Complete Genomes of Theileria orientalis Chitose and Buffeli Genotypes Reveal within Species Translocations and Differences in ABC Transporter Content
Abstract
1. Introduction
2. Results
2.1. Sequencing and Chromosomal Assembly Metrics
2.2. Apicoplast Genomes
2.3. Synteny between T. orientalis Genotypes
2.4. Ortholog Clustering
2.5. Phylogeny and Average Nucleotide Identity
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Processing
4.2. Illumina Sequencing
4.3. Genomic DNA Extraction
4.4. Pulsed-Field Gel Electrophoresis (PFGE)
4.5. Nanopore Library Preparation and Sequencing
4.6. Sequence Quality Assessment
4.7. Genome Assembly
4.8. Genome Annotation
4.9. Ortholog Clustering, Phylogeny, Gene Presence/Absence and Average Nucleotide Identity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sivakumar, T.; Hayashida, K.; Sugimoto, C.; Yokoyama, N. Evolution and genetic diversity of Theileria. Infect. Genet. Evol. 2014, 27, 250–263. [Google Scholar] [CrossRef] [PubMed]
- Kamau, J.; de Vos, A.J.; Playford, M.; Salim, B.; Kinyanjui, P.; Sugimoto, C. Emergence of new types of Theileria orientalis in Australian cattle and possible cause of theileriosis outbreaks. Parasit. Vectors 2011, 4, 22. [Google Scholar] [CrossRef] [PubMed]
- McFadden, A.M.J.; Rawdon, T.G.; Meyer, J.; Makin, J.; Morley, C.M.; Clough, R.R.; Tham, K.; Müllner, P.; Geysen, D. An outbreak of haemolytic anaemia associated with infection of Theileria orientalis in naïve cattle. N. Z. Vet. J. 2011, 59, 79. [Google Scholar] [CrossRef] [PubMed]
- Ota, N.; Mizuno, D.; Kuboki, N.; Igarashi, I.; Nakamura, Y.; Yamashina, H.; Hanzaike, T.; Fujii, K.; Onoe, S.; Hata, H.; et al. Epidemiological survey of Theileria orientalis infection in grazing cattle in the eastern part of Hokkaido, Japan. J. Vet. Med. Sci. Jpn. Soc. Vet. Sci. 2009, 71, 937–944. [Google Scholar] [CrossRef]
- Oakes, V.J.; Yabsley, M.J.; Schwartz, D.; LeRoith, T.; Bissett, C.; Broaddus, C.; Schlater, J.L.; Todd, S.M.; Boes, K.M.; Brookhart, M. Theileria orientalis ikeda genotype in cattle, Virginia, USA. Emerg. Infect. Dis. 2019, 25, 1653. [Google Scholar] [CrossRef]
- Kim, S.; Yu, D.-H.; Chae, J.-B.; Choi, K.-S.; Kim, H.-C.; Park, B.-K.; Chae, J.-S.; Park, J. Pathogenic genotype of major piroplasm surface protein associated with anemia in Theileria orientalis infection in cattle. Acta Vet. Scand. 2017, 59, 51. [Google Scholar] [CrossRef]
- Eamens, G.J.; Gonsalves, J.R.; Jenkins, C.; Collins, D.; Bailey, G. Theileria orientalis MPSP types in Australian cattle herds associated with outbreaks of clinical disease and their association with clinical pathology findings. Vet. Parasitol. 2013, 191, 209–217. [Google Scholar] [CrossRef]
- Eamens, G.J.; Bailey, G.; Jenkins, C.; Gonsalves, J.R. Significance of Theileria orientalis types in individual affected beef herds in New South Wales based on clinical, smear and PCR findings. Vet. Parasitol. 2013, 196, 96–105. [Google Scholar] [CrossRef]
- Yam, J.; Bogema, D.; Jenkins, C. Oriental Theileriosis. In Ticks and Tick–Borne Pathogens; IntechOpen: London, UK, 2018. [Google Scholar]
- Kakuda, T.; Shiki, M.; Kubota, S.; Sugimoto, C.; Brown, W.C.; Kosum, C.; Nopporn, S.; Onuma, M. Phylogeny of benign Theileria species from cattle in Thailand, China and the U.S.A. based on the major piroplasm surface protein and small subunit ribosomal RNA genes. Int. J. Parasitol. 1998, 28, 1261. [Google Scholar] [CrossRef]
- Kim, S.J.; Tsuji, M.; Kubota, S.; Wei, Q.; Lee, J.M.; Ishihara, C.; Onuma, M. Sequence analysis of the major piroplasm surface protein gene of benign bovine Theileria parasites in east Asia. Int. J. Parasitol. 1998, 28, 1219. [Google Scholar] [CrossRef]
- Chae, J.S.; Allsopp, B.A.; Waghela, S.D.; Park, J.H.; Kakuda, T.; Sugimoto, C.; Allsopp, M.T.; Wagner, G.G.; Holman, P.J. A study of the systematics of Theileria spp. based upon small–subunit ribosomal RNA gene sequences. Parasitol. Res. 1999, 85, 877–883. [Google Scholar] [CrossRef] [PubMed]
- Jeong, W.; Yoon, S.H.; An, D.J.; Cho, S.H.; Lee, K.K.; Kim, J.Y. A molecular phylogeny of the benign Theileria parasites based on major piroplasm surface protein (MPSP) gene sequences. Parasitology 2010, 137, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Khukhuu, A.; Lan, D.T.B.; Long, P.T.; Ueno, A.; Li, Y.; Luo, Y.; Macedo, A.C.C.d.; Matsumoto, K.; Inokuma, H.; Kawazu, S.-I.; et al. Molecular epidemiological survey of Theileria orientalis in Thua Thien Hue Province, Vietnam. J. Vet. Med. Sci. Jpn. Soc. Vet. Sci. 2011, 73, 701–705. [Google Scholar] [CrossRef]
- Yokoyama, N.; Ueno, A.; Mizuno, D.; Kuboki, N.; Khukhuu, A.; Igarashi, I.; Miyahara, T.; Shiraishi, T.; Kudo, R.; Oshiro, M. Genotypic diversity of Theileria orientalis detected from cattle grazing in Kumamoto and Okinawa prefectures of Japan. J. Vet. Med. Sci. 2011, 73, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Hupalo, D.N.; Luo, Z.; Melnikov, A.; Sutton, P.L.; Rogov, P.; Escalante, A.; Vallejo, A.F.; Herrera, S.; Arévalo–Herrera, M.; Fan, Q. Population genomics studies identify signatures of global dispersal and drug resistance in Plasmodium vivax. Nat. Genet. 2016, 48, 953. [Google Scholar] [CrossRef]
- Miotto, O.; Amato, R.; Ashley, E.A.; MacInnis, B.; Almagro–Garcia, J.; Amaratunga, C.; Lim, P.; Mead, D.; Oyola, S.O.; Dhorda, M. Genetic architecture of artemisinin–resistant Plasmodium falciparum. Nat. Genet. 2015, 47, 226. [Google Scholar] [CrossRef]
- Hayashida, K.; Hara, Y.; Abe, T.; Yamasaki, C.; Toyoda, A.; Kosuge, T.; Suzuki, Y.; Sato, Y.; Kawashima, S.; Katayama, T.; et al. Comparative Genome Analysis of Three Eukaryotic Parasites with Differing Abilities To Transform Leukocytes Reveals Key Mediators of Theileria–Induced Leukocyte Transformation. mBio 2012, 3, e00204-12. [Google Scholar] [CrossRef]
- Bogema, D.R.; Micallef, M.L.; Liu, M.; Padula, M.P.; Djordjevic, S.P.; Darling, A.E.; Jenkins, C. Analysis of Theileria orientalis draft genome sequences reveals potential species–level divergence of the Ikeda, Chitose and Buffeli genotypes. BMC Genom. 2018, 19, 298. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Cerdeira, L.T.; Hawkey, J.; Méric, G.; Vezina, B.; Wyres, K.L.; Holt, K.E. Trycycler: Consensus long–read assemblies for bacterial genomes. Genome Biol. 2021, 22, 266. [Google Scholar] [CrossRef]
- Minh, B.Q.; Hahn, M.W.; Lanfear, R. New Methods to Calculate Concordance Factors for Phylogenomic Datasets. Mol. Biol. Evol. 2020, 37, 2727–2733. [Google Scholar] [CrossRef]
- Kandziora, M.; Sklenář, P.; Kolář, F.; Schmickl, R. How to Tackle Phylogenetic Discordance in Recent and Rapidly Radiating Groups? Developing a Workflow Using Loricaria (Asteraceae) as an Example. Front. Plant Sci. 2022, 12, 765719. [Google Scholar] [CrossRef] [PubMed]
- DeBarry, J.D.; Kissinger, J.C. Jumbled Genomes: Missing Apicomplexan Synteny. Mol. Biol. Evol. 2011, 28, 2855–2871. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; He, L.; Hu, J.; He, P.; He, J.; Yu, L.; Malobi, N.; Zhou, Y.; Shen, B.; Zhao, J. Characterization and annotation of Babesia orientalis apicoplast genome. Parasit. Vectors 2015, 8, 543. [Google Scholar] [CrossRef] [PubMed]
- Lizundia, R.; Werling, D.; Langsley, G.; Ralph, S.A. Theileria Apicoplast as a Target for Chemotherapy. Antimicrob. Agents Chemother. 2009, 53, 1213–1217. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tran, P.N.; Brown, S.H.J.; Mitchell, T.W.; Matuschewski, K.; McMillan, P.J.; Kirk, K.; Dixon, M.W.A.; Maier, A.G. A female gametocyte–specific ABC transporter plays a role in lipid metabolism in the malaria parasite. Nat. Commun. 2014, 5, 4773. [Google Scholar] [CrossRef]
- Rijpma, S.R.; van der Velden, M.; González–Pons, M.; Annoura, T.; van Schaijk, B.C.; van Gemert, G.J.; van den Heuvel, J.J.; Ramesar, J.; Chevalley–Maurel, S.; Ploemen, I.H.; et al. Multidrug ATP–binding cassette transporters are essential for hepatic development of Plasmodium sporozoites. Cell. Microbiol. 2016, 18, 369–383. [Google Scholar] [CrossRef]
- Jalovecka, M.; Hajdusek, O.; Sojka, D.; Kopacek, P.; Malandrin, L. The Complexity of Piroplasms Life Cycles. Front. Cell. Infect. Microbiol. 2018, 8, 248. [Google Scholar] [CrossRef]
- Kappmeyer, L.S.; Thiagarajan, M.; Herndon, D.R.; Ramsay, J.D.; Caler, E.; Djikeng, A.; Gillespie, J.J.; Lau, A.O.T.; Roalson, E.H.; Silva, J.C.; et al. Comparative genomic analysis and phylogenetic position of Theileria equi. BMC Genom. 2012, 13, 603. [Google Scholar] [CrossRef]
- Jenkins, C.; Micallef, M.; Alex, S.M.; Collins, D.; Djordjevic, S.P.; Bogema, D.R. Temporal dynamics and subpopulation analysis of Theileria orientalis genotypes in cattle. Infect. Genet. Evol. 2015, 32, 199–207. [Google Scholar] [CrossRef]
- Bogema, D.R.; Deutscher, A.T.; Fell, S.; Collins, D.; Eamens, G.J.; Jenkins, C. Development and validation of a quantitative PCR assay using multiplexed hydrolysis probes for detection and quantification of Theileria orientalis isolates and differentiation of clinically relevant subtypes. J. Clin. Microbiol. 2015, 53, 941–950. [Google Scholar] [CrossRef]
- Grüning, B.; Dale, R.; Sjödin, A.; Chapman, B.A.; Rowe, J.; Tomkins-Tinch, C.H.; Valieris, R.; Köster, J. Bioconda: Sustainable and Comprehensive Software Distribution for the Life Sciences. Nat. Methods 2018, 475–476. [Google Scholar] [CrossRef] [PubMed]
- Leger, A.; Leonardi, T. pycoQC, interactive quality control for Oxford Nanopore Sequencing. J. Open Source Softw. 2019, 34, 1236. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error–prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540. [Google Scholar] [CrossRef] [PubMed]
- Vaser, R.; Šikić, M. Time–and memory–efficient genome assembly with Raven. Nat. Comput. Sci. 2021, 1, 332–336. [Google Scholar] [CrossRef]
- Chen, Y.; Nie, F.; Xie, S.-Q.; Zheng, Y.-F.; Dai, Q.; Bray, T.; Wang, Y.-X.; Xing, J.-F.; Huang, Z.-J.; Wang, D.-P.; et al. Efficient assembly of nanopore reads via highly accurate and intact error correction. Nat. Commun. 2021, 12, 60. [Google Scholar] [CrossRef]
- Shafin, K.; Pesout, T.; Lorig–Roach, R.; Haukness, M.; Olsen, H.E.; Bosworth, C.; Armstrong, J.; Tigyi, K.; Maurer, N.; Koren, S.; et al. Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes. Nat. Biotechnol. 2020, 38, 1044–1053. [Google Scholar] [CrossRef]
- Wick, R.R.; Holt, K.E. Polypolish: Short–read polishing of long–read bacterial genome assemblies. PLoS Comput. Biol. 2022, 18, e1009802. [Google Scholar] [CrossRef]
- Zimin, A.V.; Marçais, G.; Puiu, D.; Roberts, M.; Salzberg, S.L.; Yorke, J.A. The MaSuRCA genome assembler. Bioinformatics 2013, 29, 2669–2677. [Google Scholar] [CrossRef]
- Darling, A.E.; Mau, B.; Perna, N.T. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Stanke, M.; Steinkamp, R.; Waack, S.; Morgenstern, B. AUGUSTUS: A web server for gene finding in eukaryotes. Nucleic Acids Res. 2004, 32, W309–W312. [Google Scholar] [CrossRef] [PubMed]
- Korf, I. Gene finding in novel genomes. BMC Bioinform. 2004, 5, 59. [Google Scholar] [CrossRef] [PubMed]
- Majoros, W.H.; Pertea, M.; Salzberg, S.L. TigrScan and GlimmerHMM: Two open source ab initio eukaryotic gene–finders. Bioinformatics 2004, 20, 2878–2879. [Google Scholar] [CrossRef] [PubMed]
- Lomsadze, A.; Burns, P.D.; Borodovsky, M. Integration of mapped RNA–Seq reads into automatic training of eukaryotic gene finding algorithm. Nucleic Acids Res. 2014, 42, e119. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra–fast all–in–one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Slater, G.S.C.; Birney, E. Automated generation of heuristics for biological sequence comparison. BMC Bioinform. 2005, 6, 31. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full–length transcriptome assembly from RNA–Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Haas, B.J.; Salzberg, S.L.; Zhu, W.; Pertea, M.; Allen, J.E.; Orvis, J.; White, O.; Buell, C.R.; Wortman, J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008, 9, R7. [Google Scholar] [CrossRef]
- Otto, T.D.; Dillon, G.P.; Degrave, W.S.; Berriman, M. RATT: Rapid Annotation Transfer Tool. Nucleic Acids Res. 2011, 39, e57. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome–scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Huerta–Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; Von Mering, C.; Bork, P. Fast genome–wide functional annotation through orthology assignment by eggNOG–mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef] [PubMed]
- Huerta–Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández–Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef] [PubMed]
- Emms, D.M.; Kelly, S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015, 16, 157. [Google Scholar] [CrossRef]
- Brayton, K.A.; Lau, A.O.; Herndon, D.R.; Hannick, L.; Kappmeyer, L.S.; Berens, S.J.; Bidwell, S.L.; Brown, W.C.; Crabtree, J.; Fadrosh, D.; et al. Genome sequence of Babesia bovis and comparative analysis of apicomplexan hemoprotozoa. PLoS Pathog. 2007, 3, 1401–1413. [Google Scholar] [CrossRef]
- Pain, A.; Renauld, H.; Berriman, M.; Murphy, L.; Yeats, C.A.; Weir, W.; Kerhornou, A.; Aslett, M.; Bishop, R.; Bouchier, C.; et al. Genome of the host–cell transforming parasite Theileria annulata compared with T. parva. Science 2005, 309, 131–133. [Google Scholar] [CrossRef]
- Gardner, M.J.; Bishop, R.; Shah, T.; de Villiers, E.P.; Carlton, J.M.; Hall, N.; Ren, Q.; Paulsen, I.T.; Pain, A.; Berriman, M.; et al. Genome sequence of Theileria parva, a bovine pathogen that transforms lymphocytes. Science 2005, 309, 134–137. [Google Scholar] [CrossRef]
- Cornillot, E.; Hadj–Kaddour, K.; Dassouli, A.; Noel, B.; Ranwez, V.; Vacherie, B.; Augagneur, Y.; Brès, V.; Duclos, A.; Randazzo, S.; et al. Sequencing of the smallest Apicomplexan genome from the human pathogen Babesia microti. Nucleic Acids Res. 2012, 40, 9102–9114. [Google Scholar] [CrossRef]
- Jackson, A.P.; Otto, T.D.; Darby, A.; Ramaprasad, A.; Xia, D.; Echaide, I.E.; Farber, M.; Gahlot, S.; Gamble, J.; Gupta, D.; et al. The evolutionary dynamics of variant antigen genes in Babesia reveal a history of genomic innovation underlying host–parasite interaction. Nucleic Acids Res. 2014, 42, 7113–7131. [Google Scholar] [CrossRef]
- Gardner, M.J.; Hall, N.; Fung, E.; White, O.; Berriman, M.; Hyman, R.W.; Carlton, J.M.; Pain, A.; Nelson, K.E.; Bowman, S.; et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 2002, 419, 498–511. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L. –T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ–TREE: A fast and effective stochastic algorithm for estimating maximum–likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and taxonomy in diagnostics for food security: Soft–rotting enterobacterial plant pathogens. Anal. Methods 2016, 8, 12–24. [Google Scholar] [CrossRef]
- Kubota, S.; Sugimoto, C.; Kakuda, T.; Onuma, M. Analysis of immunodominant piroplasm surface antigen alleles in mixed populations of Theileria sergenti and T. buffeli. Int. J. Parasitol. 1996, 26, 741–747. [Google Scholar] [CrossRef]
- Bishop, R.; Musoke, A.; Morzaria, S.; Gardner, M.; Nene, V. Theileria: Intracellular protozoan parasites of wild and domestic ruminants transmitted by ixodid ticks. Parasitology 2004, 129, S271–S283. [Google Scholar] [CrossRef]
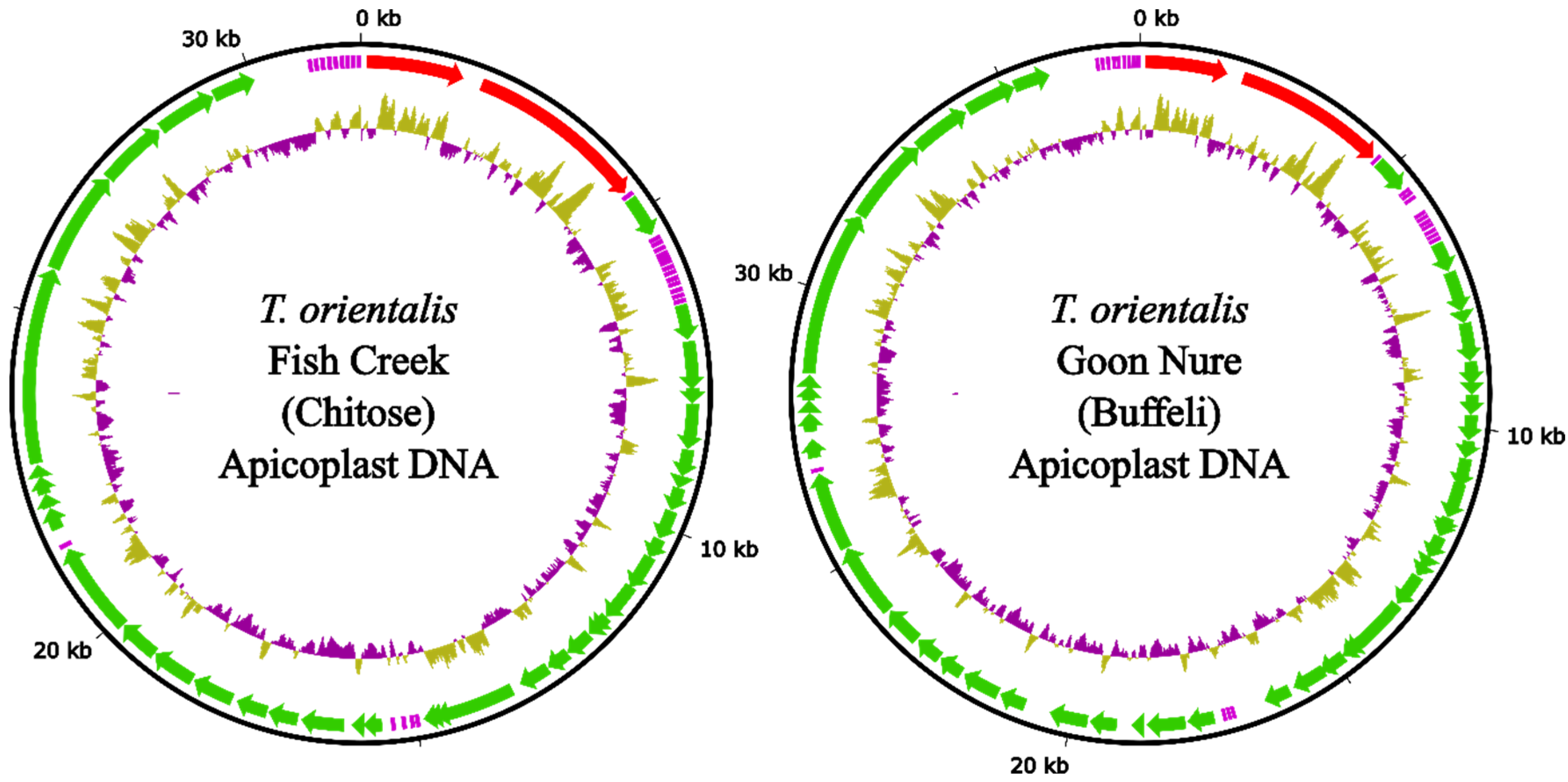
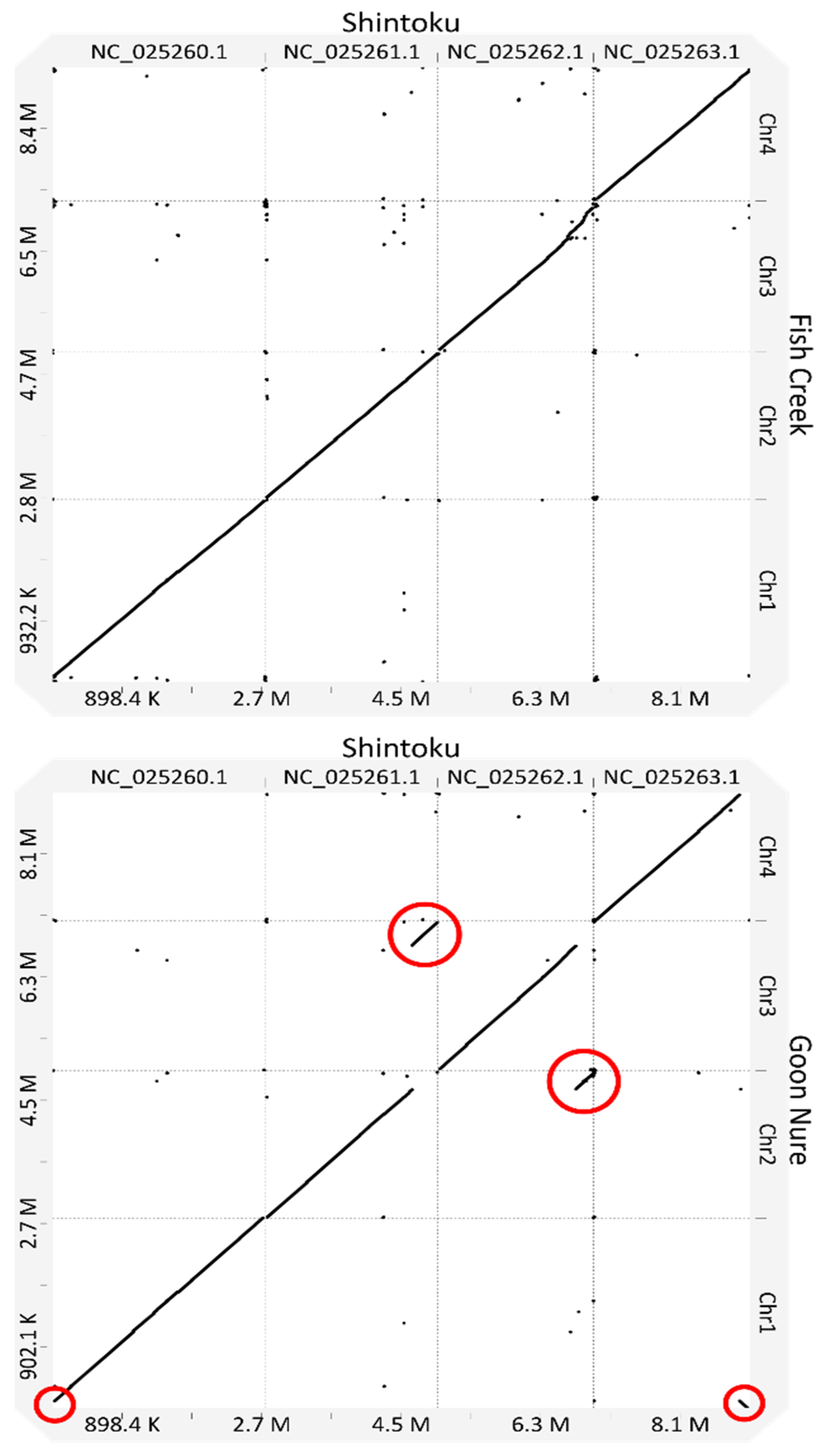




| Genotype | Assembler | Total Contigs | Contigs (≥50 kb) | Total Length | N50 | Largest Contig (bp) |
|---|---|---|---|---|---|---|
| Chitose (Fish Creek) | Flye | 14–16 | 7–8 | 9,344,963 | 2,171,492 | 2,745,486 |
| Miniasm | 7–10 | 5–6 | 9,559,641 | 2,254,955 | 2,765,560 | |
| Necat | 4–6 | 4–6 | 9,416,796 | 2,296,410 | 2,765,760 | |
| Raven | 5–6 | 4 | 9,365,432 | 2,296,609 | 2,770,085 | |
| Shasta | 9–12 | 4–5 | 9,427,530 | 2,242,537 | 2,765,028 | |
| Buffeli (Goon Nure) | Flye | 20 | 7–12 | 9,316,485 | 1,958,568 | 2,504,925 |
| Miniasm | 9–14 | 6–12 | 9,531,547 | 1,733,126 | 2,109,761 | |
| Necat | 10–19 | 10–19 | 10,547,787 | 1,967,178 | 2,912,118 | |
| Raven | 9–12 | 4–6 | 9,269,508 | 2,079,080 | 2,896,450 | |
| Shasta | 17 | 5–7 | 9,313,556 | 2,177,669 | 2,788,592 |
| Isolate | Chr 1 | Chr 2 | Chr 3 | Chr 4 | Apicoplast | Mitochondria |
|---|---|---|---|---|---|---|
| Shintoku | 2,746,313 | 2,216,979 | 2,000,793 | 2,019,511 | 24,173 * | 2595 * |
| Fish Creek | 2,765,963 | 2,233,854 | 2,297,733 | 2,024,851 | 31,688 | 6231 |
| Goon Nure | 2,785,604 | 2,153,779 | 2,196,581 | 1,884,878 | 37,498 | 5965 |
| Shintoku (Ikeda) | Fish Creek (Chitose) | Goon Nure (Buffeli) | |
|---|---|---|---|
| Genome | |||
| Total predicted genes | 4058 | 3980 | 3924 |
| Total predicted mRNA | 4002 | 3907 | 3848 |
| Total predicted tRNA | 47 | 66 | 69 |
| Total predicted rRNA | 9 | 7 | 7 |
| Total predicted CDS | 4002 | 3907 | 3848 |
| Percentage coding sequence | 68.43 | 68.46 | 68.73 |
| Total annotated sequence length | 9,010,364 | 9,360,320 | 9,064,305 |
| Percentage GC | 41.53 | 38.84 | 37.46 |
| Genes (+tRNA and rRNA) | |||
| Longest gene | 26,436 | 23,559 | 25,877 |
| Shortest gene | 39 | 24 | 33 |
| Total gene length | 7,386,640 | 7,385,994 | 7,214,907 |
| Average gene length | 1820 | 1856 | 1839 |
| Average gene coding sequence | 1541 | 1640 | 1619 |
| Gene density (per 10,000 bp) | 450.37 | 425.2 | 432.91 |
| Percentage coding genes with introns | 78.3 | 76 | 76.1 |
| Exons | |||
| Total exon length | 6,180,198 | 6,424,419 | 6,246,171 |
| Total number of exons | 16,558 | 15,809 | 15,837 |
| Longest exon | 11,241 | 16,092 | 25,364 |
| Shortest exon | 2 | 3 | 3 |
| Average exon length | 373.2 | 406.4 | 394.4 |
| Percentage GC | 46.06 | 43.21 | 41.89 |
| Introns | |||
| Total intron length | 1,206,442 | 961,575 | 968,736 |
| Total number of introns | 12,500 | 11,829 | 11,913 |
| Longest intron | 5418 | 6291 | 4043 |
| Shortest intron | 4 | 11 | 11 |
| Average intron length | 96.5 | 81.3 | 81.3 |
| Average introns per gene | 3.1 | 3 | 3 |
| Percentage GC | 34.12 | 29.54 | 27.26 |
| Intergenic regions | |||
| Total intergenic length | 1,636,669 | 1,974,520 | 1,849,793 |
| Total intergenic regions | 4020 | 3962 | 3901 |
| Longest intergenic region | 9728 | 8374 | 18,440 |
| Shortest intergenic region | 1 | 1 | 1 |
| Average intergenic length | 407.1 | 498.4 | 474.2 |
| Percentage GC | 29.9 | 29.12 | 27.86 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yam, J.; Bogema, D.R.; Micallef, M.L.; Djordjevic, S.P.; Jenkins, C. Complete Genomes of Theileria orientalis Chitose and Buffeli Genotypes Reveal within Species Translocations and Differences in ABC Transporter Content. Pathogens 2022, 11, 801. https://doi.org/10.3390/pathogens11070801
Yam J, Bogema DR, Micallef ML, Djordjevic SP, Jenkins C. Complete Genomes of Theileria orientalis Chitose and Buffeli Genotypes Reveal within Species Translocations and Differences in ABC Transporter Content. Pathogens. 2022; 11(7):801. https://doi.org/10.3390/pathogens11070801
Chicago/Turabian StyleYam, Jerald, Daniel R. Bogema, Melinda L. Micallef, Steven P. Djordjevic, and Cheryl Jenkins. 2022. "Complete Genomes of Theileria orientalis Chitose and Buffeli Genotypes Reveal within Species Translocations and Differences in ABC Transporter Content" Pathogens 11, no. 7: 801. https://doi.org/10.3390/pathogens11070801
APA StyleYam, J., Bogema, D. R., Micallef, M. L., Djordjevic, S. P., & Jenkins, C. (2022). Complete Genomes of Theileria orientalis Chitose and Buffeli Genotypes Reveal within Species Translocations and Differences in ABC Transporter Content. Pathogens, 11(7), 801. https://doi.org/10.3390/pathogens11070801







