Exploring the Host Range of Rose rosette Virus among Herbaceous Annual Plants
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and RRV-Containing Sap Inoculum
2.2. RRV Infectious Clone & Agrobacterium Infiltration
2.3. Sampling and Nucleic Acid Extraction
2.4. Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
2.5. Photography and Compiling Images
2.6. Contagious Sap Extraction and Transmission Electron Microscopy
2.7. Bioinformatics Analysis
3. Results
3.1. RT-PCR Detection of RRV in Systemically Infected Plants
3.2. Testing the Susceptibility of Seventeen Plant Species to RRV Infection
3.3. Comparing the Infectious Clone and Crude Sap Inoculum for Achieving Systemic RRV Infection
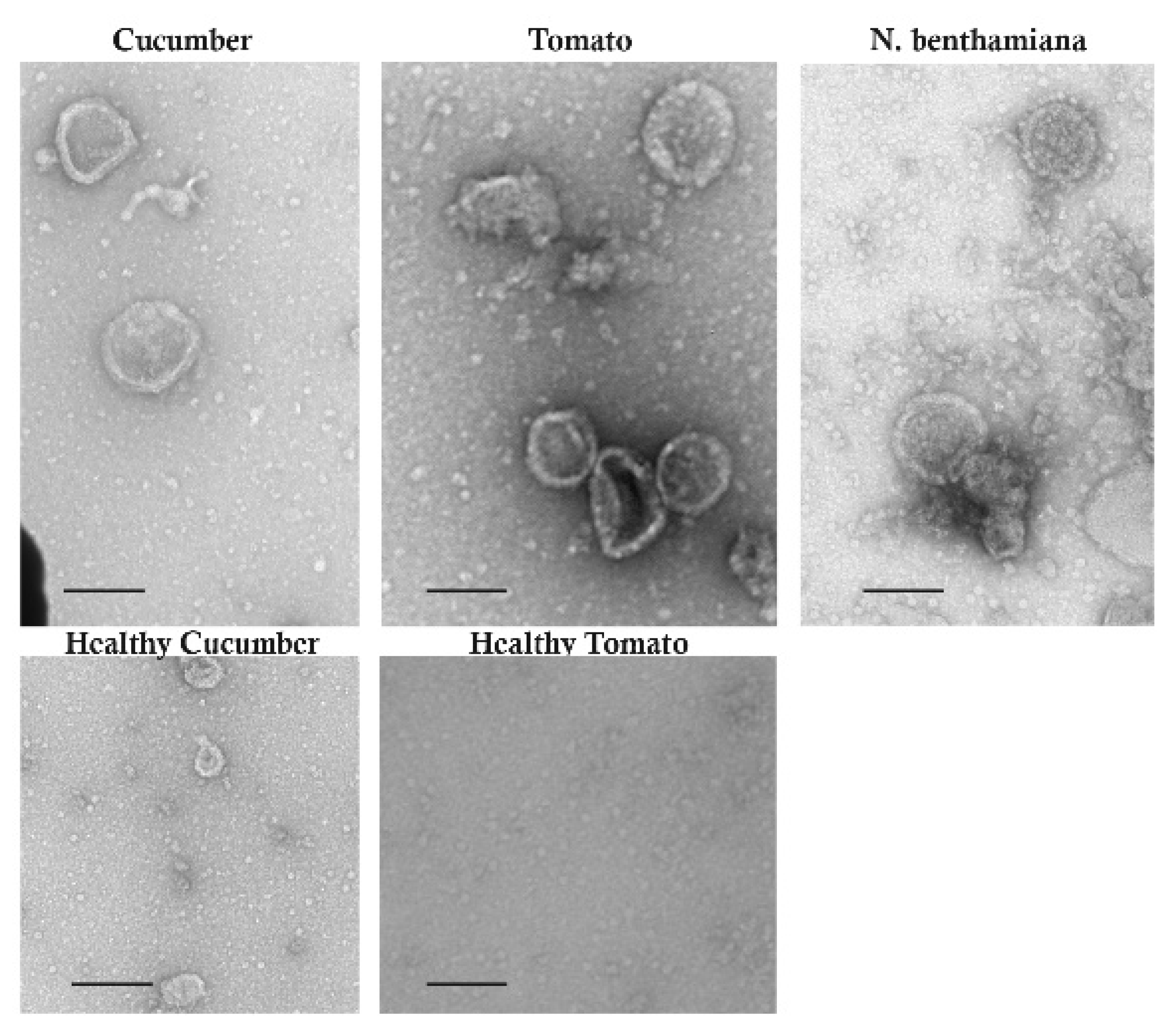
3.4. Symptoms of RRV-Infected Plants and Virions
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pemberton, H.B.; Ong, K.; Windham, M.; Olson, J.; Byrne, D.H. What is Rose rosette disease? HortScience 2018, 53, 592–595. [Google Scholar] [CrossRef]
- Crowe, F.J. Witches’ broom of rose: A new outbreak in several Central States. Plant Dis. 1983, 67, 544–546. [Google Scholar] [CrossRef]
- Waliczek, T.M.; Byrne, D.; Holeman, D. Opinions of landscape roses available for purchase and preferences for the future market. Horttechnology 2018, 28, 807–814. [Google Scholar] [CrossRef]
- Conners, I.L. Twentieth Annual Report of the Canadian Plant Disease Survey, 1940; Agriculture and Agri Food Canada: Ontario, ON, Canada, 1941; p. 98. [Google Scholar]
- Laney, A.G.; Keller, K.E.; Martin, R.R.; Tzanetakis, I.E. A discovery 70 years in the making: Characterization of the Rose rosette virus. J. Gen. Virol. 2011, 92, 1727–1732. [Google Scholar] [CrossRef]
- Bostock, R.M.; Thomas, C.S.; Hoenisch, R.W.; Golino, D.A.; Vidalakis, G. Plant health: How diagnostic networks and interagency partnerships protect plant systems from pests and pathogens. Calif. Agric. 2014, 68, 117–124. [Google Scholar] [CrossRef]
- Aparicio Claros, N.; Shires, M.; Mollov, D.; Hammond, J.; Jordan, R.; Ochoa-Corona, F.M.; Olson, J.; Ong, K.L.; Rodriguez Salamanca, L. Rose rosette Disease: A Diagnostic Guide. Plant Health Prog. 2022. [Google Scholar] [CrossRef]
- Ong, K. Rose rosette Disease Monitoring Network. Available online: https://roserosette.org (accessed on 6 November 2022).
- Di Bello, P.L.; Ho, T.; Tzanetakis, I.E. The evolution of emaraviruses is becoming more complex: Seven segments identified in the causal agent of Rose rosette disease. Virus Res. 2015, 210, 241–244. [Google Scholar] [CrossRef]
- Mielke-Ehret, N.; Mühlbach, H.-P. Emaravirus: A novel genus of multipartite, negative strand RNA plant viruses. Viruses 2012, 4, 1515–1536. [Google Scholar] [CrossRef]
- Epstein, A.H.; Hill, J.H. The biology of Rose rosette disease: A mite-associated disease of uncertain aetiology. J. Phytopathol. 1995, 143, 353–360. [Google Scholar] [CrossRef]
- Kim, K.S.; Ahn, K.K.; Gergerich, R.C. Double membrane-bound virus-like particles associated with Rose rosette and some other diseases transmitted by eriophyid mites. In Proceedings of the International Symposium: Rose Rosette Other Eriophyid Mite-Transmitted Plant Disease Agents Uncertain Etiology; Epstein, H.A., Hill, J.H., Eds.; Iowa State University: Ames, IA, USA, 1995; pp. 34–41. [Google Scholar]
- Verchot, J.; Herath, V.; Urrutia, C.D.; Gayral, M.; Lyle, K.; Shires, M.K.; Ong, K.; Byrne, D. Development of a reverse genetic system for studying Rose rosette virus in whole plants. Mol. Plant-Microbe Interact. 2020, 33, 1209–1221. [Google Scholar] [CrossRef]
- Urrutia, C.D.; Romay, G.; Shaw, B.D.; Verchot, J. Advancing the Rose rosette virus minireplicon and encapsidation system by incorporating GFP, mutations, and the CMV 2b silencing suppressor. Viruses 2022, 14, 836. [Google Scholar] [CrossRef]
- Amrine, J.W., Jr.; Hindal, D.F.; Stasny, T.A.; Williams, R.L.; Coffman, C.C. Transmission of the Rose rosette disease agent to rosa multiflora by phyllocoptes fructiphilus (Acari: Eriophyidae). Ent. News 1988, 99, 239–252. [Google Scholar]
- Amrine, J.W., Jr.; Hindal, D.F.; Williams, R.; Appel, J.; Stasny, T.; Kassar, A. Rose rosette as a biocontrol of multiflora rose, 1987-1989. In Proceedings of the 43rd Annual Meeting of the Southern Weed Science Society, Atlanta, GA, USA, 15–17 January 1990; pp. 316–320. [Google Scholar]
- Seifers, D.L.; Martin, T.J.; Harvey, T.L.; Fellers, J.P.; Michaud, J.P. Identification of the wheat curl mite as the vector of Triticum mosaic virus. Plant Dis. 2008, 93, 25–29. [Google Scholar] [CrossRef]
- Caglayan, K.; Elci, E.; Ulubas Serce, C.; Kaya, K.; Gazel, M.; Medina, V. Detection of fig mosaic virus in viruliferous eriophyid mite Aceria ficus. J. Plant Pathol. 2012, 94, 629–634. [Google Scholar] [CrossRef]
- Epstein, A.H.A.H.; Hill, J.H.; Nutter, F.W.; Epstein, A.H.A.H.; Nutter, J.F.W. Augmentation of Rose rosette disease for biocontrol of multiflora rose (Rosa multiflora). Weed Sci. 1997, 45, 172–178. [Google Scholar] [CrossRef]
- Schroeder, D. Biological control of weeds. In Recent Advances in Weed Research.; Fletcher, W.W., Ed.; Commonwealth Agricultural Bureaux: Slough, UK, 1983; pp. 41–78. [Google Scholar]
- Laney, A.G.A.G. Characterization of the Causal Agents of Rose Rosette and Redbud Yellow Ringspot; University of Arkansas: Fayetteville, AR, USA, 2010. [Google Scholar]
- Epstein, A.H.; Hill, J.H. Status of Rose rosette disease as a biological control for multiflora rose. Plant Dis. 1999, 83, 92–101. [Google Scholar] [CrossRef]
- Di Bello, P.L.; Thekke-Veetil, T.; Druciarek, T.; Tzanetakis, I.E. Transmission attributes and resistance to Rose rosette virus. Plant Pathol. 2018, 67, 499–504. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Dobhal, S.; Olson, J.D.; Arif, M.; Garcia Suarez, J.A.; Ochoa-Corona, F.M. A simplified strategy for sensitive detection of Rose rosette virus compatible with three RT-PCR chemistries. J. Virol. Methods 2016, 232, 47–56. [Google Scholar] [CrossRef]
- Rohozinski, J.; Epstein, A.H.; Hill, J.H. Probable mechanical transmission of a virus-like agent from Rose rosette disease-infected multiflora rose to Nicotiana species. Ann. Appl. Biol. 2001, 138, 181–186. [Google Scholar] [CrossRef]
- Di, R.; Hill, J.H.; Epstein, A.H. Double-stranded RNA associated with the Rose rosette disase of multiflora rose. Plant Dis. 1990, 74, 56–58. [Google Scholar] [CrossRef]
- Babu, B.; Jeyaprakash, A.; Jones, D.; Schubert, T.S.; Baker, C.; Washburn, B.K.; Miller, S.H.; Poduch, K.; Knox, G.W.; Ochoa-Corona, F.M.; et al. Development of a rapid, sensitive TaqMan real-time RT-PCR assay for the detection of Rose rosette virus using multiple gene targets. J. Virol. Methods 2016, 235, 41–50. [Google Scholar] [CrossRef]
- Shires, M.K. Study of Resistance to Rose Rosette Disease Utilizing Field Research, Molecular Tools, and Transmission Methods; Texas A&M University: College Station, TX, USA, 2020. [Google Scholar]
- Allington, W.B.; Staples, R.; Viehmeyer, G. Transmission of Rose rosette virus by the eriophyid mite Phyllocoptes fructiphilus1. J. Econ. Entomol. 1968, 61, 1137–1140. [Google Scholar] [CrossRef]
- Babu, B.; Washburn, B.K.; Miller, S.H.; Poduch, K.; Sarigul, T.; Knox, G.W.; Ochoa-Corona, F.M.; Paret, M.L. A rapid assay for detection of Rose rosette virus using reverse transcription-recombinase polymerase amplification using multiple gene targets. J. Virol. Methods 2017, 240, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Babu, B.; Knox, G.; Paret, M.L.; Ochoa-Corona, F.M. Rose rosette disease: Recent advances on molecular diagnostic tools. HortScience 2018, 53, 596–600. [Google Scholar] [CrossRef]
- Golino, D.A.; Sim, S.T.; Cunningham, M.; Rowhani, A. Transmission of Rose mosaic viruses. Acta Hortic. 2007, 751, 217–224. [Google Scholar] [CrossRef]


| Plant Family | Plant Name | Scientific Name | Cultivar or Type |
|---|---|---|---|
| Cucurbitaceae | Cucumber | Cucumis sativus | Harris |
| Yellow squash | Cucurbita pepo var. recticollis | yellow crookneck squash | |
| Zucchini | Cucurbita pepo | dark green zucchini | |
| Winter squash | Cucurbita maxima | Butter cup | |
| Pepper | Capsicum annuum | California Wonder | |
| Tomato | Solanum lycopersicum | Brandywine heirloom | |
| Solanaceae | Tobacco | N. rustica | N/A * |
| Tobacco | N. benthamiana | N/A | |
| Tobacco | N. glutinosa | N/A | |
| Soybean | Glycine max | Uidori giant | |
| Leguminosae | Pea | Pisum sativum | Lincoln |
| Lupine | Lupinus sp. | N/A | |
| Malvaceae | Okra | Abelmoschus esculentus | Clemson Spineless |
| Amaranthaceae | Spinach | Spinacia oleracea | Bloomshade Long Standing |
| Goosefoot | Chenopodium amaranticolor | N/A | |
| Quinoa | C. quinoa | N/A | |
| Brassicaceae | Thale cress | Arabidopsis thaliana | Col-0 |
| Primer Set: | Name | Sequence 5′ to 3′ | Product Size (nt) | Position | Target Gene | Reference |
|---|---|---|---|---|---|---|
| 1 | RRVF RRVR | GCACATCCAACACTCTTGCAGC CTTATTTGAAGCTGCTCCTTGATTTC | 271 | 154–176 425–399 | ORF3 (NP) | [23] |
| 2 | RRV-2F RRV-2R | TGCTATAAGTCTCATTGGAAGAGAA CCTATAGCTTCATCATTCCTCTTTG | 104 | 881–906 986–961 | ORF3 (NP) | [25] |
| 3 | RRV3F2 RRV30917R | GGCATAGCTGTTTCTTATCTTTCTAGG AGGGCGAATTCTTCTCTTCC | 551 | 366–393 917–897 | ORF3 (NP) | [13] |
| 4 | agRRV4-F1 agRRV4-R1 | AAACTCAATCTACAGCTGGATTCAT GTCCATCTCTTGAGGGATATTTTCAG | 500 | 636–661 1136–1110 | ORF4 (MP) | [13] |
| 5 | RmActin-F RmActin-R | AGGGTTTGCTGGAGATGATG CGGGTTAAGAGGTGCTTCAG | 280 | Actin * | This study |
| Plant | 2 wpi | 3 wpi | 4 wpi | 5 wpi | Total † | Mock |
|---|---|---|---|---|---|---|
| Cucumber | 3 | 2 | 2 | 2 | 5/5 | 0/3 |
| Yellow squash | 1 | 0 | 0 | 0 | 1/10 | 0/3 |
| Winter squash | 0 | 0 | 3 | 4 | 5/5 | 0/3 |
| Pumpkin | 3 | - | - | - | 3/5 | 0/3 |
| Pepper | 2 | 2 | 2 | 4 | 4/5 | 0/3 |
| Tomato | 2 | 2 | 0 | 0 | 3/10 | 0/3 |
| N. rustica | 2 | 3 | 6 | 0 | 7/10 | 0/3 |
| N. glutinosa | 2 | 0 | 0 | - | 2/6 | 0/3 |
| Pea | 2 | 1 | 0 | 0 | 2/6 | 0/3 |
| Peanut | 2 | 2 | 3 | 4 | 4/6 | 0/3 |
| Soybean | 1 | 0 | 0 | 2 | 3/10 | 0/3 |
| Lupine | 0 | 0 | 0 | 0 | 0/5 | 0/3 |
| Okra | 0 | 2 | 2 | 2 | 3/5 | 0/3 |
| Spinach | 0 | 0 | 0 | 4 | 4/5 | 0/3 |
| C. amaranticolor | 0 | 0 | 2 | 2 | 3/5 | 0/3 |
| C. quinoa | 0 | 0 | 2 | 2 | 2/5 | 0/3 |
| Arabidopsis | 0 | 0 | 0 | 0 | 0/6 | 0/3 |
| Plant Type | Infectious cDNA Clone of RRV | Crude Sap from RRV-Infected Rose | ||||
|---|---|---|---|---|---|---|
| Local (2–3 wpi) | Systemic (2–3 wpi) | Systemic (4–5 wpi) | Local (2 wpi) | Systemic (2–3 wpi) | Systemic (4–5 wpi) | |
| N. benthamiana | 5/5 | 0/5 | 4/5 | 5/5 | 0/5 | 5/5 |
| Pepper | 5/5 | 0/5 | 3/5 | 5/5 | 2/5 | 4/5 |
| Spinach | 5/5 | 0/5 | 2/5 | 1/5 | 2/5 | 4/5 |
| Okra | 5/5 | 0/5 | 2/5 | 3/5 | 0/5 | 2/5 |
| Zucchini | 5/5 | 0/5 | 1/5 | 4/5 | 2/5 | 3/5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Atallah, O.O.; Yassin, S.M.; Shirley, N.; Verchot, J. Exploring the Host Range of Rose rosette Virus among Herbaceous Annual Plants. Pathogens 2022, 11, 1514. https://doi.org/10.3390/pathogens11121514
Atallah OO, Yassin SM, Shirley N, Verchot J. Exploring the Host Range of Rose rosette Virus among Herbaceous Annual Plants. Pathogens. 2022; 11(12):1514. https://doi.org/10.3390/pathogens11121514
Chicago/Turabian StyleAtallah, Osama O., Sherin M. Yassin, Natalie Shirley, and Jeanmarie Verchot. 2022. "Exploring the Host Range of Rose rosette Virus among Herbaceous Annual Plants" Pathogens 11, no. 12: 1514. https://doi.org/10.3390/pathogens11121514
APA StyleAtallah, O. O., Yassin, S. M., Shirley, N., & Verchot, J. (2022). Exploring the Host Range of Rose rosette Virus among Herbaceous Annual Plants. Pathogens, 11(12), 1514. https://doi.org/10.3390/pathogens11121514







