Antimicrobial Resistance, Pathogenic, and Molecular Characterization of Escherichia coli from Diarrheal Patients in South Korea
Abstract
:1. Introduction
2. Results
2.1. Identification and Pathogenic Properties of E. coli
2.2. Antimicrobial Susceptibility
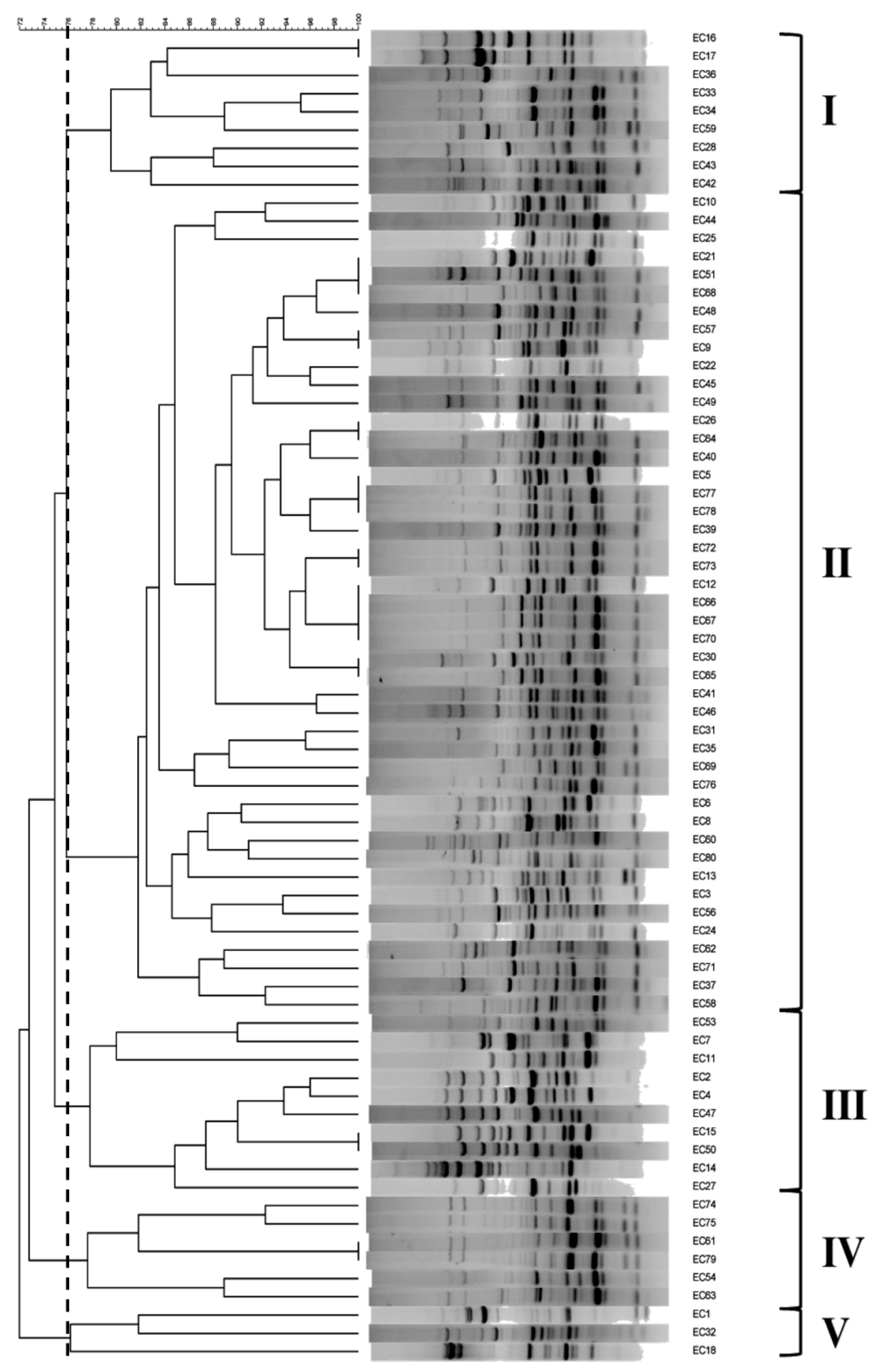
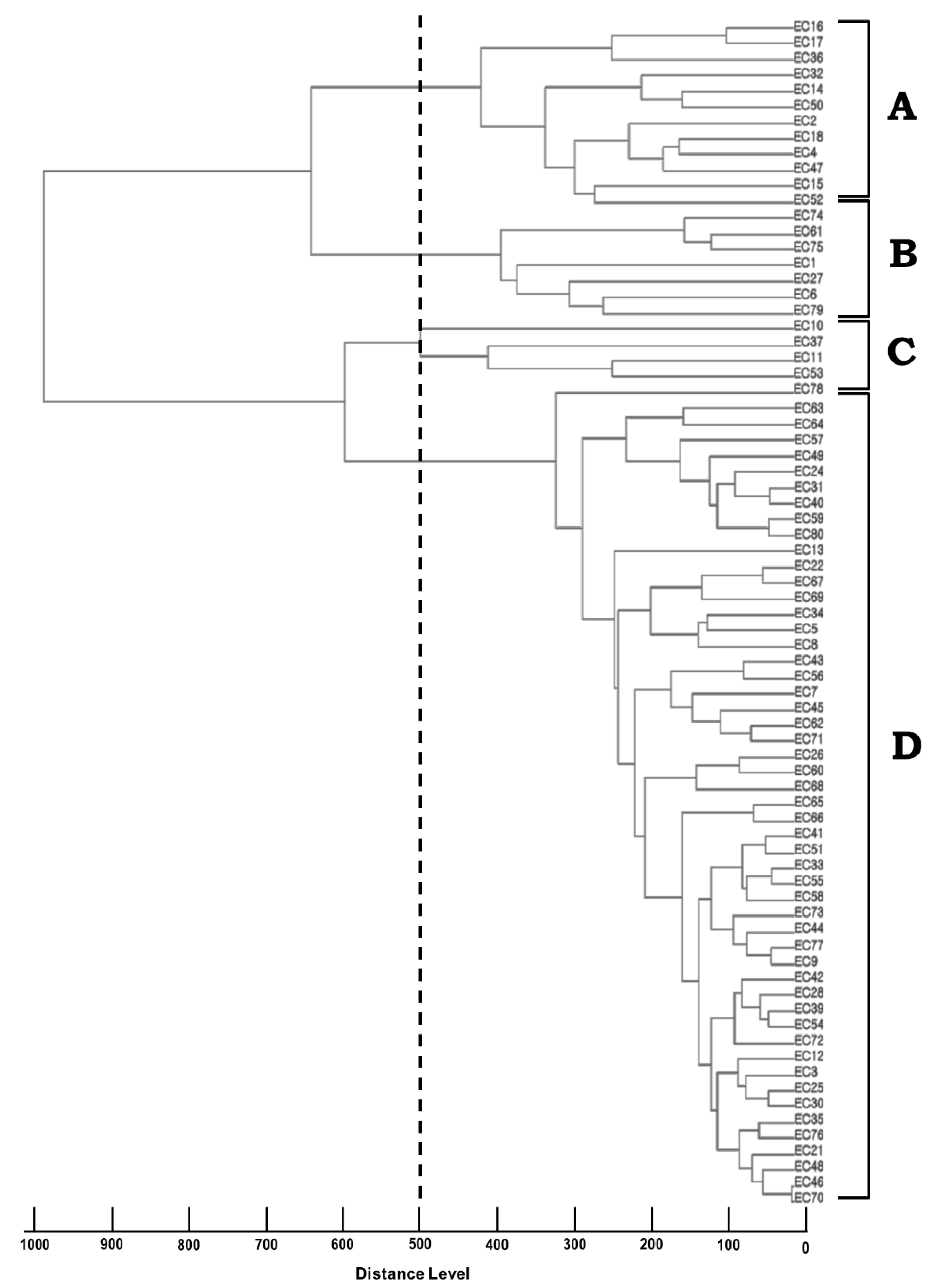
2.3. Molecular Typing Using RAPD and MALDI-TOF MS
3. Discussion
4. Materials and Methods
4.1. E. coli Isolates
4.2. O-Serotyping
4.3. Characterization of Pathogenic Properties of E. coli by PCR
4.4. Antimicrobial Susceptibility Test
4.5. Sample Preparation and Measurement of MALDI-TOF MS
4.6. Random Amplified Polymorphic DNA (RAPD) Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Lee, H.; Yoon, Y. Etiological agents implicated in foodborne illness world wide. Food Sci. Anim. Resour. 2021, 41, 1. [Google Scholar] [CrossRef] [PubMed]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.-A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States—Major pathogens. Emerg. Infect. Dis. 2011, 17, 7. [Google Scholar] [CrossRef] [PubMed]
- Bari, M.L.; Yeasmin, S. Foodborne diseases and responsible agents. In Food Safety and Preservation; Elsevier: Cambridge, MA, USA, 2018; pp. 195–229. [Google Scholar]
- Wang, J.; Song, F.; Walia, K.; Farber, J.; Dara, R. Using convolutional neural networks to extract keywords and keyphrases: A case study for foodborne illnesses. In Proceedings of the 2019 18th IEEE International Conference on Machine Learning and Applications (ICMLA), Boca Raton, FL, USA, 16–19 December 2019; pp. 1398–1403. [Google Scholar]
- Alizade, H.; Teshnizi, S.H.; Azad, M.; Shojae, S.; Gouklani, H.; Davoodian, P.; Ghanbarpour, R. An overview of diarrheagenic Escherichia coli in Iran: A systematic review and meta-analysis. J. Res. Med. Sci. Off. J. Isfahan Univ. Med. Sci. 2019, 24, 23. [Google Scholar]
- Fratamico, P.M.; DebRoy, C.; Liu, Y.; Needleman, D.S.; Baranzoni, G.M.; Feng, P. Advances in molecular serotyping and subtyping of Escherichia coli. Front. Microbiol. 2016, 7, 644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valilis, E.; Ramsey, A.; Sidiq, S.; DuPont, H.L. Non-O157 Shiga toxin-producing Escherichia coli—A poorly appreciated enteric pathogen: Systematic review. Int. J. Infect. Dis. 2018, 76, 82–87. [Google Scholar] [CrossRef] [Green Version]
- Amaretti, A.; Righini, L.; Candeliere, F.; Musmeci, E.; Bonvicini, F.; Gentilomi, G.A.; Rossi, M.; Raimondi, S. Antibiotic resistance, virulence factors, phenotyping, and genotyping of non-Escherichia coli Enterobacterales from the gut microbiota of healthy subjects. Int. J. Mol. Sci. 2020, 21, 1847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, C.G.; Kruczkiewicz, P.; Guan, C.; McCorrister, S.J.; Chong, P.; Wylie, J.; van Caeseele, P.; Tabor, H.A.; Snarr, P.; Gilmour, M.W. Evaluation of MALDI-TOF mass spectroscopy methods for determination of Escherichia coli pathotypes. J. Microbiol. Methods 2013, 94, 180–191. [Google Scholar] [CrossRef]
- El-Badawy, M.F.; Tawakol, W.M.; Maghrabi, I.A.; Mansy, M.S.; Shohayeb, M.M.; Ashour, M.S. Iodometric and molecular detection of ESBL production among clinical isolates of E. coli fingerprinted by ERIC-PCR: The first Egyptian report declares the emergence of E. coli O25b-ST131clone harboring bla GES. Microb. Drug Resist. 2017, 23, 703–717. [Google Scholar] [CrossRef]
- Książczyk, M.; Kuczkowski, M.; Dudek, B.; Korzekwa, K.; Tobiasz, A.; Korzeniowska-Kowal, A.; Paluch, E.; Wieliczko, A.; Bugla-Płoskońska, G. Application of routine diagnostic procedure, VITEK 2 compact, MALDI-TOF MS, and PCR assays in identification procedure of bacterial strain with ambiguous phenotype. Curr. Microbiol. 2016, 72, 570–582. [Google Scholar] [CrossRef]
- Pormohammad, A.; Nasiri, M.J.; Azimi, T. Prevalence of antibiotic resistance in Escherichia coli strains simultaneously isolated from humans, animals, food, and the environment: A systematic review and meta-analysis. Infect. Drug Resist. 2019, 12, 1181. [Google Scholar] [CrossRef] [Green Version]
- Magiorakos, A.; Srinivasan, A.; Carey, R.; Carmeli, Y.; Falagas, M.; Giske, C.; Harbarth, S.; Hinndler, J. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2020, 2012, 268–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farber, J. An introduction to the hows and whys of molecular typing. J. Food Prot. 1996, 59, 1091–1101. [Google Scholar] [CrossRef] [PubMed]
- Darkazanli, M.; Kiseleva, I.; Darkazanli, K. Genetic Diversity of E. coli O157: H7 Isolated from Some Leafy Greens, Irrigated by Aleppo River, Using Random Amplified Polymorphic DNA (RAPD) Marker. Russ. Agric. Sci. 2018, 44, 146–152. [Google Scholar]
- de Lagarde, M.; Vanier, G.; Desmarais, G.; Kohan-Ghadr, H.R.; Arsenault, J.; Fairbrother, J. A new multidrug-resistant enterotoxigenic Escherichia coli pulsed-field gel electrophoresis cluster associated with enrofloxacin non-susceptibility in diseased pigs. J. Appl. Microbiol. 2021, 130, 707–721. [Google Scholar] [CrossRef] [PubMed]
- Jazeela, K.; Chakraborty, G.; Shetty, S.S.; Rohit, A.; Karunasagar, I.; Vijaya Kumar, D. Comparison of mismatch amplification mutation assay PCR and PCR-restriction fragment length polymorphism for detection of major mutations in gyrA and parC of Escherichia coli associated with fluoroquinolone resistance. Microb. Drug Resist. 2019, 25, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Kazibwe, G.; Katami, P.; Alinaitwe, R.; Alafi, S.; Nanteza, A.; Nakavuma, J.L. Bacteriophage activity against and characterisation of avian pathogenic Escherichia coli isolated from colibacillosis cases in Uganda. PLoS ONE 2020, 15, e0239107. [Google Scholar] [CrossRef] [PubMed]
- Kotłowski, R.; Grecka, K.; Kot, B.; Szweda, P. New approaches for Escherichia coli Genotyping. Pathogens 2020, 9, 73. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Furevi, A.; Perepelov, A.V.; Guo, X.; Cao, H.; Wang, Q.; Reeves, P.R.; Knirel, Y.A.; Wang, L.; Widmalm, G. Structure and genetics of Escherichia coli O antigens. FEMS Microbiol. Rev. 2020, 44, 655–683. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, A.; Takahashi, H.; Arai, M.; Tsuchiya, T.; Wada, S.; Fujimoto, Y.; Shimabara, Y.; Kuda, T.; Kimura, B. Molecular subtyping for source tracking of Escherichia coli using core genome multilocus sequence typing at a food manufacturing plant. PLoS ONE 2021, 16, e0261352. [Google Scholar] [CrossRef]
- Govindarajan, D.K.; Viswalingam, N.; Meganathan, Y.; Kandaswamy, K. Adherence patterns of Escherichia coli in the intestine and its role in pathogenesis. Med. Microecol. 2020, 5, 100025. [Google Scholar] [CrossRef]
- Jeong, Y.-W.; Kim, T.-E.; Kim, J.-H.; Kwon, H.-J. Pathotyping avian pathogenic Escherichia coli strains in Korea. J. Vet. Sci. 2012, 13, 145–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ağbaş, A.; Göknar, N.; Akıncı, N.; Yıldırım, Z.Y.; Taşdemir, M.; Benzer, M.; Gökçe, İ.; Candan, C.; Küçük, N.; Uzuner, S. Outbreak of Shiga toxin-producing Escherichia coli-associated hemolytic uremic syndrome in Istanbul in 2015: Outcome and experience with eculizumab. Pediatric. Nephrol. 2018, 33, 2371–2381. [Google Scholar] [CrossRef] [PubMed]
- DebRoy, C.; Roberts, E.; Valadez, A.M.; Dudley, E.G.; Cutter, C.N. Detection of Shiga toxin–producing Escherichia coli O26, O45, O103, O111, O113, O121, O145, and O157 serogroups by multiplex polymerase chain reaction of the wzx gene of the O-antigen gene cluster. Foodborne Pathog. Dis. 2011, 8, 651–652. [Google Scholar] [CrossRef]
- Fratamico, P.M.; Bagi, L.K.; Cray, W.C., Jr.; Narang, N.; Yan, X.; Medina, M.; Liu, Y. Detection by multiplex real-time polymerase chain reaction assays and isolation of Shiga toxin–producing Escherichia coli serogroups O26, O45, O103, O111, O121, and O145 in ground beef. Foodborne Pathog. Dis. 2011, 8, 601–607. [Google Scholar] [CrossRef] [Green Version]
- Perelle, S.; Dilasser, F.; Grout, J.; Fach, P. Screening food raw materials for the presence of the world’s most frequent clinical cases of Shiga toxin-encoding Escherichia coli O26, O103, O111, O145 and O157. Int. J. Food Microbiol. 2007, 113, 284–288. [Google Scholar] [CrossRef]
- Tzschoppe, M.; Martin, A.; Beutin, L. A rapid procedure for the detection and isolation of enterohaemorrhagic Escherichia coli (EHEC) serogroup O26, O103, O111, O118, O121, O145 and O157 strains and the aggregative EHEC O104: H4 strain from ready-to-eat vegetables. Int. J. Food Microbiol. 2012, 152, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Bautista, V.A.L.M.; Mendoza, B.C. Serogroup, pathotype and multiple drug resistance of Escherichia coli strains isolated from the cloaca of layer chickens in San Jose, Batangas, Philippines. Philipp. Sci. Lett. 2018, 11, 69–77. [Google Scholar]
- Moyo, S.J.; Maselle, S.Y.; Matee, M.I.; Langeland, N.; Mylvaganam, H. Identification of diarrheagenic Escherichia coli isolated from infants and children in Dar es Salaam, Tanzania. BMC Infect. Dis. 2007, 7, 92. [Google Scholar] [CrossRef] [Green Version]
- Dubreuil, J.D.; Isaacson, R.E.; Schifferli, D.M. Animal enterotoxigenic Escherichia coli. EcoSal Plus 2016, 7, 1. [Google Scholar] [CrossRef] [Green Version]
- Mare, A.D.; Ciurea, C.N.; Man, A.; Tudor, B.; Moldovan, V.; Decean, L.; Toma, F. Enteropathogenic Escherichia coli—A Summary of the Literature. Gastroenterol. Insights 2021, 12, 28–40. [Google Scholar] [CrossRef]
- Liu, L.; Saitz-Rojas, W.; Smith, R.; Gonyar, L.; In, J.G.; Kovbasnjuk, O.; Zachos, N.C.; Donowitz, M.; Nataro, J.P.; Ruiz-Perez, F. Mucus layer modeling of human colonoids during infection with enteroaggragative E. coli. Sci. Rep. 2020, 10, 10533. [Google Scholar] [CrossRef] [PubMed]
- Zare, S.; Derakhshandeh, A.; Haghkhah, M.; Naziri, Z.; Broujeni, A.M. Molecular typing of Staphylococcus aureus from different sources by RAPD-PCR analysis. Heliyon 2019, 5, e02231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faraji, Z.; Khasheii, B.; Ghaemi, E.A.; Anvari, S.; Rajabnia, R.; Jamali, A. Molecular Typing of Uropathogenic Escherichia coli Strains Isolated from Patients by Random Amplified Polymorphic DNA-PCR (RAPD-PCR). Arch. Med. Lab. Sci. 2021, 7, e17. [Google Scholar] [CrossRef]
- Albesharat, R.; Ehrmann, M.A.; Korakli, M.; Yazaji, S.; Vogel, R.F. Phenotypic and genotypic analyses of lactic acid bacteria in local fermented food, breast milk and faeces of mothers and their babies. Syst. Appl. Microbiol. 2011, 34, 148–155. [Google Scholar] [CrossRef]
- Firacative, C.; Trilles, L.; Meyer, W. MALDI-TOF MS enables the rapid identification of the major molecular types within the Cryptococcus neoformans/C. gattii species complex. PLoS ONE 2012, 7, e37566. [Google Scholar] [CrossRef] [Green Version]
- Simmon, K.E.; Fang, D.C.; Tesic, V.; Khot, P.D.; Giangeruso, E.; Bolesta, E.S.; Wagner-Reiss, K.M.; Fisher, M.A.; Petti, C.A.; Han, X.Y. Isolation and characterization of “Pseudomonas andersonii” from four cases of pulmonary granulomas and emended species description. J. Clin. Microbiol. 2011, 49, 1518–1523. [Google Scholar] [CrossRef] [Green Version]
- Lindenstrauß, A.G.; Pavlovic, M.; Bringmann, A.; Behr, J.; Ehrmann, M.A.; Vogel, R.F. Comparison of genotypic and phenotypic cluster analyses of virulence determinants and possible role of CRISPR elements towards their incidence in Enterococcus faecalis and Enterococcus faecium. Syst. Appl. Microbiol. 2011, 34, 553–560. [Google Scholar] [CrossRef]
- More, S.J. European perspectives on efforts to reduce antimicrobial usage in food animal production. Ir. Vet. J. 2020, 73, 2. [Google Scholar] [CrossRef] [Green Version]
- Pokharel, S.; Shrestha, P.; Adhikari, B. Antimicrobial use in food animals and human health: Time to implement ‘One Health’ approach. Antimicrob. Resist. Infect. Control 2020, 9, 181. [Google Scholar] [CrossRef]
- Karama, M.; Cenci-Goga, B.T.; Malahlela, M.; Smith, A.M.; Keddy, K.H.; El-Ashram, S.; Kabiru, L.M.; Kalake, A. Virulence characteristics and antimicrobial resistance profiles of shiga toxin-producing Escherichia coli isolates from humans in South Africa: 2006–2013. Toxins 2019, 11, 424. [Google Scholar] [CrossRef] [Green Version]
- Eltai, N.O.; Al Thani, A.A.; Al Hadidi, S.H.; Al Ansari, K.; Yassine, H.M. Antibiotic resistance and virulence patterns of pathogenic Escherichia coli strains associated with acute gastroenteritis among children in Qatar. BMC Microbiol. 2020, 20, 54. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Mosci, R.E.; Anderson, C.M.; Snyder, B.A.; Collins, J.; Rudrik, J.T.; Manning, S.D. Antimicrobial drug–resistant Shiga toxin–producing Escherichia coli infections, Michigan, USA. Emerg. Infect. Dis. 2017, 23, 1609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aranda, K.R.; Fabbricotti, S.H.; Fagundes-Neto, U.; Scaletsky, I.C. Single multiplex assay to identify simultaneously enteropathogenic, enteroaggregative, enterotoxigenic, enteroinvasive and Shiga toxin-producing Escherichia coli strains in Brazilian children. FEMS Microbiol. Lett. 2007, 267, 145–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanco, M.; Blanco, J.; Mora, A.; Rey, J.; Alonso, J.; Hermoso, M.; Hermoso, J.; Alonso, M.; Dahbi, G.; González, E. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from healthy sheep in Spain. J. Clin. Microbiol. 2003, 41, 1351–1356. [Google Scholar] [CrossRef] [Green Version]
- Cho, S.-H.; Kim, J.-H.; Kim, J.-C.; Shin, H.-H.; Kang, Y.-H.; Lee, B.-K. Surveillance of bacterial pathogens associated with acute diarrheal disease in the Republic of Korea during one year, 2003. J. Microbiol. 2006, 44, 327–335. [Google Scholar]
- Jenssen, G.R.; Veneti, L.; Lange, H.; Vold, L.; Naseer, U.; Brandal, L.T. Implementation of multiplex PCR diagnostics for gastrointestinal pathogens linked to increase of notified Shiga toxin-producing Escherichia coli cases in Norway, 2007–2017. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 801–809. [Google Scholar] [CrossRef] [Green Version]
- López-Saucedo, C.; Cerna, J.F.; Villegas-Sepulveda, N.; Thompson, R.; Velazquez, F.R.; Torres, J.; Tarr, P.I.; Estrada-García, T. Single multiplex polymerase chain reaction to detect diverse loci associated with diarrheagenic Escherichia coli. Emerg. Infect. Dis. 2003, 9, 127. [Google Scholar] [CrossRef]
- Wayne, P. Clinical and laboratory standards institute. Performance standards for antimicrobial susceptibility testing. Inform. Suppl. 2011, 31, 100–121. [Google Scholar]
- Wikler, M.A. Performance Standards for Antimicrobial Susceptibility Testing; Sixteenth Informational Supplement, M 100-S 16; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2006. [Google Scholar]
| Pathotypes (%) | Virulence Genes | Total No. of Isolates | O Serotype (Total No. of Isolates) |
|---|---|---|---|
| ETEC (6.6) | st | 5 | O25(3), O159(1), UN(1) |
| ETEC and EAEC (2.7) | st, aggR | 2 | O159(1), UN(1) |
| ETEC and EPEC (6.6) | st, eaeA, bfpA | 1 | O8(1) |
| st, eaeA | 2 | UN(2) | |
| st, bfpA | 2 | UN(2) | |
| EPEC (47.0) | eaeA | 27 | O8(2), O18(2), O20(2), O55(1), O119(3), O125(1), O153(1), O166(2), O127a(2), O28ac(1), UN(10) |
| eaeA, bfpA | 2 | UN(2) | |
| bfpA | 6 | O6(1), O153(1), UN(4) | |
| EPEC and EAEC (4.0) | eaeA, aggR | 3 | O159(1), O169(1), UN(1) |
| EAEC (33.3) | aggR | 25 | O1(1), O8(1), O20(2), O25(1), O78(1), O166(4), O28ac(2), UN(13) |
| Antimicrobial (Abbreviations) | No. of Resistant Isolates (%) | No. of Intermediate Isolates (%) | No. of Susceptible Isolates (%) |
|---|---|---|---|
| Penicillins | |||
| Ampicillin (AM) | 30 (40.0) | 0 | 45 (60) |
| Ticarcillin (TIC) | 29 (38.7) | 1 (1.3) | 45 (60) |
| ß-Lactam/ß-Lactamase inhibitor combinations | |||
| Ampicillin/Sulbactam (SAM) | 12 (16.0) | 8 (10.7) | 55 (73.3) |
| Cephems | |||
| Cefazolin (CZ) | 6 (8.0) | 0 | 69 (92) |
| Cefepime (FEB) | 1 (1.3) | 3 (4.0) | 71 (94.6) |
| Cefotaxime (CTX) | 4 (5.3) | 2 (2.7) | 69 (92) |
| Cefotetan (CTT) | 1 (1.3) | 0 | 74 (98.6) |
| Cephalothin (CF) | 11 (14.7) | 19 (25.3) | 45 (60) |
| Phenicols | |||
| Chloramphenicol (C) | 4 (5.3) | 0 | 71 (94.6) |
| Fluoroquinolones | |||
| Ciprofloxacin (CIP) | 10 (13.3) | 4 (5.3) | 61 (81.3) |
| Aminoglycosides | |||
| Amikacin (AN) | 5 (6.6) | 1 (1.3) | 69 (92) |
| Gentamicin (GM) | 14 (18.7) | 3 (4.0) | 58 (77.3) |
| Carbapenems | |||
| Imipenem (IPM) | 1 (1.3) | 0 | 74 (98.6) |
| Quinolones | |||
| Nalidixic acid (NA) | 33 (44.0) | 1 (1.3) | 41 (54.6) |
| Tetracyclines | |||
| Tetracycline (TE) | 31 (41.3) | 0 | 44 (58.6) |
| Folate pathway inhibitors | |||
| Trimethoprim/sulfamethoxazole (SXT) | 26 (34.7) | 2 (2.7) | 47 (62.6) |
| No. of Antimicrobials | Resistance Patterns | No. of Isolates |
|---|---|---|
| 0 | - | 23 |
| 1 | SXT | 1 |
| CZ | 1 | |
| TE | 1 | |
| NA | 8 | |
| TIC | 2 | |
| AM | 1 | |
| 2 | TE, NA | 1 |
| TE, TIC | 1 | |
| AN, TE | 1 | |
| AN, NA | 1 | |
| 3 | AM, CF, SAM | 1 |
| TE, NA, TIC | 2 | |
| CF, NA, TIC | 1 | |
| C, TE, NA | 1 | |
| 4 | AM, SXT, NA, TIC | 1 |
| AM, SXT, TE, SAM | 1 | |
| AM, NA, SAM, TIC | 1 | |
| AM, SXT, TE, TIC | 3 | |
| AM, SXT, TE, NA | 1 | |
| 5 | AM, SXT, TE, NA, TIC | 2 |
| AM, CF, C, TE, TIC | 1 | |
| GM, SXT, C, NA, TIC | 1 | |
| AM, CF, CIP, IPM, TE | 1 | |
| AM, GM, CTT, SAM, TIC | 1 | |
| 6 | AM, CF, SXT, C, TE, TIC | 1 |
| AM, CIP, SXT, TE, NA, SAM | 1 | |
| AM, GM, SXT, TE, NA, TIC | 3 | |
| AM, CZ, CF, SXT, TE, NA | 1 | |
| AM, CF, SXT, TE, NA, TIC | 1 | |
| 7 | AM, GM, CIP, SXT, TE, NA, TIC | 3 |
| 8 | AM, GM, CIP, SXT, TE, NA, SAM, TIC | 2 |
| AM, GM, AN, CTX, CIP, SXT, TE, SAM | 1 | |
| AM, CZ, CF, AN, SXT, NA, SAM, TIC | 1 | |
| 10 | AM, CZ, CF, GM, AN, FEP, CTX, SXT, SAM, TIC | 1 |
| 11 | AM, CZ, CF, GM, CTX, CIP, SXT, TE, NA, SAM, TIC | 2 |
| Total | 35 patterns | 75 strains |
| No. (%) of Strains | MALDI-TOF MS | ||||
|---|---|---|---|---|---|
| RAPD | A | B | C | D | Total N (%) |
| I | 3 (4.0) | - | - | 6 (8.0) | 9 (12.0) |
| II | - | 1 (1.3) | 2 (2.7) | 41 (54.6) * | 44 (58.7) * |
| III | 6 (8.0) | 1 (1.3) | 3 (4.0) | 1 (1.3) | 11 (14.7) |
| IV | - | 4 (5.3) | - | 2 (2.7) | 6 (8.0) |
| V | 2 (2.7) | 1 (1.3) | - | - | 3 (4.0) |
| None | 1 (1.3) | - | - | 1 (1.3) | 2 (2.7) |
| Total N (%) | 12 (16.0) | 7 (9.3) | 5 (6.7) | 51 (68.0) * | 75 (100) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, S.B.; Park, Y.K.; Ha, M.W.; Thompson, K.D.; Jung, T.S. Antimicrobial Resistance, Pathogenic, and Molecular Characterization of Escherichia coli from Diarrheal Patients in South Korea. Pathogens 2022, 11, 385. https://doi.org/10.3390/pathogens11040385
Park SB, Park YK, Ha MW, Thompson KD, Jung TS. Antimicrobial Resistance, Pathogenic, and Molecular Characterization of Escherichia coli from Diarrheal Patients in South Korea. Pathogens. 2022; 11(4):385. https://doi.org/10.3390/pathogens11040385
Chicago/Turabian StylePark, Seong Bin, Yon Kyoung Park, Min Woo Ha, Kim D. Thompson, and Tae Sung Jung. 2022. "Antimicrobial Resistance, Pathogenic, and Molecular Characterization of Escherichia coli from Diarrheal Patients in South Korea" Pathogens 11, no. 4: 385. https://doi.org/10.3390/pathogens11040385
APA StylePark, S. B., Park, Y. K., Ha, M. W., Thompson, K. D., & Jung, T. S. (2022). Antimicrobial Resistance, Pathogenic, and Molecular Characterization of Escherichia coli from Diarrheal Patients in South Korea. Pathogens, 11(4), 385. https://doi.org/10.3390/pathogens11040385






