Antagonism of Rhizosphere Streptomyces yangpuensis CM253 against the Pathogenic Fungi Causing Corm Rot in Saffron (Crocus sativus L.)
Abstract
1. Introduction
2. Results
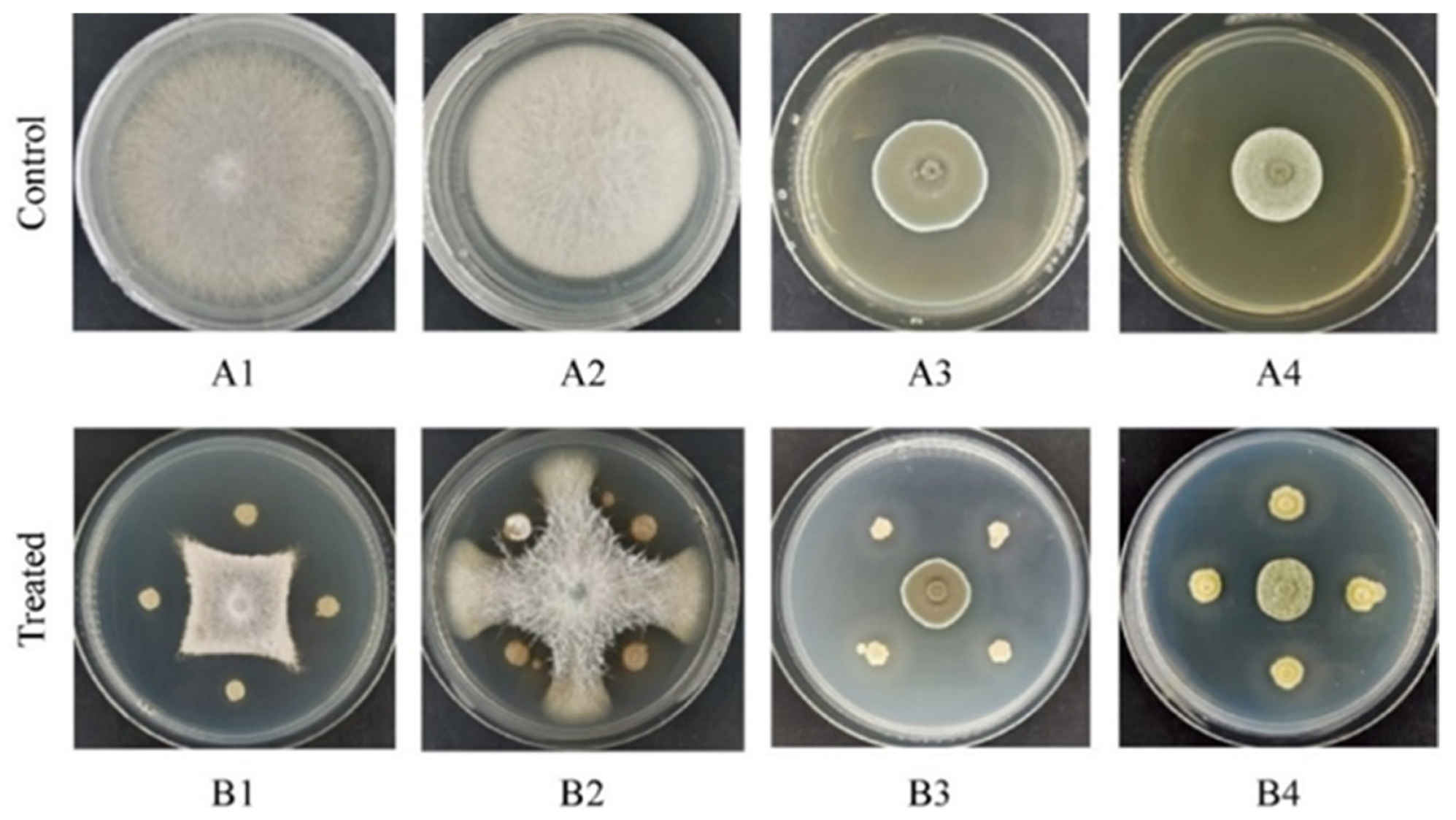
2.1. Screening of Biocontrol Actinomyces
2.2. Effect of Biocontrol Actinomycete CM253 on Mycelium Morphology
2.3. Analysis of Degrading Enzyme Production and Growth-Promoting Performance of Biocontrol Strain CM253
2.4. Identification and Whole Genome Sequencing of the CM253 Biocontrol Strain
3. Discussion
4. Materials and Methods
4.1. Indicative Pathogenic Fungi
4.2. Collection of Soil and Isolation and Purification of Biocontrol Actinomycetes
4.3. In Vitro Screening for Antibacterial Activity
4.4. Morphological Characteristics, Physiology, and Biochemistry of Rhizosphere Actinomycete Strain CM253
4.5. Molecular Identification of Rhizosphere Actinomycete Strain CM253
4.6. Effect of Rhizosphere Actinomycete Strain CM253 on Mycelia Morphology of F. oxysporum
4.7. Determination of Hydrolase Activity
4.8. In Vitro Assessment of Plant Growth Promotion (PGP) Traits
4.9. Whole Genome Sequencing (WGS) of Biocontrol Actinomycete CM253
4.9.1. Sample Preparation
4.9.2. DNA Extraction, Genome Sequencing, and Assembly
4.9.3. Gene Prediction and Annotation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Pitsikas, N. Crocus sativus L. extracts and its constituents crocins and safranal; Potential candidates for schizophrenia treatment? Molecules 2021, 26, 1237. [Google Scholar] [CrossRef] [PubMed]
- Cardone, L.; Castronuovo, D.; Perniola, M.; Cicco, N.; Candido, V. Saffron (Crocus sativus L.), the king of spices: An overview. Sci. Hortic. 2020, 272, 109560. [Google Scholar] [CrossRef]
- Moratalla-López, N.; Bagur, M.J.; Lorenzo, C.; Martínez-Navarro, M.E.; Salinas, M.R.; Alonso, G.L. Bioactivity and bioavailability of the major metabolites of Crocus sativus L. flower. Molecules 2019, 24, 2827. [Google Scholar] [CrossRef]
- Srivastava, R.; Ahmed, H.; Dixit, R.K.; Dharamveer; Saraf, S.A. Crocus sativus L.: A comprehensive review. Pharm. Rev. 2010, 4, 200–208. [Google Scholar] [CrossRef]
- Mzabri, I.; Addi, M.; Berrichi, A. Traditional and Modern Uses of Saffron (Crocus sativus). Cosmetics 2019, 6, 63. [Google Scholar] [CrossRef]
- Commission, C.P. Pharmacopoeia of the People’s Republic of China 2020; China Medical Science Press: Beijing, China, 2020. [Google Scholar]
- Liu, B.; Dong, Y.; Yao, C.; Jiang, F. Survey on Resource Development of Saffron in China. Chin. J. Mod. Appl. Pharm. 2022, 39, 1783–1788. [Google Scholar] [CrossRef]
- Najari, G.; Nourollahi, K.; Piri, M. The first report of Fusarium oxysporum causal agent of wild saffron corm rot disease in Iran. Saffron Agron. Technol. 2018, 6, 119–123. [Google Scholar] [CrossRef]
- Kafi, M.; Kamili, A.N.; Husaini, A.M.; Ozturk, M.; Altay, V. An Expensive Spice Saffron (Crocus sativus L.): A Case Study from Kashmir, Iran, and Turkey. In Global Perspectives on Underutilized Crops; Springer: Berlin/Heidelberg, Germany, 2018; pp. 109–149. [Google Scholar]
- Muñoz, R.M.; Lerma, M.L.; Castillo, P.; Armengol, J.; Somoza, E.; Woodhall, J.W. First report of stromatinia gladioli causing neck and corm rot of Crocus sativus in Spain. Plant Dis. 2020, 104, 282–283. [Google Scholar] [CrossRef]
- Sutton, M.W.; Wale, S.J. The control of Penicillium corymbiferum on crocus and its effect on corm production. Plant Pathol. 1985, 34, 566–570. [Google Scholar] [CrossRef]
- Kalha, C.; Gupta, V.; Gupta, D.; Priya, S. First report of sclerotial rot of saffron caused by Sclerotium rolfsii in India. Plant Dis. 2007, 91, 1203. [Google Scholar] [CrossRef]
- Gupta, R.; Vakhlu, J. Native Bacillus amyloliquefaciens W2 as a potential biocontrol for Fusarium oxysporum R1 causing corm rot of Crocus sativus. Eur. J. Plant Pathol. 2015, 143, 123–131. [Google Scholar] [CrossRef]
- Wani, Z.A.; Mirza, D.N.; Arora, P.; Riyaz-Ul-Hassan, S. Molecular phylogeny, diversity, community structure, and plant growth promoting properties of fungal endophytes associated with the corms of saffron plant: An insight into the microbiome of Crocus sativus Linn. Fungal Biol. 2016, 120, 1509–1524. [Google Scholar] [CrossRef] [PubMed]
- Belfiori, B.; Rubini, A.; Riccioni, C. Diversity of endophytic and pathogenic fungi of saffron (Crocus sativus) plants from cultivation sites in Italy. Diversity 2021, 13, 535. [Google Scholar] [CrossRef]
- Feizi, H.; Moradi, R. Assessing involved managing factors in gap yield between traditional and ideal saffron cultivating systems in Razavi and South Khorasan provinces. J. Saffron Res. 2020, 7, 283–298. [Google Scholar] [CrossRef]
- Aghhavani Shajari, M.; Rezvani Moghaddam, P.; Ghorbani, R.; Koocheki, A. The possibility of improving saffron (Crocus sativus L.) flower and corm yield through the irrigation and soil texture managements. Sci. Hortic. 2020, 271, 109485. [Google Scholar] [CrossRef]
- Zhou, T.; Qiu, X.; Zhao, L.; Yang, W.; Wen, F.; Wu, Q.; Yan, J.; Xu, B.; Chen, J.; Ma, Y.; et al. Optimal light intensity and quality increased the saffron daughter corm yield by inhibiting the degradation of reserves in mother corms during the reproductive stage. Ind. Crops Prod. 2022, 176, 114396. [Google Scholar] [CrossRef]
- Zhang, T.; Huang, C.; Deng, C.; Zhang, Y.; Feng, Y.; Hu, J.; Wang, R.; Zhao, L.; Wang, Y.; Kai, G. First report of corm rot on saffron caused by Penicillium solitum in China. Plant Dis. 2020, 104, 579. [Google Scholar] [CrossRef]
- Wei, L.; Duan, X.; Lu, G.; Chang, J.; Zhou, X.; Ma, H.; Qi, H. ldentification of saffron corm rot disease pathogen and laboratory screening of chemical agents. Plant Prot. 2021, 47, 139–145. [Google Scholar] [CrossRef]
- Jiening, W.; Siqi, G.; Jiajia, C.; Xue, D.; Junbo, L.; Xiujuan, L.; Guoyin, K.; Wei, Z. Isolation and identification of pathogenic fungi of stem rot in Crocus sativus. J. Zhejiang A&F Univ. 2022, 39, 1080–1086. [Google Scholar] [CrossRef]
- Hu, S.; Sun, W.; Wang, X.; Wang, L.; Li, W. First report of black spot caused by Penicillium citreosulfuratum on saffron in Chongming Island, China. Plant Dis. 2021, 106, 760. [Google Scholar] [CrossRef]
- Ali, M.A.; Naveed, M.; Mustafa, A.; Abbas, A. The good, the bad, and the ugly of rhizosphere microbiome. In Probiotics and Plant Health; Springer: Berlin/Heidelberg, Germany, 2017; pp. 253–290. [Google Scholar]
- Oleńska, E.; Małek, W.; Wójcik, M.; Swiecicka, I.; Thijs, S.; Vangronsveld, J. Beneficial features of plant growth-promoting rhizobacteria for improving plant growth and health in challenging conditions: A methodical review. Sci. Total Environ. 2020, 743, 140682. [Google Scholar] [CrossRef] [PubMed]
- Bhanse, P.; Kumar, M.; Singh, L.; Awasthi, M.K.; Qureshi, A. Role of plant growth-promoting rhizobacteria in boosting the phytoremediation of stressed soils: Opportunities, challenges, and prospects. Chemosphere 2022, 303, 134954. [Google Scholar] [CrossRef] [PubMed]
- Azizi, S.; Tabari Kouchaksaraei, M.; Hadian, J.; Fallah Nosrat Abad, A.R.; Modarres Sanavi, S.A.M.; Ammer, C.; Bader, M.K.F. Dual inoculations of arbuscular mycorrhizal fungi and plant growth-promoting rhizobacteria boost drought resistance and essential oil yield of common myrtle. For. Ecol. Manag. 2021, 497, 119478. [Google Scholar] [CrossRef]
- Yuan, Y.; Zuo, J.; Zhang, H.; Zu, M.; Liu, S. The Chinese medicinal plants rhizosphere: Metabolites, microorganisms, and interaction. Rhizosphere 2022, 22, 100540. [Google Scholar] [CrossRef]
- Madhurama, G.; Sonam, D.; Urmil, P.G.; Ravindra, N.K. Diversity and biopotential of endophytic actinomycetes from three medicinal plants in India. Afr. J. Microbiol. Res. 2014, 8, 184–191. [Google Scholar] [CrossRef]
- Djemouai, N.; Meklat, A.; Gaceb-Terrak, R.; Youcef, K.O.H.; Nacer, A.; Saadi, S.A.; Saad, S.; Verheecke-Vaessen, C.; Bouras, N. Streptomyces species from the rhizosphere of the medicinal plant Artemisia herba-alba Asso: Screening for biological activities biological activities. Biologia 2022, 77, 2281–2299. [Google Scholar] [CrossRef]
- Selim, M.S.M.; Abdelhamid, S.A.; Mohamed, S.S. Secondary metabolites and biodiversity of actinomycetes. J. Genet. Eng. Biotechnol. 2021, 19, 72. [Google Scholar] [CrossRef]
- Vurukonda, S.S.K.P.; Giovanardi, D.; Stefani, E. Plant growth promoting and biocontrol activity of Streptomyces spp. as endophytes. Int. J. Mol. Sci. 2018, 19, 952. [Google Scholar] [CrossRef]
- Qi, Y.; Li, X.; Wang, J.; Wang, C.; Zhao, S. Efficacy of plant growth-promoting bacteria Streptomyces werraensis F3 for chemical modifications of diseased soil of ginseng. Biocontrol Sci. Technol. 2021, 31, 219–233. [Google Scholar] [CrossRef]
- Faheem, M.; Raza, W.; Zhong, W.; Nan, Z.; Shen, Q.; Xu, Y. Evaluation of the biocontrol potential of Streptomyces goshikiensis YCXU against Fusarium oxysporum f. sp. niveum. Biol. Control 2015, 81, 101–110. [Google Scholar] [CrossRef]
- Yun, T.; Zhang, M.; Zhou, D.; Jing, T.; Zang, X.; Qi, D.; Chen, Y.; Li, K.; Zhao, Y.; Tang, W. Anti-Foc RT4 activity of a newly isolated Streptomyces sp. 5–10 from a medicinal plant (Curculigo capitulata). Front. Microbiol. 2021, 11, 610698. [Google Scholar] [CrossRef] [PubMed]
- Chaiharn, M.; Theantana, T.; Pathom-Aree, W. Evaluation of biocontrol activities of Streptomyces spp. against rice blast disease fungi. Pathogens 2020, 9, 126. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Wang, X.; Sun, W.; Wang, L.; Li, W. In vitro study of biocontrol potential of rhizospheric Pseudomonas aeruginosa against pathogenic fungi of saffron (Crocus sativus L.). Pathogens 2021, 10, 1423. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, T.; Bashir, A.; Farooq, S.; Riyaz-Ul-Hassan, S. Burkholderia gladioli E39CS3, an endophyte of Crocus sativus Linn., induces host resistance against corm-rot caused by Fusarium oxysporum. J. Appl. Microbiol. 2022, 132, 495–508. [Google Scholar] [CrossRef] [PubMed]
- Diez-Mendez, A.; Rivas, R. Improvement of saffron production using Curtobacterium herbarum as a bioinoculant under greenhouse conditions. AIMS Microbiol. 2017, 3, 354–364. [Google Scholar] [CrossRef]
- Zhang, H.; Xi, H.; Zhao, Y.; Wan, X.; Zhang, Y.; Zhao, Y.; Wen, C. Screening, identification of Streptomyces yangpuensis WLU210 and its biocontrol effect on Fusarium wilt of bitter gourd. Chin. J. Biol. Control 2021, 37, 1058–1065. [Google Scholar] [CrossRef]
- Shen, X.; Hu, H.; Peng, H.; Wang, W.; Zhang, X. Comparative genomic analysis of four representative plant growth-promoting rhizobacteria in Pseudomonas. BMC Genom. 2013, 14, 271. [Google Scholar] [CrossRef]
- Zaid, D.S.; Cai, S.; Hu, C.; Li, Z.; Li, Y. Comparative genome analysis reveals phylogenetic identity of Bacillus velezensis HNA3 and genomic insights into its plant growth promotion and biocontrol effects. Microbiol. Spectr. 2022, 10, e02169-21. [Google Scholar] [CrossRef]
- Khasin, M.; Cahoon, R.R.; Nickerson, K.W.; Riekhof, W.R. Molecular machinery of auxin synthesis, secretion, and perception in the unicellular chlorophyte alga Chlorella sorokiniana UTEX 1230. PLoS ONE 2018, 13, e0205227. [Google Scholar] [CrossRef]
- Goswami, D.; Thakker, J.N.; Dhandhukia, P.C. Portraying mechanics of plant growth promoting rhizobacteria (PGPR): A review. Cogent Food Agric. 2016, 2, 1127500. [Google Scholar] [CrossRef]
- Guo, D.-J.; Singh, R.K.; Singh, P.; Li, D.-P.; Sharma, A.; Xing, Y.-X.; Song, X.-P.; Yang, L.-T.; Li, Y.-R. Complete genome sequence of Enterobacter roggenkampii ED5, a nitrogen fixing plant growth promoting endophytic bacterium with biocontrol and stress tolerance properties, isolated from sugarcane root. Front. Microbiol. 2020, 11, 580081. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Martinez-Hidalgo, P.; Ice, T.A.; Maymon, M.; Humm, E.A.; Nejat, N.; Sanders, E.R.; Kaplan, D.; Hirsch, A.M. Antifungal activity of bacillus species against Fusarium and analysis of the potential mechanisms used in biocontrol. Front. Microbiol. 2018, 9, 2363. [Google Scholar] [CrossRef] [PubMed]
- Yeo, W.-S.; Anokwute, C.; Marcadis, P.; Levitan, M.; Ahmed, M.; Bae, Y.; Kim, K.; Kostrominova, T.; Liu, Q.; Bae, T. A membrane-bound transcription factor is proteolytically regulated by the AAA+ protease FtsH in Staphylococcus aureus. J. Bacteriol. 2020, 202, e00019-20. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, J.; Liu, J.; Liu, T.; Xue, S. Hydrogen sulfide (H2S) signaling in plant development and stress responses. aBIOTECH 2021, 2, 32–63. [Google Scholar] [CrossRef]
- Iqbal, S.; Ullah, N.; Janjua, H.A. In vitro evaluation and genome mining of Bacillus subtilis strain RS10 reveals its biocontrol and plant growth-promoting potential. Agriculture 2021, 11, 1273. [Google Scholar] [CrossRef]
- Khezri, M.; Jouzani, G.S.; Ahmadzadeh, M. Fusarium culmorum affects expression of biofilm formation key genes in Bacillus subtilis. Braz. J. Microbiol. 2016, 47, 47–54. [Google Scholar] [CrossRef]
- Knights, H.E.; Jorrin, B.; Haskett, T.L.; Poole, P.S. Deciphering bacterial mechanisms of root colonization. Environ. Microbiol. Rep. 2021, 13, 428–444. [Google Scholar] [CrossRef]
- Ciccoritti, R.; Pasquini, M.; Sgrulletta, D.; Nocente, F. Effect of 5-n-alkylresorcinol extracts from durum wheat whole grain on the growth of fusarium head blight (FHB) causal agents. J. Agric. Food Chem. 2015, 63, 43–50. [Google Scholar] [CrossRef]
- Williams, J.C.; Sheldon, J.R.; Imlay, H.D.; Dutter, B.F.; Draelos, M.M.; Skaar, E.P.; Sulikowski, G.A. Synthesis of the siderophore coelichelin and its utility as a probe in the study of bacterial metal sensing and response. Org. Lett. 2019, 21, 679–682. [Google Scholar] [CrossRef]
- Benaud, N.; Edwards, R.J.; Amos, T.G.; D’Agostino, P.M.; Gutiérrez-Chávez, C.; Montgomery, K.; Nicetic, I.; Ferrari, B.C. Antarctic desert soil bacteria exhibit high novel natural product potential, evaluated through long-read genome sequencing and comparative genomics. Environ. Microbiol. 2021, 23, 3646–3664. [Google Scholar] [CrossRef]
- Aeny, T.N.; Prasetyo, J.; Suharjo, R.; Dirmawati, S.R.; Efri, E.; Niswati, A. Isolation and identification of actinomycetes potential as the antagonist of Dickeya zeae pineapple soft rot in Lampung, Indonesia. Biodiversitas J. Biol. Divers. 2018, 19, 2052–2058. [Google Scholar] [CrossRef]
- Sadeghian, M.; Shahidi Bonjar, G.H.; Sharifi Sirchi, G.R. Post harvest biological control of apple bitter rot by soil-borne actinomycetes and molecular identification of the active antagonist. Postharvest Biol. Technol. 2016, 112, 46–54. [Google Scholar] [CrossRef]
- Anggelina, A.; Pringgenies, D.; Setyati, W. Presence of biosynthetic gene clusters (NRPS/PKS) in actinomycetes of mangrove sediment in Semarang and Karimunjawa, Indonesia. Environ. Nat. Resour. J. 2021, 19, 391–401. [Google Scholar] [CrossRef]
- Silveira, M.A.V.; Batista dos Santos, S.M.; Okamoto, D.N.; de Melo, I.S.; Juliano, M.A.; Ribeiro Chagas, J.; Vasconcellos, S.P. Atlantic Forest’s and Caatinga’s semiarid soils and their potential as a source for halothermotolerant actinomycetes and proteolytic enzymes. Environ. Technol. 2021, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Shafi, J.; Li, M.; Fu, D.; Ji, M. Insecticidal activity of endophytic actinomycetes isolated from Azadirachta indica against Myzus persicae. Arch. Biol. Sci. 2018, 70, 349–357. [Google Scholar] [CrossRef]
- Song, L.; Jiang, N.; Wei, S.; Lan, Z.; Pan, L. Isolation, screening, and identification of actinomycetes with antifungal and enzyme activity assays against Colletotrichum dematium of Sarcandra glabra. Mycobiology 2020, 48, 37–43. [Google Scholar] [CrossRef]
- Xiong, Z.-Q.; Tu, X.-R.; Wei, S.-J.; Huang, L.; Li, X.-H.; Lu, H.; Tu, G.-Q. In vitro antifungal activity of antifungalmycin 702, a new polyene macrolide antibiotic, against the rice blast fungus Magnaporthe grisea. Biotechnol. Lett. 2013, 35, 1475–1479. [Google Scholar] [CrossRef]
- Zhang, H.; Zhou, H.; Bai, J.; Li, Y.; Yang, J.; Ma, Q.; Qu, Y. Biosynthesis of selenium nanoparticles mediated by fungus Mariannaea sp. HJ and their characterization. Colloids Surf. A Physicochem. Eng. Asp. 2019, 571, 9–16. [Google Scholar] [CrossRef]
- Dong, X.; Cai, M. Handbook of Identification of Common Bacterial Systems, 1st ed.; Science Press: Beijing, China, 2001; pp. 370–387. [Google Scholar]
- Passari, A.K.; Mishra, V.K.; Leo, V.V.; Gupta, V.K.; Singh, B.P. Phytohormone production endowed with antagonistic potential and plant growth promoting abilities of culturable endophytic bacteria isolated from Clerodendrum colebrookianum Walp. Microbiol. Res. 2016, 193, 57–73. [Google Scholar] [CrossRef]
- Ait Bahadou, S.; Ouijja, A.; Karfach, A.; Tahiri, A.; Lahlali, R. New potential bacterial antagonists for the biocontrol of fire blight disease (Erwinia amylovora) in Morocco. Microb. Pathog. 2018, 117, 7–15. [Google Scholar] [CrossRef]
- Agarwal, M.; Dheeman, S.; Dubey, R.C.; Kumar, P.; Maheshwari, D.K.; Bajpai, V.K. Differential antagonistic responses of Bacillus pumilus MSUA3 against Rhizoctonia solani and Fusarium oxysporum causing fungal diseases in Fagopyrum esculentum Moench. Microbiol. Res. 2017, 205, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Aboelnour, A.; Noreldin, A.E.; Massoud, D.; Abumandour, M.M. Retinal characterization in the eyes of two bats endemic in the Egyptian fauna, the Egyptian fruit bat (Rousettus aegyptiacus) and insectivorous bat (Pipistrellus kuhlii), using the light microscope and transmission electron microscope. Microsc. Res. Tech. 2020, 83, 1391–1400. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Mishra, J.; Dwivedi, S.K.; Arora, N.K. Microbial enzymes in biocontrol of phytopathogens. In Microbial Enzymes: Roles and Applications in Industries; Springer: Singapore, 2020; pp. 259–285. [Google Scholar]
- Lau, E.T.; Tani, A.; Khew, C.Y.; Chua, Y.Q.; Hwang, S.S. Plant growth-promoting bacteria as potential bio-inoculants and biocontrol agents to promote black pepper plant cultivation. Microbiol. Res. 2020, 240, 126549. [Google Scholar] [CrossRef] [PubMed]
- Putri, R.E.; Mubarik, N.R.; Ambarsari, L.; Wahyudi, A.T. Antagonistic activity of glucanolytic bacteria Bacillus subtilis W3. 15 against Fusarium oxysporum and its enzyme characterization. Biodiversitas J. Biol. Divers. 2021, 22, 4067–4077. [Google Scholar] [CrossRef]
- Prajapati, V.; Patel, S.; Ray, S.; Patel, K.C. Identification of protease enzymes involved in biocontrol activity. In Practical Handbook on Agricultural Microbiology; Springer: Berlin/Heidelberg, Germany, 2022; pp. 323–329. [Google Scholar]
- Sukmawati, D.; Sondana, G.A.; Fikriyyah, N.N.; Afifah, Z.N.; Firhandini, A.; Khumaiya, U.; Komsiatun, D.A.; Asmara, Y.T.; Supiyani, A.; Puspitaningrum, R.; et al. Cellulase-producing yeast isolated from fermented cocoa beans as biocontrol for pathogenic mold chocolate fruit collected from Sentul, Jawa Barat, Indonesia. J. Phys. Conf. Ser. 2021, 1869, 012043. [Google Scholar] [CrossRef]
- Suárez-Moreno, Z.R.; Vinchira-Villarraga, D.M.; Vergara-Morales, D.I.; Castellanos, L.; Ramos, F.A.; Guarnaccia, C.; Degrassi, G.; Venturi, V.; Moreno-Sarmiento, N. Plant-growth promotion and biocontrol properties of three Streptomyces spp. isolates to control bacterial rice pathogens. Front. Microbiol. 2019, 10, 290. [Google Scholar] [CrossRef]
- Chengqun, L.; Baoling, H. Isolation and characterization of Azotobacteria from pine rhizosphere. Afr. J. Microbiol. Res. 2010, 4, 1299–1306. [Google Scholar] [CrossRef]
- Dubey, A.; Saiyam, D.; Kumar, A.; Hashem, A.; Abd_Allah, E.F.; Khan, M.L. Bacterial root endophytes: Characterization of their competence and plant growth promotion in Soybean (Glycine max (L.) Merr.) under drought stress. Int. J. Environ. Res. Public Health 2021, 18, 931. [Google Scholar] [CrossRef]
- Sheng, M.M.; Jia, H.K.; Zhang, G.Y.; Zeng, L.N.; Zhang, T.T.; Long, Y.H.; Lan, J.; Hu, Z.Q.; Zeng, Z.; Wang, B.; et al. Siderophore production by rhizosphere biological control bacteria Brevibacillus brevis GZDF3 of Pinellia ternata and its antifungal effects on Candida albicans. J. Microbiol. Biotechnol. 2020, 30, 689–699. [Google Scholar] [CrossRef]
- Mohammed, B.L.; Hussein, R.A.; Toama, F.N. Biological control of Fusarium wilt in tomato by endophytic rhizobactria. Energy Procedia 2019, 157, 171–179. [Google Scholar] [CrossRef]
- Chin, C.-S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E.; et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Bateman, A.; Marshall, M.; Khanna, A.; Eddy, S.R. Rfam: An RNA family database. Nucleic Acids Res. 2003, 31, 439–441. [Google Scholar] [CrossRef]
- Alkhnbashi, O.S.; Meier, T.; Mitrofanov, A.; Backofen, R.; Voss, B. CRISPR-Cas bioinformatics. Methods 2020, 172, 3–11. [Google Scholar] [CrossRef]
- Xu, W.; Zhang, L.; Goodwin, P.H.; Xia, M.; Zhang, J.; Wang, Q.; Liang, J.; Sun, R.; Wu, C.; Yang, L. Isolation, identification, and complete genome assembly of an endophytic Bacillus velezensis YB-130, potential biocontrol agent against Fusarium graminearum. Front. Microbiol. 2020, 11, 598285. [Google Scholar] [CrossRef]
- Park, B.H.; Karpinets, T.V.; Syed, M.H.; Leuze, M.R.; Uberbacher, E.C. CAZymes Analysis Toolkit (CAT): Web service for searching and analyzing carbohydrate-active enzymes in a newly sequenced organism using CAZy database. Glycobiology 2010, 20, 1574–1584. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef]








| Hydrolytic Enzyme and PGP Activities | Result |
|---|---|
| Chitinase | − |
| Cellulase | + |
| Protease | + |
| Glucanase | + |
| IAA production | + |
| Phosphate solubilization | − |
| NH3 production | + |
| ACC deaminase enzyme | + |
| Potassium dissolution | − |
| Siderophore production | + |
| Physiological and Biochemical Characterization | Result |
|---|---|
| Gram reaction | + |
| Growth at 4 °C | − |
| Growth at 4 °C | − |
| Oxidase | − |
| Catalase | − |
| Urease | + |
| Lipase | + |
| Voges–Proskauer | − |
| Methyl red | − |
| Starch hydrolysis | − |
| Gelatin liquefaction | − |
| Nitrate reduction | + |
| Citrate utilization | − |
| H2S production | + |
| Milk coagulation | − |
| Region | Type | Start | End | Similar Cluster | Similarity |
|---|---|---|---|---|---|
| 1 | Lanthipeptide | 23898 | 45961 | Venezuelin | 100% |
| 2 | Lanthipeptide | 52651 | 77063 | - | – |
| 3 | Terpene | 427823 | 452809 | Carotenoid | 63% |
| 4 | NRPS | 475101 | 526320 | Deimino-antipain | 66% |
| 5 | T2PKS | 529223 | 601766 | Spore pigment | 66% |
| 6 | NRPS | 897035 | 964537 | JBIR-126 | 100% |
| 7 | Siderophore | 2716464 | 2727579 | Desferrioxamin B/Desferrioxamine E | 83% |
| 8 | NRPS | 4016003 | 4057260 | Phosphonoglycans | 3% |
| 9 | Siderophore | 5560082 | 5573594 | - | - |
| 10 | Bacteriocin | 5869688 | 5881185 | - | - |
| 11 | Terpene | 5964827 | 5986769 | Toxoflavin/fervenulin | 14% |
| 12 | T1PKS | 6014531 | 6059440 | Herboxidiene | 9% |
| 13 | Terpene | 6308285 | 6335005 | Hopene | 61% |
| 14 | NRPS-like | 6526608 | 6569022 | Polyketomycin | 4% |
| 15 | NRPS | 6704968 | 6745944 | Streptothricin | 87% |
| 16 | Terpene | 6838051 | 6857502 | – | – |
| 17 | NRPS | 6860761 | 6911849 | Coelichelin | 100% |
| 18 | Terpene | 6937552 | 6957566 | Ebelactone | 5% |
| 19 | Terpene | 7007207 | 7028140 | Monensin | 5% |
| 20 | Melanin | 7031380 | 7057085 | Istamycin | 4% |
| 21 | T1PKS | 7057399 | 7102148 | Daptomycin | 4% |
| 22 | Siderophore | 7138154 | 7151437 | – | – |
| 23 | T3PKS | 7184657 | 7225715 | Alkylresorcinol | 100% |
| 24 | CDPS | 7237760 | 7287223 | Cosmomycin C | 5% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, L.; Hu, S.; Wang, X.; Guo, Y.; Huang, L.; Wang, L.; Li, W. Antagonism of Rhizosphere Streptomyces yangpuensis CM253 against the Pathogenic Fungi Causing Corm Rot in Saffron (Crocus sativus L.). Pathogens 2022, 11, 1195. https://doi.org/10.3390/pathogens11101195
Tian L, Hu S, Wang X, Guo Y, Huang L, Wang L, Li W. Antagonism of Rhizosphere Streptomyces yangpuensis CM253 against the Pathogenic Fungi Causing Corm Rot in Saffron (Crocus sativus L.). Pathogens. 2022; 11(10):1195. https://doi.org/10.3390/pathogens11101195
Chicago/Turabian StyleTian, Li, Shuang Hu, Xingxing Wang, Yingqiu Guo, Luyang Huang, Lili Wang, and Wankui Li. 2022. "Antagonism of Rhizosphere Streptomyces yangpuensis CM253 against the Pathogenic Fungi Causing Corm Rot in Saffron (Crocus sativus L.)" Pathogens 11, no. 10: 1195. https://doi.org/10.3390/pathogens11101195
APA StyleTian, L., Hu, S., Wang, X., Guo, Y., Huang, L., Wang, L., & Li, W. (2022). Antagonism of Rhizosphere Streptomyces yangpuensis CM253 against the Pathogenic Fungi Causing Corm Rot in Saffron (Crocus sativus L.). Pathogens, 11(10), 1195. https://doi.org/10.3390/pathogens11101195





