah, H.; Lemrani, M.; Akarid, K.; Martin-Sanchez, J.; Chiheb, S.; et al. Possible introduction of Leishmania Tropica to urban areas determined by pidemiological and clinical profiles of patients with cutaneous ceishmaniasis in Casablanca (Morocco). Parasite Epidemiol. Control 2020, 9, e00129. [Google Scholar] [CrossRef]
- Jaouadi, K.; Depaquit, J.; Haouas, N.; Chaara, D.; Gorcii, M.; Chargui, N.; Dedet, J.P.; Pratlong, F.; Boubabous, R.; Babba, H. Twenty-four new human cases of cutaneous leishmaniasis due to Leishmania killicki in Metlaoui, Southwestern Tunisia. Probable role of Phlebotomus sergenti in the transmission. Acta Trop. 2012, 122, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Remadi, L.; Chargui, N.; Jiménez, M.; Molina, R.; Haouas, N.; González, E.; Chaabane-Banaouas, R.; Ben Salah, E.; Haddaji, M.; Chaabouni, Y.; et al. Molecular detection and identification of Leishmania DNA and blood meal analysis in Phlebotomus (Larroussius) species. PLoS Negl. Trop. Dis. 2020, 14, e0008077. [Google Scholar] [CrossRef]
- Zhioua, E.; Kaabi, B.; Chelbi, I. Entomological investigations following the spread of visceral leishmaniasis in Tunisia. J. Vector Ecol. 2007, 32, 371–374. [Google Scholar] [CrossRef]
- Es-Sette, N.; Ajaoud, M.; Laamrani-Idrissi, A.; Mellouki, F.; Lemrani, M. Molecular detection and identification of Leishmania infection in naturally infected sand flies in a focus of cutaneous leishmaniasis in Northern Morocco. Parasite Vectors 2014, 7, 305. [Google Scholar] [CrossRef]
- Mhaidi, I.; El Kacem, S.; Ait Kbaich, M.; El Hamouchi, A.; Sarih, M.; Akarid, K.; Lemrani, M. Molecular identification of Leishmania infection in the most relevant sand fly species and in patient skin samples from a cutaneous leishmaniasis focus, in Morocco. PLoS Negl. Trop. Dis. 2018, 12, e0006315. [Google Scholar] [CrossRef] [PubMed]
- Berdjane-Brouk, Z.; Charrel, R.N.; Hamrioui, B.; Izri, A. First detection of Leishmania infantum DNA in Phlebotomus longicuspis Nitzulescu, 1930 from visceral leishmaniasis endemic focus in Algeria. Parasitol. Res. 2012, 111, 419–422. [Google Scholar] [CrossRef]
- Jaouadi, K.; Ghawar, W.; Salem, S.; Gharbi, M.; Bettaieb, J.; Yazidi, R.; Harrabi, M.; Hamarsheh, O.; Ben Salah, A. First report of naturally infected Sergentomyia minuta with Leishmania major in Tunisia. Parasites Vectors 2015, 8, 649. [Google Scholar] [CrossRef]



| Species (Subgenus) | ST ♂/♀ | Subtotal | LT ♂/♀ | Subtotal | Total | (%) |
|---|---|---|---|---|---|---|
| P. (Phlebotomus) papatasi | 1874/474 | 2348 | 3364/1897 | 5261 | 7609 | (44.30) |
| P. (Paraphlebotomus) sergenti | 68/19 | 87 | 70/89 | 159 | 246 | (1.43) |
| P. (Paraphlebotomus) alexandri | 15/8 | 23 | 67/44 | 111 | 134 | (0.78) |
| P. (Paraphlebotomus) chabaudi | 1/2 | 3 | 0/3 | 3 | 6 | (0.03) |
| P. (Paraphlebotomus) riouxi | 2/0 | 2 | 2 | (0.01) | ||
| P. (Larroussius) ariasi | 8/0 | 8 | 1/6 | 7 | 15 | (0.09) |
| P. (Larroussius) longicuspis | 29/0 | 29 | 1210/769 | 1979 | 2008 | (11.69) |
| P. (Larroussius) perfiliewi | 3/1 | 4 | 10/8 | 18 | 22 | (0.13) |
| P. (Larroussius) perniciosus | 40/2 | 42 | 14/17 | 31 | 73 | (0.43) |
| P. (Larroussius) langeroni | 0/2 | 2 | 2 | (0.01) | ||
| S. (Sergentomyia) fallax | 994/471 | 1465 | 1506/1195 | 2701 | 4166 | (24.26) |
| S. (Sergentomyia) minuta | 767/320 | 1087 | 765/376 | 1141 | 2228 | (12.97) |
| S. (Sergentomyia) antennata | 40/21 | 61 | 82/247 | 329 | 390 | (2.27) |
| S. (Grassomyia) dreyfussi | 2/1 | 3 | 53/191 | 244 | 247 | (1.44) |
| S. (Sintonius) christophersi | 1/1 | 2 | 2/19 | 21 | 23 | (0.13) |
| S. (Sintonius) clydei | 0/2 | 2 | 0/2 | 2 | 4 | (0.02) |
| Total | 3844/1322 | 5166 | 7144/4865 | 12,009 | 17,175 |
| G.B | Bed | A.S | R.H | BMs | BPo | Total | |
|---|---|---|---|---|---|---|---|
| Species | Su.T (%) | Su.T (%) | Su.T (%) | Su.T (%) | Su.T (%) | Su.T (%) | Total (%) |
| P. papatasi | 63 (2.1) | 1747 (33.13) | 1623 (39.13) | 1866 (88.31) | 1799 (88.36) | 511 (84.6) | 7609 (44.3) |
| P. sergenti | 100 (3.33) | 77 (1.46) | 55 (1.33) | 14 (0.66) | 246 (1.43) | ||
| P. chabaudi | 5 (0.17) | 1 (0.02) | 6 (0.03) | ||||
| P. alexandri | 26 (0.87) | 56 (1.06) | 47 (1.13) | 2 (0.99) | 2 (0.1) | 1 (0.17) | 134 (0.78) |
| P. riouxi | 2 (0.07) | 2 (0.01) | |||||
| S. fallax | 1487 (49.55) | 1895 (35.94) | 502 (12.1) | 105 (4.97) | 118 (5.8) | 59 (9.77) | 4166 (24.26) |
| S. minuta | 1145 (38.15) | 659 (12.5) | 277 (6.68) | 39 (1.85) | 90 (4.42) | 18 (2.98) | 2228 (12.97) |
| S. antennata | 63 (2.10) | 208 (3.94) | 85 (2.05) | 12 (0.57) | 16 (0.79) | 6 (0.99) | 390 (2.27) |
| S. dreyfussi | 24 (0.8) | 115 (2.18) | 103 (2.48) | 3 (0.14) | 1 (0.05) | 1 (0.17) | 247 (1.44) |
| S. clydei | 1 (0.02) | 2 (0.1) | 4 (0.02) | ||||
| S. christophersi | 1 (0.03) | 9 (0.17) | 13 (0.31) | 1 (0.05) | 23 (0.13) | ||
| P. longicuspis | 25 (0.83) | 487 (9.24) | 1416 (34.14) | 67 (3.17) | 6 (0.29) | 7 (1.16) | 2008 (11.69) |
| P. perniciosus | 47 (1.57) | 10 (0.19) | 12 (0.29) | 3 (0.14) | 1 (0.05) | 73 (0.43) | |
| P. perfiliewi | 3 (0.1) | 10 (0.19) | 7 (0.17) | 1 (0.05) | 1 (0.17) | 22 (0.13) | |
| P. langeroni | 2 (0.05) | 2 (0.01) | |||||
| P. ariasi | 10 (0.33) | 4 (0.1) | 1 (0.05) | 15 (0.04) | |||
| Total | 3001 | 5273 | 4148 | 2113 | 2036 | 604 | 17,175 |
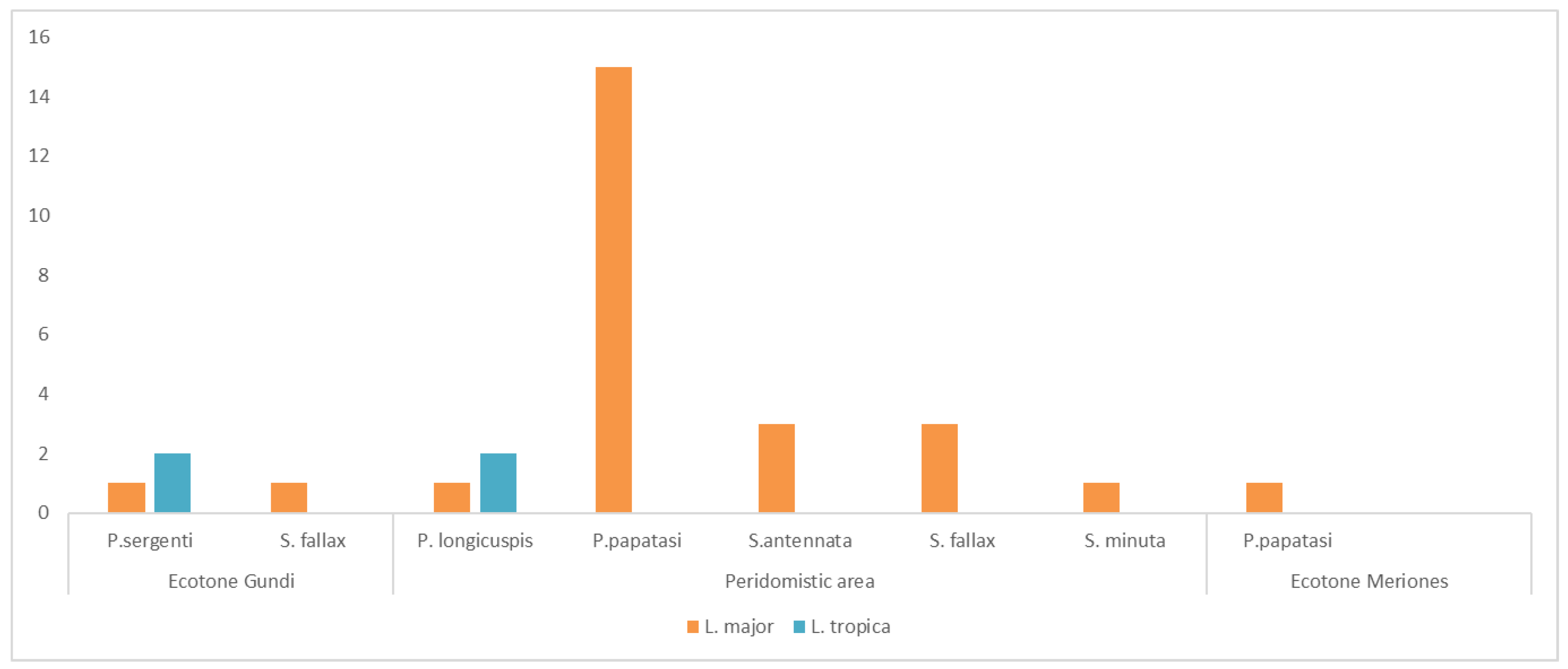
| Date | Biotype | Sandfly/Pool (Total) | Sandfly Species | Leishmania Species |
|---|---|---|---|---|
| 13 July 2017 | A. S | 1 (435) | P. longicuspis | L. tropica |
| Bed | 4 (494) | P. papatasi | L. major | |
| A. S | 24 (435) | P. papatasi | L. major | |
| Bed | 3 (494) | P. papatasi | L. major | |
| A. S | 1 (435) | S. antennata | L. major | |
| 5 September 2017 | A. S | 1 (136) | P. papatasi | L. major |
| A. S | 1 (136) | S. antennata | L. major | |
| R.H | 19 (1062) | P. papatasi | L. major | |
| 19 September 2017 | R.H | 2 (277) | P. papatasi | L. major |
| Bed | 9 (154) | S. fallax | L. major | |
| 26 September 2017 | Bed | 3 (485) | S. antennata | L. major |
| A. S | 2 (215) | S. fallax | L. major | |
| 23 August 2018 | A.S | 1 (39) | P. longicuspis | L. tropica |
| 7 September 2018 | A. S | 3 (82) | P. papatasi | L. major |
| 12 September 2018 | A. S | 6 (260) | S. fallax | L. major |
| R.H | 8 (59) | P. papatasi | L. major | |
| Bed | 2 (445) | P. papatasi | L. major | |
| A. S | 1 (260) | P. papatasi | L. major | |
| 10 October 2018 | A. S | 1 (127) | S. minuta | L. major |
| 25 September 2019 | G.B | 14 (210) | S. fallax | L. major |
| 2 October 2019 | R.B | 6 (24) | P. papatasi | L. major |
| 2 October 2019 | A. S | 3 (39) | P. papatasi | L. major |
| 25 September 2019 | Bed | 2 (170) | P. papatasi | L. major |
| 3 October 2019 | A. S | 5 (276) | P. longicuspis | L. major |
| 25 September 2019 | A. S | 30 (279) | P. papatasi | L. major |
| 24 September 2019 | A. S | 30 (695) | P. papatasi | L. major |
| 24 September 2019 | Bed | 1 (219) | P. papatasi | L. major |
| 17 October 2019 | G.B | 2 (389) | P. sergenti | L. tropica |
| 23 October 2019 | G.B | 5 (181) | P. sergenti | L. tropica |
| 2 October 2019 | G.B | 2 (224) | P. sergenti | L. major |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abbas, M.A.S.; Lachheb, J.; Chelbi, I.; Louati, D.; Dachraoui, K.; Ben Miled, S.; Zhioua, E. Independent Circulation of Leishmania major and Leishmania tropica in Their Respective Sandfly Vectors for Transmission of Zoonotic and Chronic Cutaneous Leishmaniasis Co-Existing in a Mixed Focus of Central Tunisia. Pathogens 2022, 11, 855. https://doi.org/10.3390/pathogens11080855
Abbas MAS, Lachheb J, Chelbi I, Louati D, Dachraoui K, Ben Miled S, Zhioua E. Independent Circulation of Leishmania major and Leishmania tropica in Their Respective Sandfly Vectors for Transmission of Zoonotic and Chronic Cutaneous Leishmaniasis Co-Existing in a Mixed Focus of Central Tunisia. Pathogens. 2022; 11(8):855. https://doi.org/10.3390/pathogens11080855
Chicago/Turabian StyleAbbas, Mohammed Abdo Saghir, Jihene Lachheb, Ifhem Chelbi, Dorra Louati, Khalil Dachraoui, Slimene Ben Miled, and Elyes Zhioua. 2022. "Independent Circulation of Leishmania major and Leishmania tropica in Their Respective Sandfly Vectors for Transmission of Zoonotic and Chronic Cutaneous Leishmaniasis Co-Existing in a Mixed Focus of Central Tunisia" Pathogens 11, no. 8: 855. https://doi.org/10.3390/pathogens11080855
APA StyleAbbas, M. A. S., Lachheb, J., Chelbi, I., Louati, D., Dachraoui, K., Ben Miled, S., & Zhioua, E. (2022). Independent Circulation of Leishmania major and Leishmania tropica in Their Respective Sandfly Vectors for Transmission of Zoonotic and Chronic Cutaneous Leishmaniasis Co-Existing in a Mixed Focus of Central Tunisia. Pathogens, 11(8), 855. https://doi.org/10.3390/pathogens11080855






