The Role of Dendritic Cells in the Host Response to Marek’s Disease Virus (MDV) as Shown by Transcriptomic Analysis of Susceptible and Resistant Birds
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Birds
2.2. Marek’s Disease Virus
2.3. Cell Cultures
2.4. Co-Culture Infection Experiments and FACS (Fluorescence-Activated Cell Sorting)
2.5. Cell and Sample Preparation for RNA-Sequencing (RNA-seq)
2.6. RNA-seq and Analysis
2.7. In Silico Functional Analysis of DE Genes
2.8. Determination of Potential MDV QTL Candidate Genes
3. Results
3.1. BMDC Infection in Resistant and Susceptible Lines
3.2. Analysis of Gene Expression in BMDCs of Resistant and Susceptible Lines
3.3. Inherent Differences Exist between the Two Inbred Lines
3.4. Host Response to MDV Infection in Susceptible Birds
3.5. Host Response to MDV Infection in Resistant Birds
3.6. Comparison of Host Response to Infection between the Two Lines
3.7. Expression Profiles of MicroRNAs
3.8. Viral Gene Expression
3.9. Downstream Analysis of Differentially Expressed Genes
3.10. Identification of Putative Candidate Genes for Resistance/Susceptibility to MD
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Haunshi, S.; Cheng, H.H. Differential expression of Toll-like receptor pathway genes in chicken embryo fibroblasts from chickens resistant and susceptible to Marek’s disease. Poult. Sci. 2014, 93, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Jeurissen, S.H.; Scholten, R.; Hilgers, L.A.; Pol, J.M.; De Boer, G.F. In situ detection by monoclonal antibody D-35.1 of cells infected with Marek’s disease virus that interact with splenic ellipsoid-associated reticulum cells. Avian Dis. 1989, 33, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, P.; Vervelde, L.; Dalziel, R.G.; Wasson, P.S.; Nair, V.; Dutia, B.M.; Kaiser, P. Marek’s disease virus infection of phagocytes: A de novo in vitro infection model. J. Gen. Virol. 2017, 98, 1080–1088. [Google Scholar] [CrossRef]
- Davison, T.F.; Kaiser, P. Immunity to Marek’s disease. In Marek’s Disease: An Evolving Problem; Davison, F., Nair, V., Eds.; Elsevier Academic Press: London, UK, 2004; pp. 126–139. [Google Scholar]
- Haq, K.; Schat, K.A.; Sharif, S. Immunity to Marek’s disease: Where are we now? Dev. Comp. Immunol. 2013, 41, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Boodhoo, N.; Gurung, A.; Sharif, S.; Behboudi, S. Marek’s disease in chickens: A review with focus on immunology. Vet. Res. 2016, 47, 119. [Google Scholar] [CrossRef] [Green Version]
- Gimeno, I.M. Marek’s disease vaccines: A solution for today but a worry for tomorrow? Vaccine 2008, 26 (Suppl. 3), C31–C41. [Google Scholar] [CrossRef] [PubMed]
- Nair, V. Latency and Tumorigenesis in Marek’s Disease. Avian Dis. 2013, 57 (Suppl. 2), 360–365. [Google Scholar] [CrossRef]
- Yang, Y.; Dong, M.; Hao, X.; Qin, A.; Shang, S. Revisiting cellular immune response to oncogenic Marek’s disease virus: The rising of avian T-cell immunity. Cell. Mol. Life Sci. 2020, 77, 3103–3116. [Google Scholar] [CrossRef] [Green Version]
- Zmrhal, V.; Slama, P. Current knowledge about interactions between avian dendritic cells and poultry pathogens. Dev. Comp. Immunol. 2020, 104, 103565. [Google Scholar] [CrossRef]
- Calnek, B.W. Pathogenesis of Marek’s Disease Virus Infection. Curr. Top. Microbiol. Immunol. 2000, 255, 25–55. [Google Scholar] [CrossRef]
- Wu, Z.; Rothwell, L.; Young, J.R.; Kaufman, J.; Butter, C.; Kaiser, P. Generation and characterization of chicken bone marrow-derived dendritic cells. Immunology 2010, 129, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, P.; Kuo, R.; Vervelde, L.; Dutia, B.M.; Kaiser, P.; Smith, J. Macrophages from Susceptible and Resistant Chicken Lines have Different Transcriptomes following Marek’s Disease Virus Infection. Genes 2019, 10, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cole, R.K. Studies on Genetic Resistance to Marek’s Disease. Avian Dis. 1968, 12, 9. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.F.; Powell, P.; Rennie, M.; Ross, L.; Payne, L. Nature of Genetic Resistance to Marek’s Disease in Chickens2. JNCI J. Natl. Cancer Inst. 1981, 66, 789–796. [Google Scholar] [CrossRef] [PubMed]
- Bacon, L.D.; Hunt, H.D.; Cheng, H.H. A Review of the Development of Chicken Lines to Resolve Genes Determining Resistance to Diseases. Poult. Sci. 2000, 79, 1082–1093. [Google Scholar] [CrossRef] [PubMed]
- Wasson, P. Development of Novel Virus Vectors for Influenza Vaccination. Ph.D. Thesis, University of Edinburgh, Edinburgh, UK, July 2011. [Google Scholar]
- Gimeno, I.; Silva, R.F. Deletion of the Marek’s disease virus UL41 gene (vhs) has no measurable effect on latency or pathogenesis. Virus Genes 2008, 36, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Petherbridge, L.; Howes, K.; Baigent, S.J.; Sacco, M.A.; Evans, S.; Osterrieder, N.; Nair, V. Replication-Competent Bacterial Artificial Chromosomes of Marek’s Disease Virus: Novel Tools for Generation of Molecularly Defined Herpesvirus Vaccines. J. Virol. 2003, 77, 8712–8718. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 3 November 2014).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; Smyth, G.K. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics 2007, 23, 2881–2887. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. feature Counts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Kirov, S.; Snoddy, J. WebGestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005, 33 (Suppl. 2), W741–W748. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, J.; Sadeyen, J.-R.; Paton, I.R.; Hocking, P.M.; Salmon, N.; Fife, M.; Nair, V.; Burt, D.W.; Kaiser, P. Systems Analysis of Immune Responses in Marek’s Disease Virus-Infected Chickens Identifies a Gene Involved in Susceptibility and Highlights a Possible Novel Pathogenicity Mechanism. J. Virol. 2011, 85, 11146–11158. [Google Scholar] [CrossRef] [Green Version]
- Smith, J.; Lipkin, E.; Soller, M.; Fulton, J.; Burt, D. Mapping QTL Associated with Resistance to Avian Oncogenic Marek’s Disease Virus (MDV) Reveals Major Candidate Genes and Variants. Genes 2020, 11, 1019. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Li, Y.; Ding, M.; Zhang, H.; Xu, X.; Tang, J. Molecular mechanisms and clinical applications of miR-22 in regulating malignant progression in human cancer (Review). Int. J. Oncol. 2017, 50, 345–355. [Google Scholar] [CrossRef]
- Jia, X.; Wang, X.; Guo, X.; Ji, J.; Lou, G.; Zhao, J.; Zhou, W.; Guo, M.; Zhang, M.; Li, C.; et al. MicroRNA-124: An emerging therapeutic target in cancer. Cancer Med. 2019, 8, 5638–5650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, Q.; Ruan, H.; Gilbert, J.; Wang, G.; Ma, Q.; Yao, W.-D.; Man, H.-Y. MicroRNA miR124 is required for the expression of homeostatic synaptic plasticity. Nat. Commun. 2015, 6, 10045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhoumik, A.; Ronai, Z. ATF2: A transcription factor that elicits oncogenic or tumor suppressor activities. Cell Cycle 2008, 7, 2341–2345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reith, W.; Herrero-Sanchez, C.; Kobr, M.; Silacci, P.; Berte, C.; Barras, E.; Fey, S.; Mach, B. MHC class II regulatory factor RFX has a novel DNA-binding domain and a functionally independent dimerization domain. Genes Dev. 1990, 4, 1528–1540. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Li, Y.; Dai, W.; Mo, S.; Li, Q.; Cai, S. Clinicopathologic Features and Prognostic Factors in Alpha-Fetoprotein-Producing Colorectal Cancer: Analysis of 78 Cases. Cell. Physiol. Biochem. 2018, 51, 2052–2064. [Google Scholar] [CrossRef]
- Marei, H.E.; Althani, A.; Afifi, N.; Hasan, A.; Caceci, T.; Pozzoli, G.; Morrione, A.; Giordano, A.; Cenciarelli, C. p53 signaling in cancer progression and therapy. Cancer Cell Int. 2021, 21, 703. [Google Scholar] [CrossRef]
- Xiao, L.; Zhang, C.; Li, X.; Jia, C.; Chen, L.; Yuan, Y.; Gao, Q.; Lu, Z.; Feng, Y.; Zhao, R.; et al. LEF1 Enhances the Progression of Colonic Adenocarcinoma via Remodeling the Cell Motility Associated Structures. Int. J. Mol. Sci. 2021, 22, 10870. [Google Scholar] [CrossRef]
- Qian, D.; Chen, K.; Deng, H.; Rao, H.; Huang, H.; Liao, Y.; Sun, X.; Lu, S.; Yuan, Z.; Xie, D.; et al. MicroRNA-374b Suppresses Proliferation and Promotes Apoptosis in T-cell Lymphoblastic Lymphoma by Repressing AKT1 and Wnt-16. Clin. Cancer Res. 2015, 21, 4881–4891. [Google Scholar] [CrossRef] [Green Version]
- Bumstead, N.; Kaufman, J. Genetic resistance to Marek’s disease. In Marek’s Disease: An Evolving Problem; Davison, F., Nair, V., Eds.; Elsevier Academic Press: London, UK, 2004; pp. 112–125. [Google Scholar]
- De Geus, E.D.; Vervelde, L. Regulation of macrophage and dendritic cell function by pathogens and through immunomodulation in the avian mucosa. Dev. Comp. Immunol. 2013, 41, 341–351. [Google Scholar] [CrossRef]
- Savina, A.; Amigorena, S. Phagocytosis and antigen presentation in dendritic cells. Immunol. Rev. 2007, 219, 143–156. [Google Scholar] [CrossRef]
- Kaiser, P.; Underwood, G.; Davison, F. Differential Cytokine Responses following Marek’s Disease Virus Infection of Chickens Differing in Resistance to Marek’s Disease. J. Virol. 2003, 77, 762–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akkoc, A.; Yilmaz, R.; Sonmez, G. Expression of Matrix Metalloproteinases in Marek’S Disease Tumours. Avian Biol. Res. 2012, 5, 93–98. [Google Scholar] [CrossRef]
- Muir, A.M.; Gardner, J.F.; van Jaarsveld, R.H.; de Lange, I.M.; van der Smagt, J.J.; Wilson, G.N.; Dubbs, H.; Goldberg, E.M.; Zitano, L.; Bupp, C.; et al. Variants in GNAI1 cause a syndrome associated with variable features including developmental delay, seizures, and hypotonia. Genet. Med. 2021, 23, 881–887. [Google Scholar] [CrossRef] [PubMed]
- Semina, E.V.; Rysenkova, K.D.; Troyanovskiy, K.E.; Shmakova, A.A.; Rubina, K.A. MicroRNAs in Cancer: From Gene Expression Regulation to the Metastatic Niche Reprogramming. Biochemistry 2021, 86, 785–799. [Google Scholar] [CrossRef] [PubMed]
- Payne, L.; Rennie, M. The proportions of b and t lymphocytes in lymphomas, peripheral nerves and lymphoid organs in Marek’s disease. Avian Pathol. 1976, 5, 147–154. [Google Scholar] [CrossRef] [Green Version]
- Tsuchiya, N.; Izumiya, M.; Ogata-Kawata, H.; Okamoto, K.; Fujiwara, Y.; Nakai, M.; Okabe, A.; Schetter, A.J.; Bowman, E.D.; Midorikawa, Y.; et al. Tumor Suppressor miR-22 Determines p53-Dependent Cellular Fate through Post-transcriptional Regulation of p21. Cancer Res. 2011, 71, 4628–4639. [Google Scholar] [CrossRef] [Green Version]
- Jovicic, A.; Zaldivar Jolissaint, J.F.; Moser, R.; Silva Santos, M.D.; Luthi-Carter, R. MicroRNA-22 (miR-22) Overexpression Is Neuroprotective via General Anti-Apoptotic Effects and May also Target Specific Huntington’s Disease-Related Mechanisms. PLoS ONE 2013, 8, e54222. [Google Scholar] [CrossRef] [Green Version]
- Witter, R.L.; Schat, K.A. Marek’s Disease. In Diseases of Poultry; Saif, Y.M., Ed.; Iowa State University Press: Ames, IA, USA, 2003; pp. 407–465. [Google Scholar]
- Payne, L.N. Pathological responses to infection. In Marek’s Disease: An Evolving Problem; Davison, F., Nair, V., Eds.; Elsevier Academic Press: London, UK, 2004; pp. 78–97. [Google Scholar]
- Qin, Z.; Wang, P.-Y.; Su, D.-F.; Liu, X. miRNA-124 in Immune System and Immune Disorders. Front. Immunol. 2016, 7, 406. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Luo, Z.-M.; Guo, X.-M.; Su, D.-F.; Liu, X. An updated role of microRNA-124 in central nervous system disorders: A review. Front. Cell. Neurosci. 2015, 9, 193. [Google Scholar] [CrossRef] [Green Version]
- Hess, J.; Angel, P.; Schorpp-Kistner, M. AP-1 subunits: Quarrel and harmony among siblings. J. Cell Sci. 2004, 117, 5965–5973. [Google Scholar] [CrossRef]
- Shaulian, E.; Karin, M. AP-1 as a regulator of cell life and death. Nature 2002, 4, E131–E136. [Google Scholar] [CrossRef] [PubMed]
- Shaulian, E. AP-1—The Jun proteins: Oncogenes or tumor suppressors in disguise? Cell. Signal. 2010, 22, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, M.L.; Kaygun, H.; Erkmann, J.A.; Townley-Tilson, W.H.D.; Dominski, Z.; Marzluff, W.F. SLBP is associated with histone mRNA on polyribosomes as a component of the histone mRNP. Nucleic Acids Res. 2004, 32, 4833–4842. [Google Scholar] [CrossRef] [PubMed]
- Dimitroff, C.J. Galectin-Binding O-Glycosylations as Regulators of Malignancy. Cancer Res. 2015, 75, 3195–3202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Starzyk, R.M.; Rosenow, C.; Frye, J.; Leismann, M.; Rodzinski, E.; Putney, S.; Tuomanen, E.I. Cerebral Cell Adhesion Molecule: A Novel Leukocyte Adhesion Determinant on Blood-Brain Barrier Capillary Endothelium. J. Infect. Dis. 2000, 181, 181–187. [Google Scholar] [CrossRef]
- Chong, Y.K.; Sandanaraj, E.; Koh, L.W.H.; Thangaveloo, M.; Tan, M.S.Y.; Koh, G.R.H.; Toh, T.B.; Lim, G.G.Y.; Holbrook, J.D.; Kon, O.L.; et al. ST3GAL1-Associated Transcriptomic Program in Glioblastoma Tumor Growth, Invasion, and Prognosis. JNCI J. Natl. Cancer Inst. 2016, 108. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Huang, R.; Tong, X.; Zhang, K.; Liu, Y.; Zeng, F.; Hu, H.; Jiang, T. Molecular and clinical characterization of TMEM71 expression at the transcriptional level in glioma. CNS Neurosci. Ther. 2019, 25, 965–975. [Google Scholar] [CrossRef] [Green Version]
- Chesi, M.; Brents, L.A.; Ely, S.A.; Bais, C.; Robbiani, D.F.; Mesri, E.A.; Kuehl, W.M.; Bergsagel, P.L. Activated fibroblast growth factor receptor 3 is an oncogene that contributes to tumor progression in multiple myeloma. Blood 2001, 97, 729–736. [Google Scholar] [CrossRef] [Green Version]
- Lafitte, M.; Moranvillier, I.; Garcia, S.; Peuchant, E.; Iovanna, J.; Rousseau, B.; Dubus, P.; Guyonnet-Dupérat, V.; Belleannée, G.; Ramos, J.; et al. FGFR3 has tumor suppressor properties in cells with epithelial phenotype. Mol. Cancer 2013, 12, 83. [Google Scholar] [CrossRef]
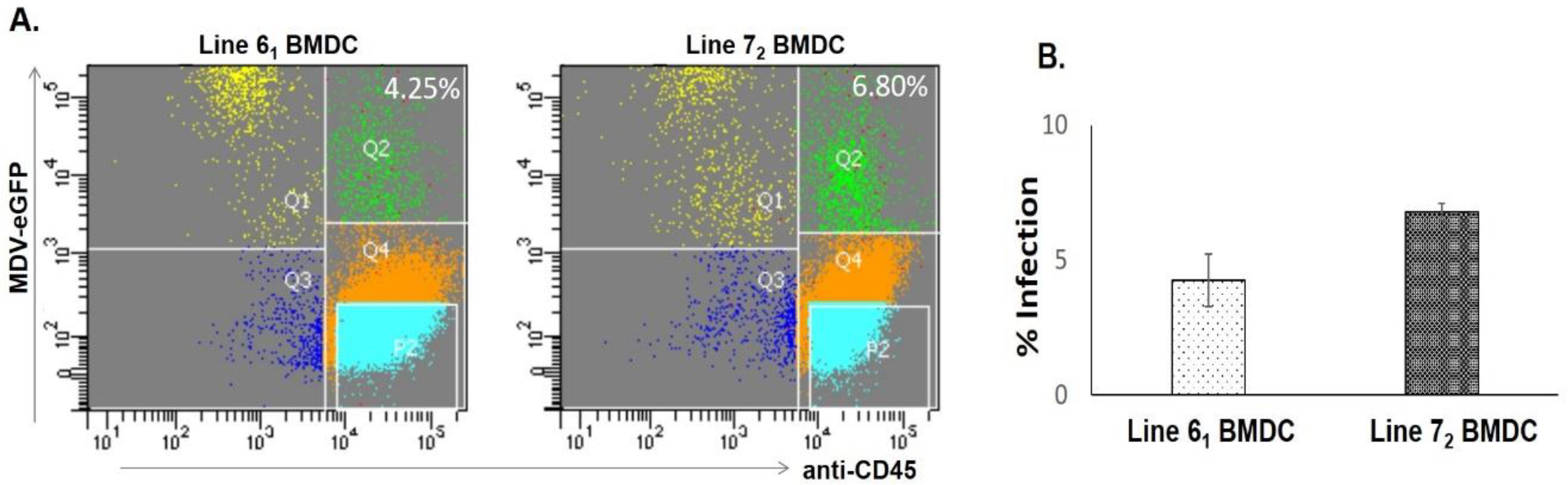
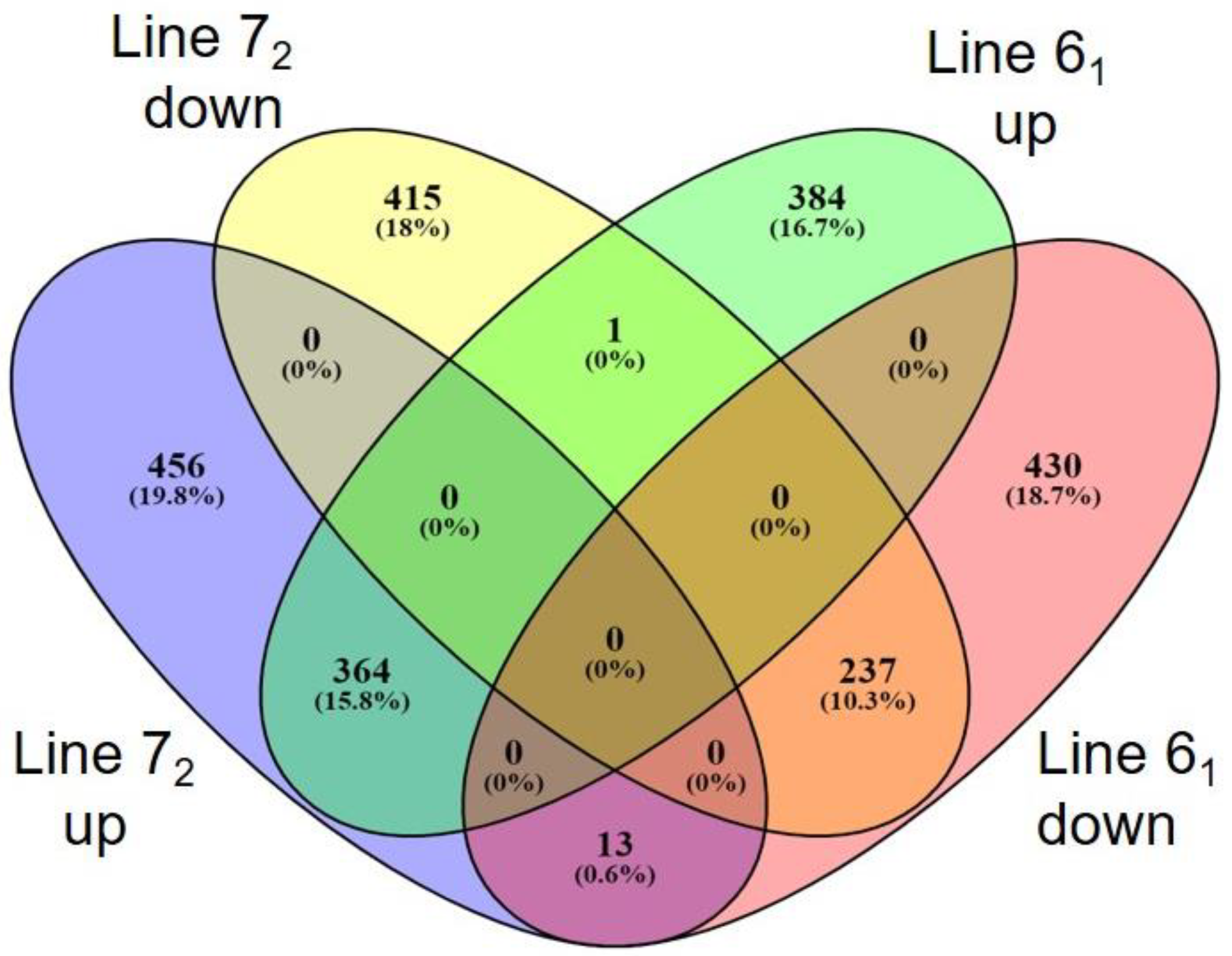
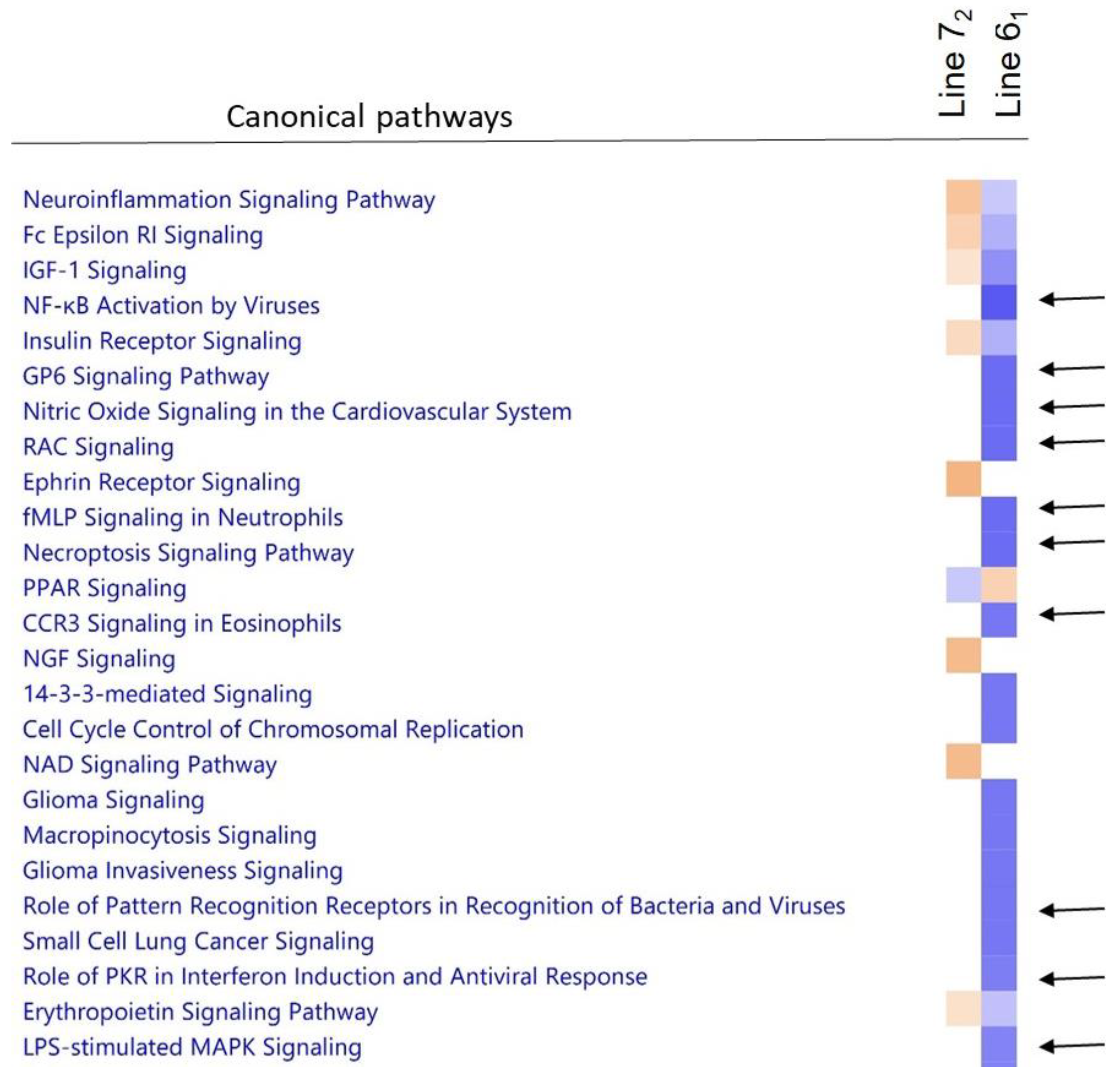
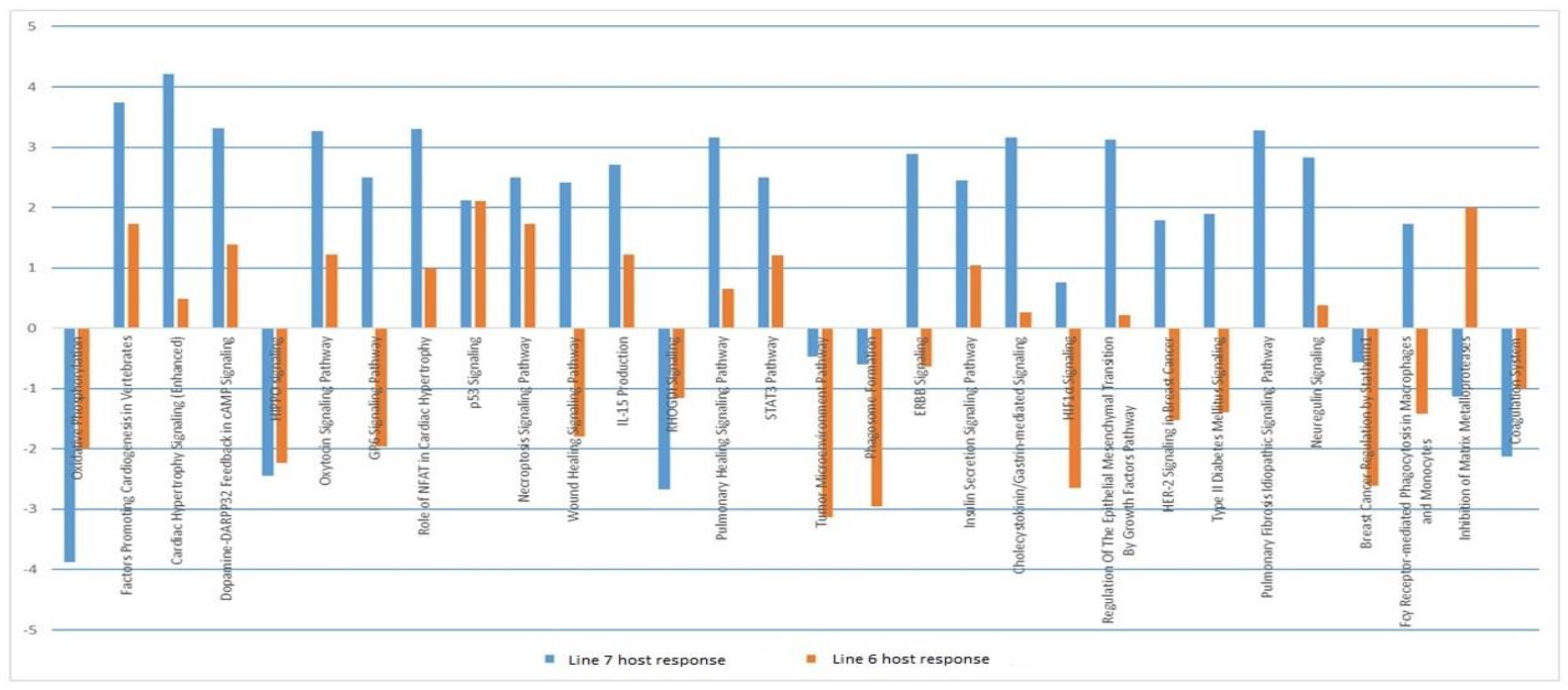
| Category | Genes More Highly Expressed in BMDCs of Line 61 | Genes More Highly Expressed in BMDCs of Line 72 |
|---|---|---|
| Host response in line 72 compared to controls | / | 843 ↑ and 678 ↓ |
| Host response in line 61 compared to controls | 760 ↑ and 722 ↓ | / |
| Inherent difference between lines | 477 | 478 |
| Difference between lines upon infection | 317 | 668 |
| Gene Set | Size | Expect | Ratio | p-Value | FDR |
|---|---|---|---|---|---|
| TGANTCA_AP1_C | 1590 | 38.364 | 1.6682 | 0.00002 | 0.009 |
| AP1_C | 409 | 9.8685 | 2.432 | 0.00005 | 0.009 |
| GGGTGGRR_PAX4_03 | 1853 | 44.71 | 1.5656 | 0.00006 | 0.009 |
| AP1_Q4_01 | 412 | 9.9409 | 2.4143 | 0.00006 | 0.009 |
| CTTTGA_LEF1_Q2 | 1683 | 40.608 | 1.576 | 0.00011 | 0.012 |
| SRY_01 | 324 | 7.8176 | 2.5583 | 0.00012 | 0.012 |
| AP1_Q2 | 380 | 9.1688 | 2.3994 | 0.00013 | 0.012 |
| CTTTAAR_UNKNOWN | 1408 | 33.973 | 1.6189 | 0.00019 | 0.013 |
| STAT4_01 | 389 | 9.386 | 2.3439 | 0.00019 | 0.013 |
| CAGGTA_AREB6_01 | 1162 | 28.037 | 1.6763 | 0.00027 | 0.017 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chakraborty, P.; Kuo, R.I.; Wu, Z.; Morris, K.M.; Dutia, B.M.; Kaiser, P.; Smith, J. The Role of Dendritic Cells in the Host Response to Marek’s Disease Virus (MDV) as Shown by Transcriptomic Analysis of Susceptible and Resistant Birds. Pathogens 2022, 11, 1340. https://doi.org/10.3390/pathogens11111340
Chakraborty P, Kuo RI, Wu Z, Morris KM, Dutia BM, Kaiser P, Smith J. The Role of Dendritic Cells in the Host Response to Marek’s Disease Virus (MDV) as Shown by Transcriptomic Analysis of Susceptible and Resistant Birds. Pathogens. 2022; 11(11):1340. https://doi.org/10.3390/pathogens11111340
Chicago/Turabian StyleChakraborty, Pankaj, Richard I. Kuo, Zhou Wu, Katrina M. Morris, Bernadette M. Dutia, Pete Kaiser, and Jacqueline Smith. 2022. "The Role of Dendritic Cells in the Host Response to Marek’s Disease Virus (MDV) as Shown by Transcriptomic Analysis of Susceptible and Resistant Birds" Pathogens 11, no. 11: 1340. https://doi.org/10.3390/pathogens11111340
APA StyleChakraborty, P., Kuo, R. I., Wu, Z., Morris, K. M., Dutia, B. M., Kaiser, P., & Smith, J. (2022). The Role of Dendritic Cells in the Host Response to Marek’s Disease Virus (MDV) as Shown by Transcriptomic Analysis of Susceptible and Resistant Birds. Pathogens, 11(11), 1340. https://doi.org/10.3390/pathogens11111340







