The Influence of the Degree of Thermal Inactivation of Probiotic Lactic Acid Bacteria and Their Postbiotics on Aggregation and Adhesion Inhibition of Selected Pathogens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Thermal Inactivation of Bacterial Strains
2.3. Cell Surface Hydrophobicity
2.4. Auto-Aggregation Assay
2.5. Co-Aggregation Assays with Pathogens
2.6. Inhibition of Pathogens Adhesion
2.7. Statistical Analysis
3. Results
3.1. Cell Surface Hydrophobicity
3.2. Auto-Aggregation Assay
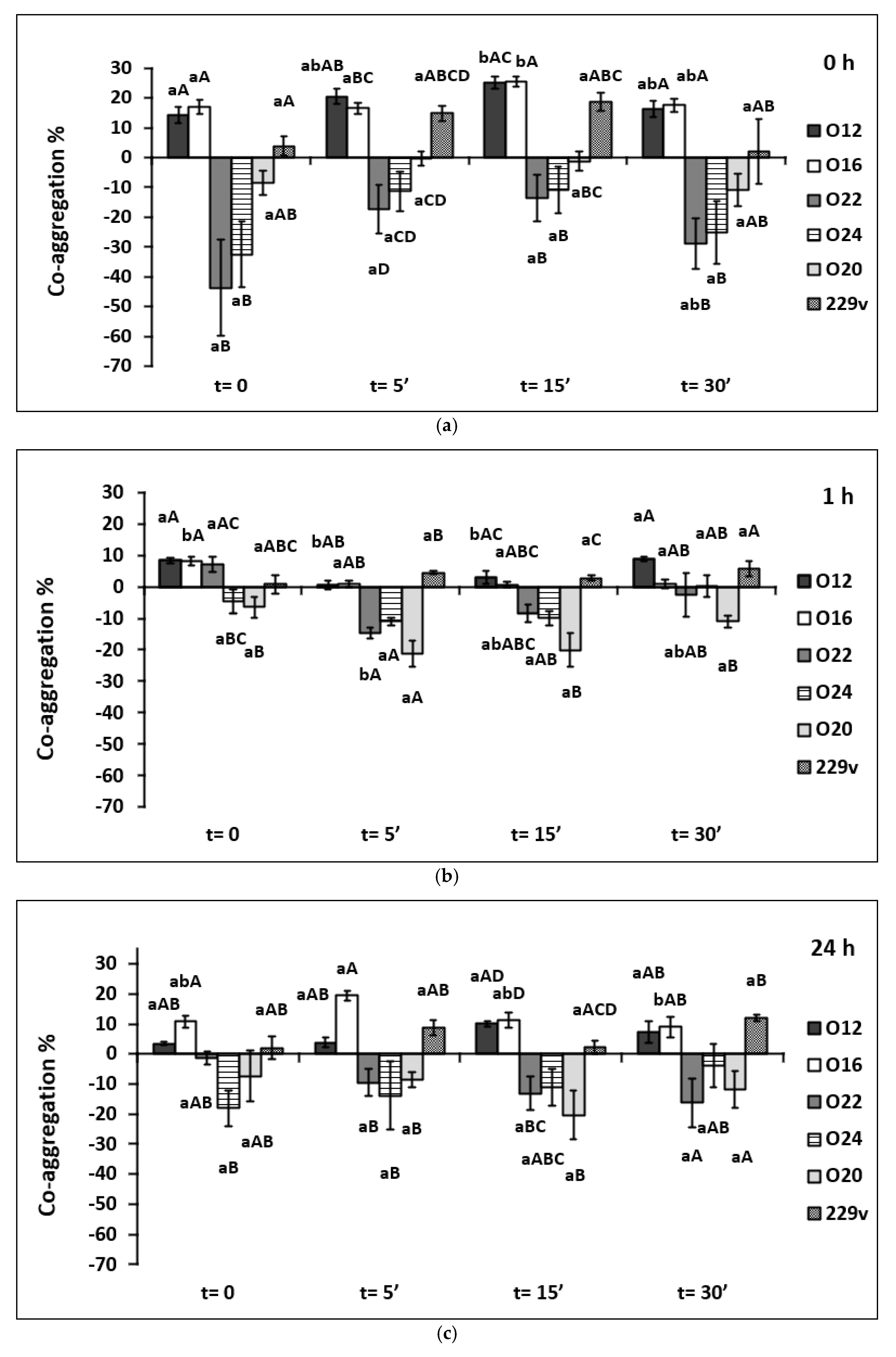
3.3. Co-Aggregation with Listeria monocytogenes
3.4. Co-Aggregation with Staphylococcus aureus
3.5. Inhibition of Pathogens Adhesion to the Mucin
4. Discussion
4.1. Cell Surface Hydrophobicity
4.2. Auto-Aggregation Assay
4.3. Co-Aggregation Assays with Pathogens
4.4. Inhibition of Pathogens Adhesion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schirone, M.; Visciano, P. Trends of Major Foodborne Outbreaks in the European Union during the Years 2015–2019. Hygiene 2021, 1, 106–119. [Google Scholar] [CrossRef]
- Bearth, A.; Cousin, M.-E.; Siegrist, M. The consumer’s perception of artificial food additives: Influences on acceptance, risk and benefit perceptions. Food Qual. Prefer. 2014, 38, 14–23. [Google Scholar] [CrossRef]
- Guimarães, J.T.; Balthazar, C.F.; Silva, R.; Rocha, R.S.; Graça, J.S.; Esmerino, E.A.; Silva, M.C.; Sant’Ana, A.S.; Duarte, M.C.K.H.; Freitas, M.Q.; et al. Impact of Probiotics and Prebiotics on Food Texture. Curr. Opin. Food Sci. 2020, 33, 38–44. [Google Scholar] [CrossRef]
- Łojewska, E.; Sakowicz, T. An Alternative to Antibiotics: Selected Methods to Combat Zoonotic Foodborne Bacterial Infections. Curr. Microbiol. 2021, 78, 4037–4049. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Song, J.; Xie, M.; Song, D. The Bidirectional Relationship between Host Physiology and Microbiota and Health Benefits of Probiotics: A Review. Trends Food Sci. Technol. 2019, 91, 426–435. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A Taxonomic Note on the Genus Lactobacillus: Description of 23 Novel Genera, Emended Description of the Genus Lactobacillus Beijerinck 1901, and Union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert Consensus Document. The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic: Expert Consensus Document. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [Green Version]
- Maldonado Galdeano, C.; Cazorla, S.I.; Lemme Dumit, J.M.; Vélez, E.; Perdigón, G. Beneficial Effects of Probiotic Consumption on the Immune System. Ann. Nutr. Metab. 2019, 74, 115–124. [Google Scholar] [CrossRef] [Green Version]
- Sadowska-Krawczenko, I.; Paprzycka, M.; Korbal, P.; Wiatrzyk, A.; Krysztopa-Grzybowska, K.; Polak, M.; Czajka, U.; Lutyńska, A. Lactobacillus Rhamnosus GG Suspected Infection in a Newborn with Intrauterine Growth Restriction. Benef. Microbes 2014, 5, 397–402. [Google Scholar] [CrossRef]
- Land, M.H.; Rouster-Stevens, K.; Woods, C.R.; Cannon, M.L.; Cnota, J.; Shetty, A.K. Lactobacillus Sepsis Associated with Probiotic Therapy. Pediatrics 2005, 115, 178–181. [Google Scholar] [CrossRef]
- Vahabnezhad, E.; Mochon, A.B.; Wozniak, L.J.; Ziring, D.A. Lactobacillus Bacteremia Associated with Probiotic Use in a Pediatric Patient with Ulcerative Colitis. J. Clin. Gastroenterol. 2013, 47, 437–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salminen, M.K.; Rautelin, H.; Tynkkynen, S.; Poussa, T.; Saxelin, M.; Valtonen, V.; Järvinen, A. Lactobacillus Bacteremia, Clinical Significance, and Patient Outcome, with Special Focus on Probiotic L. Rhamnosus GG. Clin. Infect. Dis. 2004, 38, 62–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salvana, E.M.T.; Frank, M. Lactobacillus Endocarditis: Case Report and Review of Cases Reported since 1992. J. Infect. 2006, 53, e5–e10. [Google Scholar] [CrossRef] [PubMed]
- Besselink, M.G.; van Santvoort, H.C.; Buskens, E.; Boermeester, M.A.; van Goor, H.; Timmerman, H.M.; Nieuwenhuijs, V.B.; Bollen, T.L.; van Ramshorst, B.; Witteman, B.J.; et al. Probiotic Prophylaxis in Predicted Severe Acute Pancreatitis: A Randomised, Double-Blind, Placebo-Controlled Trial. Lancet 2008, 371, 651–659. [Google Scholar] [CrossRef] [Green Version]
- Salminen, S.; Collado, M.C.; Endo, A.; Hill, C.; Lebeer, S.; Quigley, E.M.M.; Sanders, M.E.; Shamir, R.; Swann, J.R.; Szajewska, H.; et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) Consensus Statement on the Definition and Scope of Postbiotics. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 649–667. [Google Scholar] [CrossRef]
- Cuevas-González, P.F.; Liceaga, A.M.; Aguilar-Toalá, J.E. Postbiotics and Paraprobiotics: From Concepts to Applications. Food Res. Int. 2020, 136, 109502. [Google Scholar] [CrossRef]
- Aguilar-Toalá, J.E.; Garcia-Varela, R.; Garcia, H.S.; Mata-Haro, V.; González-Córdova, A.F.; Vallejo-Cordoba, B.; Hernández-Mendoza, A. Postbiotics: An Evolving Term within the Functional Foods Field. Trends Food Sci. Technol. 2018, 75, 105–114. [Google Scholar] [CrossRef]
- Nataraj, B.H.; Ali, S.A.; Behare, P.V.; Yadav, H. Postbiotics-Parabiotics: The New Horizons in Microbial Biotherapy and Functional Foods. Microb. Cell Fact. 2020, 19, 168. [Google Scholar] [CrossRef]
- Liévin-Le Moal, V.; Sarrazin-Davila, L.E.; Servin, A.L. An Experimental Study and a Randomized, Double-Blind, Placebo-Controlled Clinical Trial to Evaluate the Antisecretory Activity of Lactobacillus Acidophilus Strain LB against Nonrotavirus Diarrhea. Pediatrics 2007, 120, e795–e803. [Google Scholar] [CrossRef]
- Mehling, H.; Busjahn, A. Non-Viable Lactobacillus Reuteri DSMZ 17648 (PylopassTM) as a New Approach to Helicobacter Pylori Control in Humans. Nutrients 2013, 5, 3062–3073. [Google Scholar] [CrossRef]
- Buckley, M.; Lacey, S.; Doolan, A.; Goodbody, E.; Seamans, K. The Effect of Lactobacillus Reuteri Supplementation in Helicobacter Pylori Infection: A Placebo-Controlled, Single-Blind Study. BMC Nutr. 2018, 4, 48. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.; Sawant, S.; Hauff, K.; Hampp, G. Validated Postbiotic Screening Confirms Presence of Physiologically-Active Metabolites, Such as Short-Chain Fatty Acids, Amino Acids and Vitamins in Hylak® Forte. Probiotics Antimicrob. Proteins 2019, 11, 1124–1131. [Google Scholar] [CrossRef] [PubMed]
- Markowicz, C.; Kubiak, P.; Grajek, W.; Schmidt, M.T. Inactivation of Lactobacillus Rhamnosus GG by Fixation Modifies Its Probiotic Properties. Can. J. Microbiol. 2016, 62, 72–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siciliano, R.A.; Reale, A.; Mazzeo, M.F.; Morandi, S.; Silvetti, T.; Brasca, M. Paraprobiotics: A New Perspective for Functional Foods and Nutraceuticals. Nutrients 2021, 13, 1225. [Google Scholar] [CrossRef]
- Teame, T.; Wang, A.; Xie, M.; Zhang, Z.; Yang, Y.; Ding, Q.; Gao, C.; Olsen, R.E.; Ran, C.; Zhou, Z. Paraprobiotics and Postbiotics of Probiotic Lactobacilli, Their Positive Effects on the Host and Action Mechanisms: A Review. Front. Nutr. 2020, 7, 570344. [Google Scholar] [CrossRef]
- Thanh, N.T.; Chwen, L.T.; Foo, H.L.; Hair-bejo, M.; Kasim, A.B. Inhibitory activity of metabolites produced by strains of Lactobacillus plantarum isolated from Malaysian fermented food. Int. J. Probiot. Prebiot. 2010, 5, 37–44. [Google Scholar]
- Kareem, K.Y.; Hooi Ling, F.; Teck Chwen, L.; May Foong, O.; Anjas Asmara, S. Inhibitory Activity of Postbiotic Produced by Strains of Lactobacillus Plantarum Using Reconstituted Media Supplemented with Inulin. Gut Pathog. 2014, 6, 23. [Google Scholar] [CrossRef] [Green Version]
- Thu, T.V.; Foo, H.L.; Loh, T.C.; Bejo, M.H. Inhibitory activity and organic acid concentrations of metabolite combinations produced by various strains of Lactobacillus plantarum. Afr. J. Biotechnol. 2011, 10, 1359–1363. [Google Scholar]
- Choe, D.W.; Loh, T.C.; Foo, H.L.; Hair-Bejo, M.; Awis, Q.S. Egg Production, Faecal PH and Microbial Population, Small Intestine Morphology, and Plasma and Yolk Cholesterol in Laying Hens given Liquid Metabolites Produced by Lactobacillus Plantarum Strains. Br. Poult. Sci. 2012, 53, 106–115. [Google Scholar] [CrossRef]
- Zielińska, D.; Rzepkowska, A.; Radawska, A.; Zieliński, K. In Vitro Screening of Selected Probiotic Properties of Lactobacillus Strains Isolated from Traditional Fermented Cabbage and Cucumber. Curr. Microbiol. 2015, 70, 183–194. [Google Scholar] [CrossRef]
- Zielińska, D.; Długosz, E.; Zawistowska-Deniziak, A. Functional Properties of Food Origin Lactobacillus in the Gastrointestinal Ecosystem-in Vitro Study. Probiotics Antimicrob. Proteins 2019, 11, 820–829. [Google Scholar] [CrossRef] [PubMed]
- Pérez, P.F.; Minnaard, Y.; Disalvo, E.A.; De Antoni, G.L. Surface Properties of Bifidobacterial Strains of Human Origin. Appl. Environ. Microbiol. 1998, 64, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Kos, B.; Susković, J.; Vuković, S.; Simpraga, M.; Frece, J.; Matosić, S. Adhesion and Aggregation Ability of Probiotic Strain Lactobacillus Acidophilus M92. J. Appl. Microbiol. 2003, 94, 981–987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moisés Laparra, J.; Corzo-Martinez, M.; Villamiel, M.; Javier Moreno, F.; Sanz, Y. Maillard-Type Glycoconjugates from Dairy Proteins Inhibit Adhesion of Escherichia Coli to Mucin. Food Chem. 2011, 129, 1435–1443. [Google Scholar] [CrossRef] [Green Version]
- Rabiei, P.; Mohabatkar, H.; Behbahani, M. Studying the Effects of Several Heat-Inactivated Bacteria on Colon and Breast Cancer Cells. Mol. Biol. Res. Commun. 2019, 8, 91–98. [Google Scholar] [CrossRef]
- Dawan, J.; Ahn, J. Bacterial Stress Responses as Potential Targets in Overcoming Antibiotic Resistance. Microorganisms 2022, 10, 1385. [Google Scholar] [CrossRef]
- Sebastian, A.P.; Keerthi, T.R. Adhesion and Cell surface Properties of Wild Species of Spore Formers against Enteric Pathogens. Asian Pac. J. Trop. Med. 2013, 6, 110–114. [Google Scholar] [CrossRef] [Green Version]
- Sharma, K.; Sharma, N.; Sharma, R. Identification and Evaluation of in Vitro Probiotic Attributes of Novel and Potential Strains of Lactic Acid Bacteria Isolated from Traditional Dairy Products of North-West Himalayas. J Clin Microbiol Biochem Technol 2016, 2, 018–025. [Google Scholar] [CrossRef]
- Collado, M.C.; Surono, I.S.; Meriluoto, J.; Salminen, S. Potential Probiotic Characteristics of Lactobacillus and Enterococcus Strains Isolated from Traditional Dadih Fermented Milk against Pathogen Intestinal Colonization. J. Food Prot. 2007, 70, 700–705. [Google Scholar] [CrossRef]
- De Souza, B.M.S.; Borgonovi, T.F.; Casarotti, S.N.; Todorov, S.D.; Penna, A.L.B. Lactobacillus Casei and Lactobacillus Fermentum Strains Isolated from Mozzarella Cheese: Probiotic Potential, Safety, Acidifying Kinetic Parameters and Viability under Gastrointestinal Tract Conditions. Probiotics Antimicrob. Proteins 2019, 11, 382–396. [Google Scholar] [CrossRef]
- Bellon-Fontaine, M.-N.; Rault, J.; van Oss, C.J. Microbial Adhesion to Solvents: A Novel Method to Determine the Electron-Donor/Electron-Acceptor or Lewis Acid-Base Properties of Microbial Cells. Colloids Surf. B Biointerfaces 1996, 7, 47–53. [Google Scholar] [CrossRef]
- Alameri, F.; Tarique, M.; Osaili, T.; Obaid, R.; Abdalla, A.; Masad, R.; Al-Sbiei, A.; Fernandez-Cabezudo, M.; Liu, S.-Q.; Al-Ramadi, B.; et al. Lactic Acid Bacteria Isolated from Fresh Vegetable Products: Potential Probiotic and Postbiotic Characteristics Including Immunomodulatory Effects. Microorganisms 2022, 10, 389. [Google Scholar] [CrossRef] [PubMed]
- Balakrishna, A. In Vitro Evaluation of Adhesion and Aggregation Abilities of Four Potential Probiotic Strains Isolated from Guppy (Poecilia Reticulata). Braz. Arch. Biol. Technol. 2013, 56, 793–800. [Google Scholar] [CrossRef] [Green Version]
- Ciandrini, E.; Campana, R.; Baffone, W. Live and Heat-Killed Lactobacillus Spp. Interfere with Streptococcus Mutans and Streptococcus Oralis during Biofilm Development on Titanium Surface. Arch. Oral Biol. 2017, 78, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.; Surono, I.; Meriluoto, J.; Salminen, S. Indigenous Dadih Lactic Acid Bacteria: Cell-Surface Properties and Interactions with Pathogens. J. Food Sci. 2007, 72, M89–M93. [Google Scholar] [CrossRef]
- Dlamini, Z.C.; Langa, R.L.S.; Aiyegoro, O.A.; Okoh, A.I. Safety Evaluation and Colonisation Abilities of Four Lactic Acid Bacteria as Future Probiotics. Probiotics Antimicrob. Proteins 2019, 11, 397–402. [Google Scholar] [CrossRef]
- Falah, F.; Vasiee, A.; Behbahani, B.A.; Yazdi, F.T.; Moradi, S.; Mortazavi, S.A.; Roshanak, S. Evaluation of Adherence and Anti-Infective Properties of Probiotic Lactobacillus Fermentum Strain 4-17 against Escherichia Coli Causing Urinary Tract Infection in Humans. Microb. Pathog. 2019, 131, 246–253. [Google Scholar] [CrossRef]
- Gharbi, Y.; Fhoula, I.; Ruas-Madiedo, P.; Afef, N.; Boudabous, A.; Gueimonde, M.; Ouzari, H.-I. In-Vitro Characterization of Potentially Probiotic Lactobacillus Strains Isolated from Human Microbiota: Interaction with Pathogenic Bacteria and the Enteric Cell Line HT29. Ann. Microbiol. 2019, 69, 61–72. [Google Scholar] [CrossRef]
- Gómez, N.C.; Ramiro, J.M.P.; Quecan, B.X.V.; de Melo Franco, B.D.G. Use of Potential Probiotic Lactic Acid Bacteria (LAB) Biofilms for the Control of Listeria Monocytogenes, Salmonella Typhimurium, and Escherichia Coli O157:H7 Biofilms Formation. Front. Microbiol. 2016, 7, 863. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Xu, X.; Zhang, F.; Xu, D.; Liu, Z.; Tao, X.; Wei, H. Anti-Adhesion of Probiotic Enterococcus Faecium WEFA23 against Five Pathogens and the Beneficial Effect of Its S-Layer Proteins against Listeria Monocytogenes. Can. J. Microbiol. 2019, 65, 175–184. [Google Scholar] [CrossRef] [Green Version]
- Khojah, E.; Gomaa, M.S.; Elsherbiny, E.G.; Elawady, A.; Darwish, M.S. The in Vitro Analysis of Postbiotics in Functional Labneh to Be Used as Powerful Tool to Improve Cell Surfaces Properties and Adherence Potential of Probiotic Strains. Fermentation 2022, 8, 122. [Google Scholar] [CrossRef]
- Krausova, G.; Hyrslova, I.; Hynstova, I. In Vitro Evaluation of Adhesion Capacity, Hydrophobicity, and Auto-Aggregation of Newly Isolated Potential Probiotic Strains. Fermentation 2019, 5, 100. [Google Scholar] [CrossRef]
- Ning, Z.; Xue, B.; Wang, H. Evaluation of the Adhesive Potential of Bacteria Isolated from Meat-Related Sources. Appl. Sci. 2021, 11, 10652. [Google Scholar] [CrossRef]
- Orłowski, A.; Bielecka, M. Preliminary Characteristics of Lactobacillus and Bifidobacterium Strains as Probiotic Candidates. Pol. J. Food Nutr. Sci. 2006, 56, 269–275. [Google Scholar]
- Polak-Berecka, M.; Waśko, A.; Paduch, R.; Skrzypek, T.; Sroka-Bartnicka, A. The Effect of Cell Surface Components on Adhesion Ability of Lactobacillus Rhamnosus. Antonie Van Leeuwenhoek 2014, 106, 751–762. [Google Scholar] [CrossRef] [Green Version]
- Shin, J.-H.; Lee, J.-S.; Seo, J.-G. Assessment of Cell Adhesion, Cell Surface Hydrophobicity, Autoaggregation, and Lipopolysaccharide-Binding Properties of Live and Heat-Killed Lactobacillus Acidophilus CBT LA1. Misainmurhag. Hoiji. 2015, 51, 241–248. [Google Scholar] [CrossRef] [Green Version]
- Singh, V.; Ganger, S.; Patil, S. Characterization of Lactobacillus Brevis with Potential Probiotic Properties and Biofilm Inhibition against Pseudomonas Aeruginosa. Proceedings 2020, 66, 14. [Google Scholar] [CrossRef]
- Somashekaraiah, R.; Shruthi, B.; Deepthi, B.V.; Sreenivasa, M.Y. Probiotic Properties of Lactic Acid Bacteria Isolated from Neera: A Naturally Fermenting Coconut Palm Nectar. Front. Microbiol. 2019, 10, 1382. [Google Scholar] [CrossRef]
- Tuo, Y.; Yu, H.; Ai, L.; Wu, Z.; Guo, B.; Chen, W. Aggregation and Adhesion Properties of 22 Lactobacillus Strains. J. Dairy Sci. 2013, 96, 4252–4257. [Google Scholar] [CrossRef] [Green Version]
- Zakaria Gomaa, E. Antimicrobial and Anti-Adhesive Properties of Biosurfactant Produced by Lactobacilli Isolates, Biofilm Formation and Aggregation Ability. J. Gen. Appl. Microbiol. 2013, 59, 425–436. [Google Scholar] [CrossRef] [Green Version]
- Habimana, O.; Semião, A.J.C.; Casey, E. The Role of Cell-Surface Interactions in Bacterial Initial Adhesion and Consequent Biofilm Formation on Nanofiltration/Reverse Osmosis Membranes. J. Memb. Sci. 2014, 454, 82–96. [Google Scholar] [CrossRef] [Green Version]
- Sorroche, F.G.; Spesia, M.B.; Zorreguieta, A.; Giordano, W. A Positive Correlation between Bacterial Autoaggregation and Biofilm Formation in Native Sinorhizobium Meliloti Isolates from Argentina. Appl. Environ. Microbiol. 2012, 78, 4092–4101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rickard, A.H.; Gilbert, P.; High, N.J.; Kolenbrander, P.E.; Handley, P.S. Bacterial Coaggregation: An Integral Process in the Development of Multi-Species Biofilms. Trends Microbiol. 2003, 11, 94–100. [Google Scholar] [CrossRef]
- Li, M.; Wang, Y.; Cui, H.; Li, Y.; Sun, Y.; Qiu, H.-J. Characterization of Lactic Acid Bacteria Isolated from the Gastrointestinal Tract of a Wild Boar as Potential Probiotics. Front. Vet. Sci. 2020, 7, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zielińska, D.; Łepecka, A.; Ołdak, A.; Długosz, E.; Kołożyn-Krajewska, D. Growth and Adhesion Inhibition of Pathogenic Bacteria by Live and Heat-Killed Food-Origin Lactobacillus Strains or Their Supernatants. FEMS Microbiol. Lett. 2021, 368. [Google Scholar] [CrossRef]
- Tareb, R.; Bernardeau, M.; Gueguen, M.; Vernoux, J.-P. In Vitro Characterization of Aggregation and Adhesion Properties of Viable and Heat-Killed Forms of Two Probiotic Lactobacillus Strains and Interaction with Foodborne Zoonotic Bacteria, Especially Campylobacter Jejuni. J. Med. Microbiol. 2013, 62, 637–649. [Google Scholar] [CrossRef]
- Kim, M.-S.; Yoon, Y.-S.; Seo, J.-G.; Lee, H.-G.; Chung, M.-J.; Yum, D.-Y. A Study on the Prevention of Salmonella Infection by Using the Aggregation Characteristics of Lactic Acid Bacteria. Toxicol. Res. 2013, 29, 129–135. [Google Scholar] [CrossRef] [Green Version]
- Ding, Q.; Sun, X.; Cao, S.; Zhao, C.; Wang, Y.; Wang, X. Heat-Killed Lactobacillus Acidophilus Mediates Fusobacterium Nucleatum Induced pro-Inflammatory Responses in Epithelial Cells. FEMS Microbiol. Lett. 2021, 368, fnaa160. [Google Scholar] [CrossRef]
- Salminen, S.; Bouley, C.; Boutron-Ruault, M.C.; Cummings, J.H.; Franck, A.; Gibson, G.R.; Isolauri, E.; Moreau, M.C.; Roberfroid, M.; Rowland, I. Functional Food Science and Gastrointestinal Physiology and Function. Br. J. Nutr. 1998, 80, S147–S171. [Google Scholar] [CrossRef] [Green Version]
- Ostad, S.N.; Salarian, A.A.; Ghahramani, M.H.; Fazeli, M.R.; Samadi, N.; Jamalifar, H. Live and Heat-Inactivated Lactobacilli from Feces Inhibit Salmonella Typhi and Escherichia Coli Adherence to Caco-2 Cells. Folia Microbiol. (Praha) 2009, 54, 157–160. [Google Scholar] [CrossRef]
- Singh, T.P.; Kaur, G.; Kapila, S.; Malik, R.K. Antagonistic Activity of Lactobacillus Reuteri Strains on the Adhesion Characteristics of Selected Pathogens. Front. Microbiol. 2017, 8, 486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarkar, A.; Mandal, S. Bifidobacteria—Insight into Clinical Outcomes and Mechanisms of Its Probiotic Action. Microbiol. Res. 2016, 192, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Castro-Bravo, N.; Wells, J.M.; Margolles, A.; Ruas-Madiedo, P. Interactions of Surface Exopolysaccharides from Bifidobacterium and Lactobacillus within the Intestinal Environment. Front. Microbiol. 2018, 9, 2426. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-Y.; Tsen, H.-Y.; Lin, C.-L.; Lin, C.-K.; Chuang, L.-T.; Chen, C.-S.; Chiang, Y.-C. Enhancement of the Immune Response against Salmonella Infection of Mice by Heat-Killed Multispecies Combinations of Lactic Acid Bacteria. J. Med. Microbiol. 2013, 62, 1657–1664. [Google Scholar] [CrossRef]


| Solvent | Heat Inactivation Time | Bacterial Strains | Heat Inactivation Time × Bacterial Strains | |||||
|---|---|---|---|---|---|---|---|---|
| O12 | O16 | O22 | O24 | O20 | 299v | |||
| p-Xylene, % | t = 0 | 58.33 aAD ± 2.93 | 55.30 aAB ± 3.76 | 54.20 aAB ± 3.29 | 50.60 aB ± 2.36 | 64.10 aD ± 1.41 | 36.03 aC ± 2.30 | |
| t = 5′ | 58.47 aA ± 5.71 | 64.07 abAB ± 1.39 | 58.90 abA ± 3.74 | 64.73 bAB ± 5.26 | 72.97 aB ± 5.45 | 43.27 bC ± 1.67 | ||
| t = 15′ | 58.83 aA ± 3.10 | 67.27 bAB ± 7.09 | 59.70 abA ± 1.01 | 65.37 bAB ± 4.32 | 70.80 aB ± 3.40 | 39.53 abC ± 2.31 | ||
| t = 30′ | 52.57 aAC ± 7.26 | 68.70 bB ± 0.69 | 62.70 bAB ± 1.68 | 64.67 bB ± 5.45 | 69.50 aB ± 3.48 | 42.33 bC ± 0.21 | ||
| Two-way ANOVA, p | *** | *** | * | |||||
| n-Hexadecane, % | t = 0 | 15.87 aA ± 3.87 | 12.87 aA ± 1.96 | 17.43 aA ± 5.36 | 16.13 aA ± 2.98 | 13.70 aA ± 2.30 | 16.73 aA ± 2.35 | |
| t = 5′ | 18.13 aAB ± 1.26 | 17.23 bAB ± 0.23 | 24.13 aB ± 3.39 | 21.70 aAB ± 1.40 | 15.57 aA ± 2.18 | 14.60 aA ± 5.98 | ||
| t = 15′ | 16.53 aAC ± 1.99 | 15.03 abA ± 1.20 | 24.73 aB ± 3.18 | 23.30 aBC ± 5.30 | 11.00 aA ± 2.35 | 14.70 aA ± 0.96 | ||
| t = 30′ | 16.13 aAC ± 2.31 | 15.53 abAC ± 0.40 | 24.80 aB ± 2.78 | 20.67 aAB ± 2.96 | 16.80 aAC ± 2.40 | 14.03 aC ± 0.91 | ||
| Two-way ANOVA, p | * | *** | NS | |||||
| Pathogen | Heat Inactivation Time | Bacterial Strains | Heat Inactivation Time × Bacterial Strains | |||||
|---|---|---|---|---|---|---|---|---|
| O12 | O16 | O22 | O24 | O20 | 299v | |||
| L. monocytogenes, % | t = 0 | 98.60 aA ± 0.33 | 99.11 aA ± 0.29 | 96.68 aB ± 1.23 | 95.59 aB ± 0.42 | 95.96 aB ± 0.56 | 99.39 aA ± 0.27 | |
| t = 5′ | 98.17 aA ± 0.59 | 98.78 aA ± 0.27 | 94.10 bB ± 0.30 | 98.24 bA ± 0.20 | 98.13 bA ± 0.30 | 98.84 aA ± 0.76 | ||
| t = 15′ | 98.24 aA ± 1.07 | 98.97 aA ± 0.30 | 96.32 abB ± 0.77 | 99.05 bA ± 0.63 | 98.56 bA ± 0.34 | 99.18 aA ± 0.52 | ||
| t = 30′ | 98.25 aA ± 0.63 | 98.86 aA ± 0.32 | 96.22 abB ± 1.61 | 98.74 bA ± 0.55 | 98.59 bA ± 0.42 | 98.86 aA ± 0.53 | ||
| Two-way ANOVA, p | *** | *** | *** | |||||
| S. aureus, % | t = 0 | 87.68 aAB ± 8.95 | 95.17 aA ± 0.40 | 52.59 aB ± 1.18 | 86.66 aAB ± 4.20 | 88.75 aAB ± 1.08 | 93.13 aA ± 0.21 | |
| t = 5′ | 88.30 aAB ± 7.12 | 95.43 aA ± 0.30 | 56.23 aB ± 1.12 | 87.68 aAB ± 1.81 | 90.85 aAB ± 5.77 | 91.29 aA ± 12.10 | ||
| t = 15′ | 95.24 aAB ± 0.75 | 99.13 bB ± 0.66 | 81.99 bA ± 6.66 | 97.70 bAB ± 0.72 | 91.25 aA ± 0.49 | 97.13 aAB ± 0.31 | ||
| t= 30′ | 84.17 aAC ± 4.02 | 97.96 bB ± 1.67 | 80.29 bC ± 5.03 | 90.37 aAD ± 3.02 | 90.87 aABD ± 3.19 | 95.46 aBD ± 0.23 | ||
| Two-way ANOVA, p | *** | *** | *** | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karbowiak, M.; Gałek, M.; Szydłowska, A.; Zielińska, D. The Influence of the Degree of Thermal Inactivation of Probiotic Lactic Acid Bacteria and Their Postbiotics on Aggregation and Adhesion Inhibition of Selected Pathogens. Pathogens 2022, 11, 1260. https://doi.org/10.3390/pathogens11111260
Karbowiak M, Gałek M, Szydłowska A, Zielińska D. The Influence of the Degree of Thermal Inactivation of Probiotic Lactic Acid Bacteria and Their Postbiotics on Aggregation and Adhesion Inhibition of Selected Pathogens. Pathogens. 2022; 11(11):1260. https://doi.org/10.3390/pathogens11111260
Chicago/Turabian StyleKarbowiak, Marcelina, Michał Gałek, Aleksandra Szydłowska, and Dorota Zielińska. 2022. "The Influence of the Degree of Thermal Inactivation of Probiotic Lactic Acid Bacteria and Their Postbiotics on Aggregation and Adhesion Inhibition of Selected Pathogens" Pathogens 11, no. 11: 1260. https://doi.org/10.3390/pathogens11111260
APA StyleKarbowiak, M., Gałek, M., Szydłowska, A., & Zielińska, D. (2022). The Influence of the Degree of Thermal Inactivation of Probiotic Lactic Acid Bacteria and Their Postbiotics on Aggregation and Adhesion Inhibition of Selected Pathogens. Pathogens, 11(11), 1260. https://doi.org/10.3390/pathogens11111260







