Potential SARS-CoV-2 Susceptibility of Cetaceans Stranded along the Italian Coastline
Abstract
1. Introduction
2. Results
2.1. Detection of SARS-CoV-2 in Marine Mammals Stranded along Italian Coastline
2.1.1. SARS-CoV-2 RT-PCR
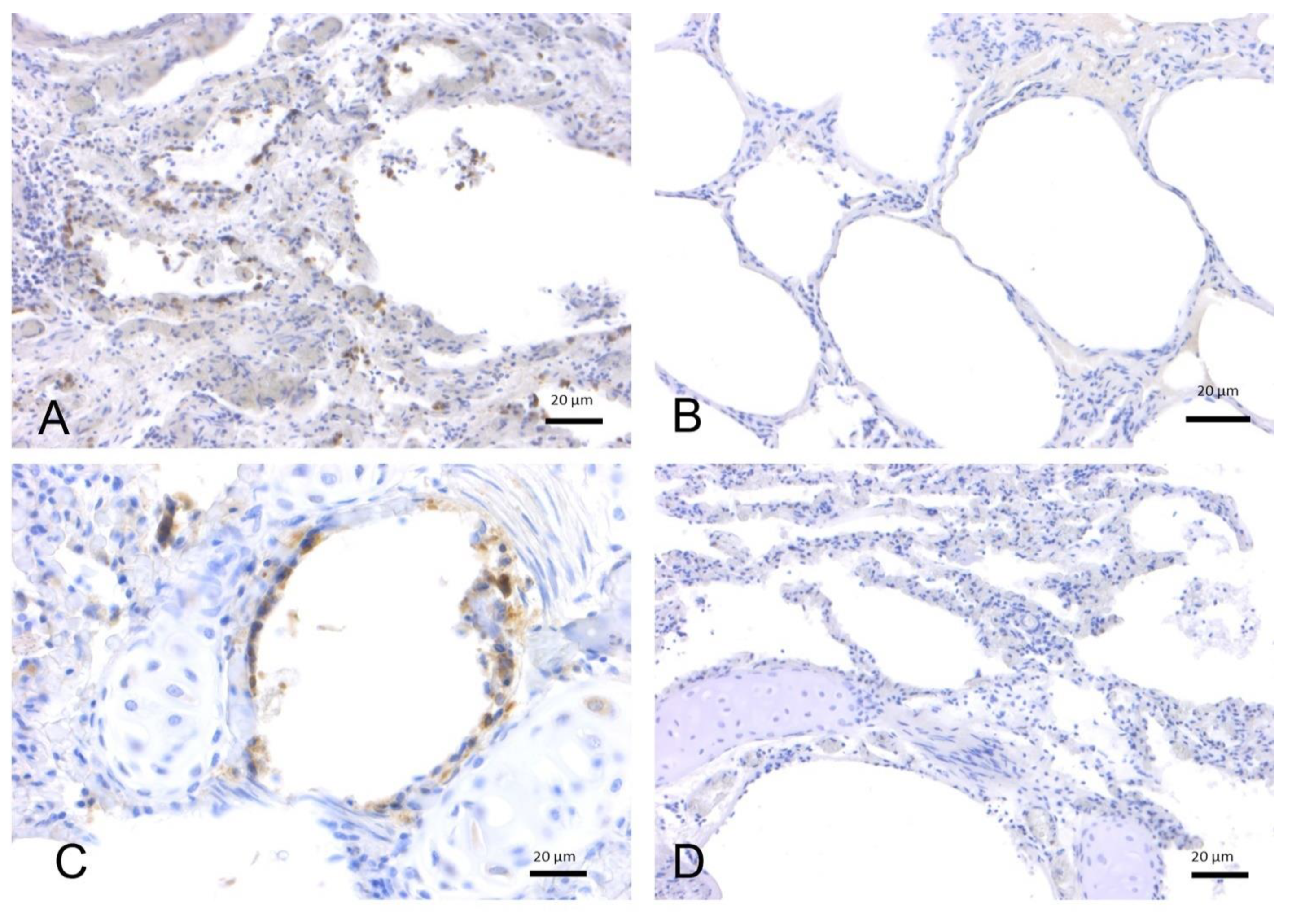
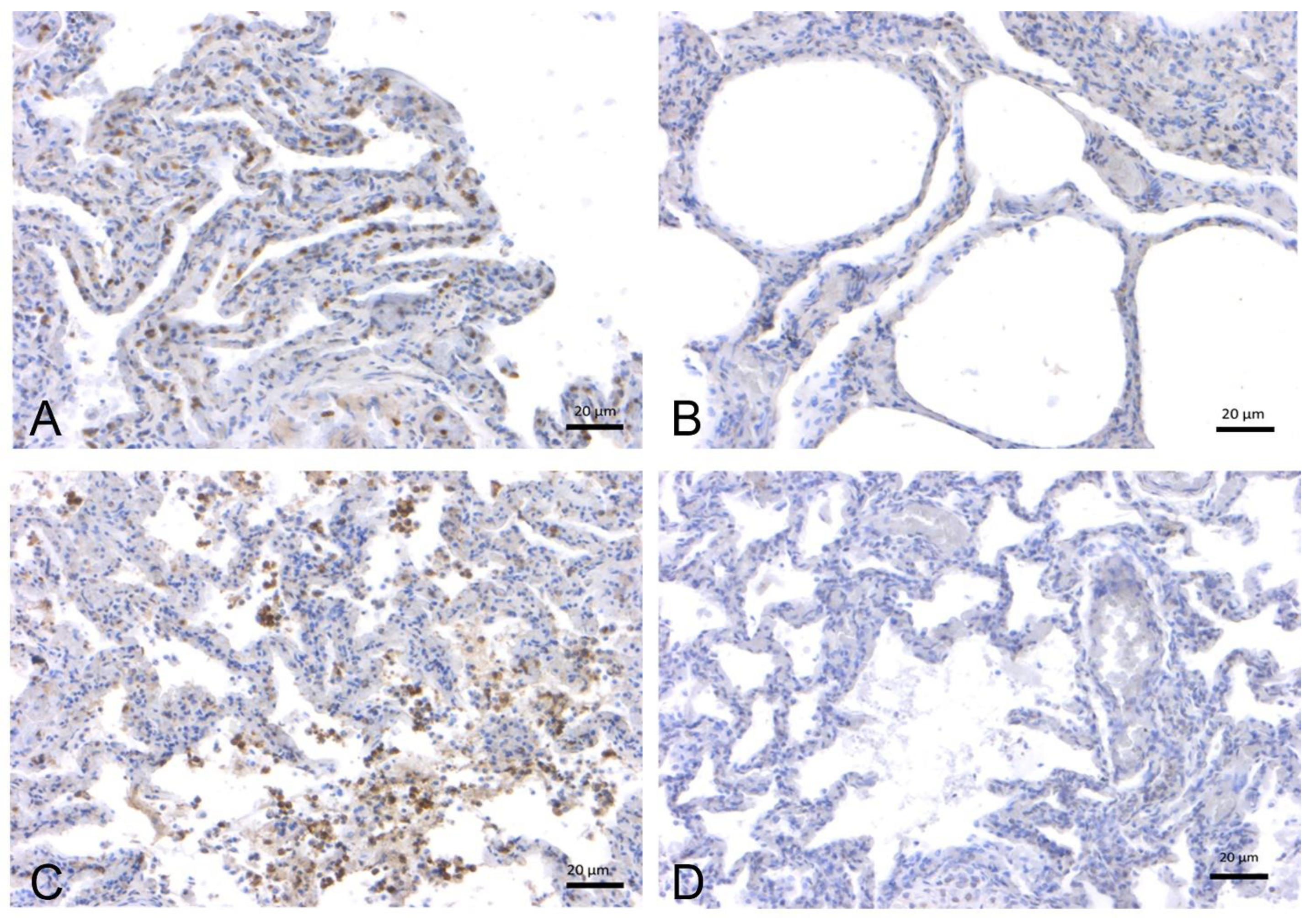
2.1.2. SARS-CoV-2 Immunohistochemistry
2.2. In-Depth Study of ACE2 Receptor
2.2.1. Effects of Age, Sex, and Origin (Captivity vs. Wild) on ACE2 Expression in Cetacean Lung Tissues
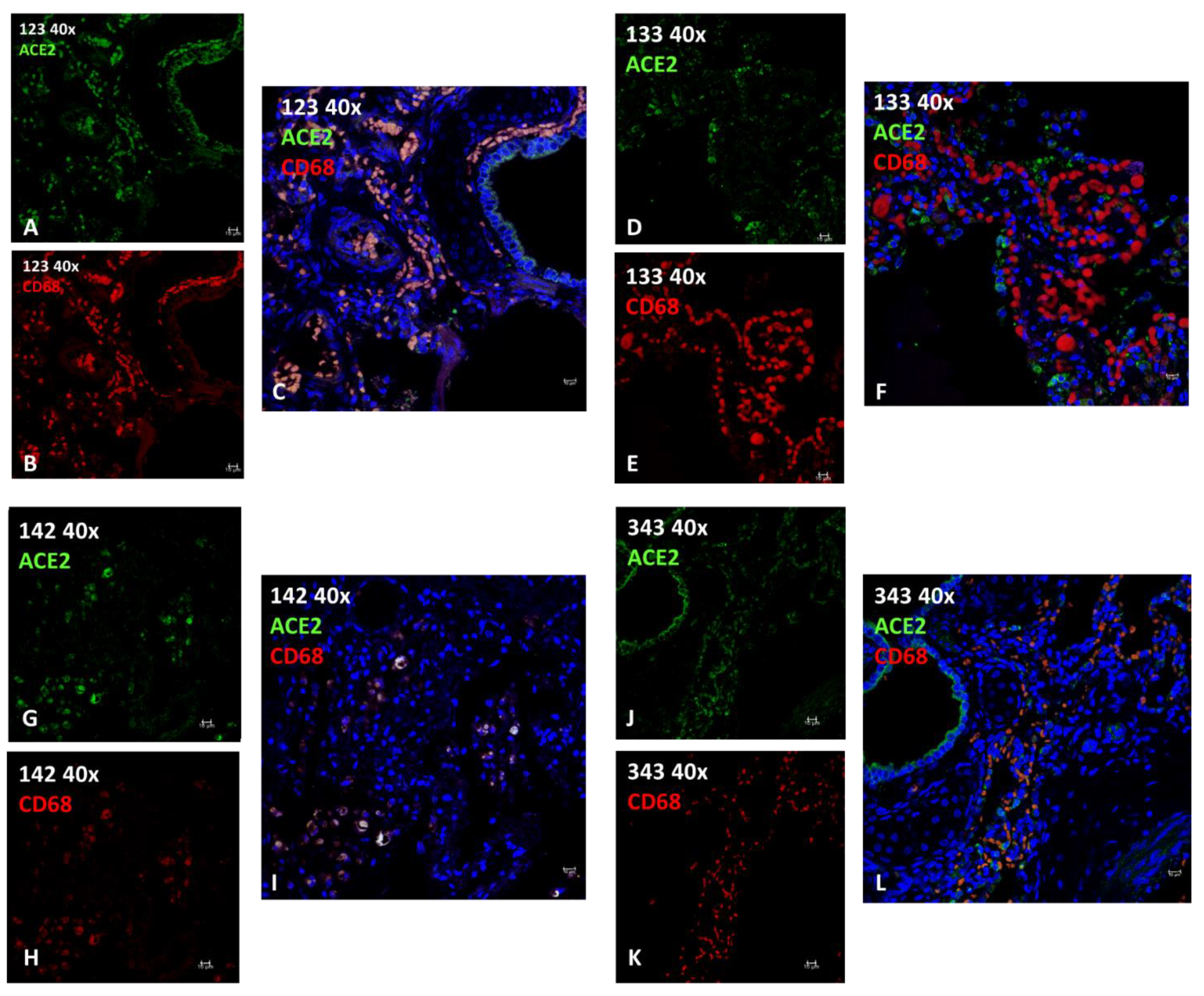
2.2.2. Immunofluorescence (IF)
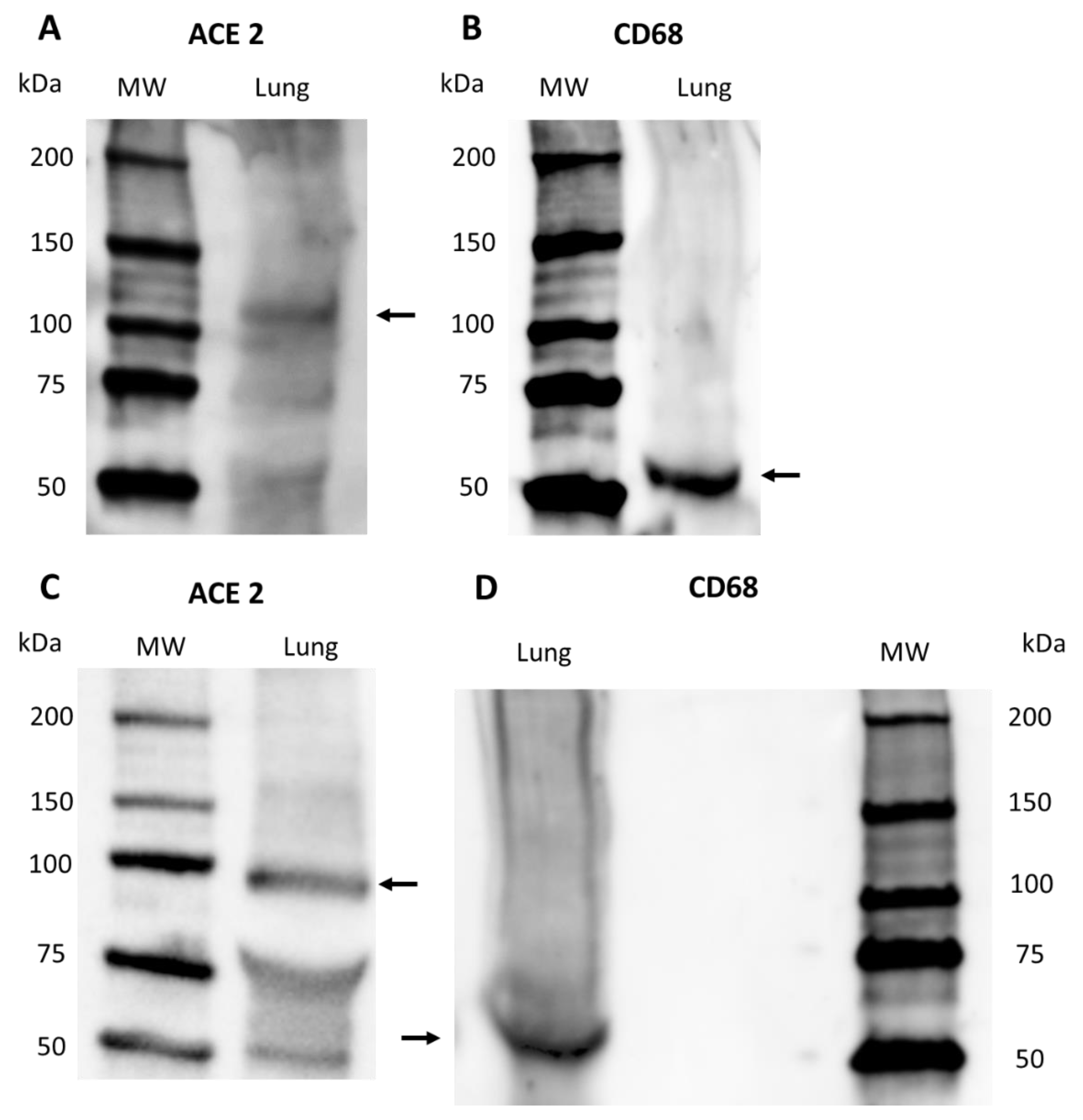
2.2.3. Western Blot
3. Discussion
4. Materials and Methods
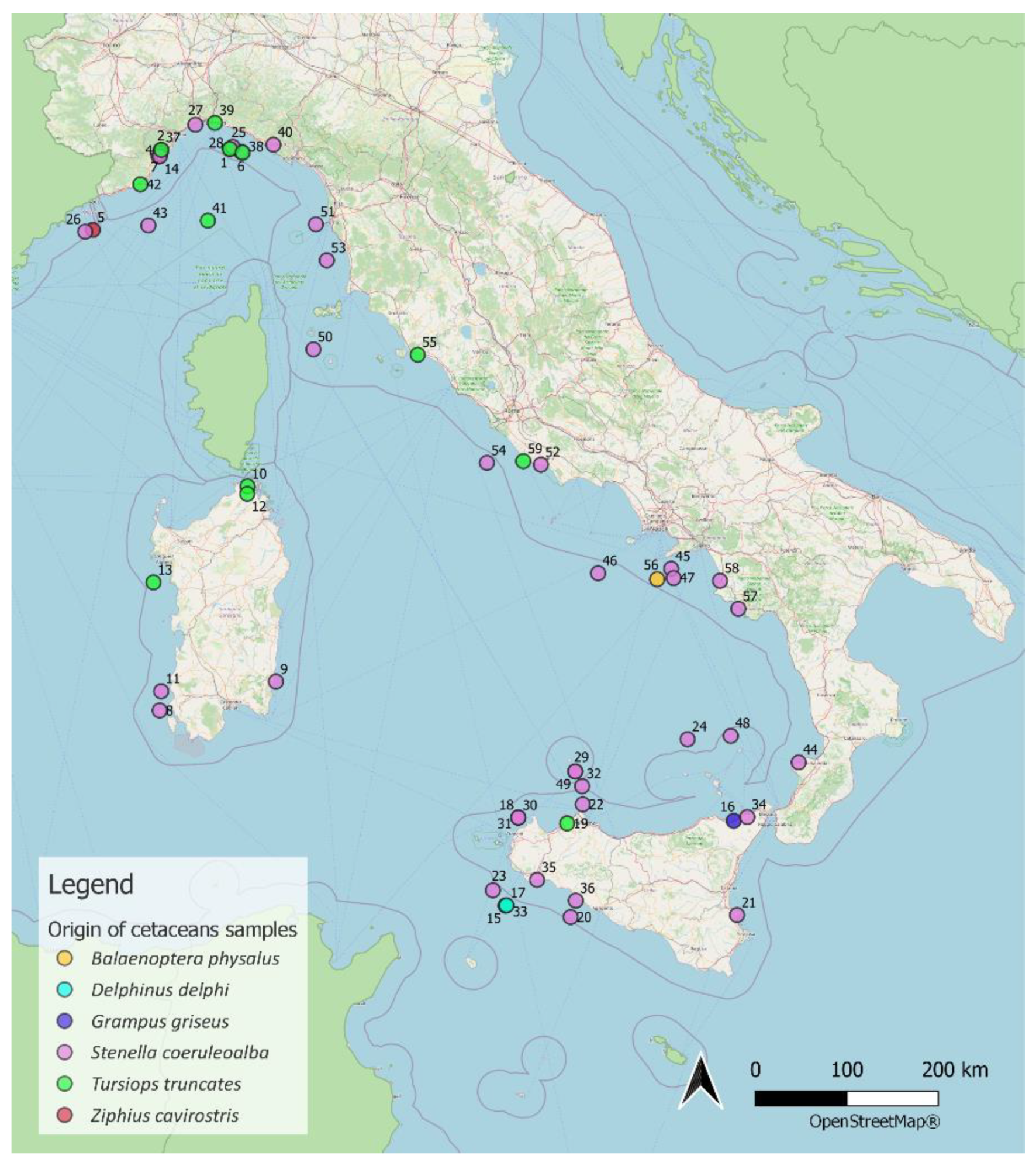
4.1. Sampling
4.2. SARS-CoV-2 Real Time-PCR (RT-PCR)
4.3. RNA Extraction and SARS-CoV-2 RT-PCR Detection
4.4. SARS-CoV 2 Immunohistochemistry
| Year | ID (Case PCR/IHC) | Organ | Swab | Species |
|---|---|---|---|---|
| 2020 | 1 | Lung | T. truncatus | |
| 2 | Lung | S. coeruleoalba | ||
| 3 | Lung | S. coeruleoalba | ||
| 4 | Lung | S. coeruleoalba | ||
| 5 | Lung | Z. cavirostris | ||
| 6 | Lung | S. coeruleoalba | ||
| 7 | Lung | S. coeruleoalba | ||
| 8 | Lung | S. coeruleoalba | ||
| 9 | Lung | S. coeruleoalba | ||
| 10 | Lung | T. truncatus | ||
| 11 | Lung | S. coeruleoalba | ||
| 12 | Lung | T. truncatus | ||
| 13 | Lung | T. truncatus | ||
| 14 | Lung | S. coeruleoalba | ||
| 15 | Lung | S. coeruleoalba | ||
| 16 | Lung | G. griseus | ||
| 17 | Lung | T. truncatus | ||
| 18 | Lung | S. coeruleoalba | ||
| 19 | Lung | T. truncatus | ||
| 20 | Lung | S. coeruleoalba | ||
| 21 | Lung | S. coeruleoalba | ||
| 22 | Lung | S. coeruleoalba | ||
| 23 | Lung | S. coeruleoalba | ||
| 24 | Lung | S. coeruleoalba | ||
| 44 * | Lung | S. coeruleoalba | ||
| 45 * | Lung | S. coeruleoalba | ||
| 46 * | Lung | S. coeruleoalba | ||
| 47 * | Lung | S. coeruleoalba | ||
| 48 * | Lung | S. coeruleoalba | ||
| 49 * | Lung | S. coeruleoalba | ||
| 50 * | Lung | S. coeruleoalba | ||
| 51 * | Lung | S. coeruleoalba | ||
| 52 * | Lung | S. coeruleoalba | ||
| 53 * | Lung | S. coeruleoalba | ||
| 54 * | Lung | S. coeruleoalba | ||
| 55 * | Lung | T. truncatus | ||
| 56 * | Lung | Balaenoptera physalus | ||
| 57 * | Lung | S. coeruleoalba | ||
| 58 * | Lung | S. coeruleoalba | ||
| 59 * | Lung | T. truncatus | ||
| 2021 | 25 | Lung | S. coeruleoalba | |
| Intestine | ||||
| CNS | ||||
| 26 | Lung | Rectum | S. coeruleoalba | |
| Intestine | ||||
| CNS | ||||
| 27 | Lung | S. coeruleoalba | ||
| CNS | ||||
| 28 | Lung | Oropharynx | S. coeruleoalba | |
| Intestine | Rectum | |||
| CNS | Trachea | |||
| Blowhole | ||||
| 29 | Lung | S. coeruleoalba | ||
| 30 | Lung | S. coeruleoalba | ||
| 31 | Lung | S. coeruleoalba | ||
| 32 | Lung | S. coeruleoalba | ||
| 33 | Lung | S. coeruleoalba | ||
| 34 | Lung | S. coeruleoalba | ||
| 35 | Lung | S. coeruleoalba | ||
| 36 | Lung | S. coeruleoalba | ||
| 37 | Lung | Oropharynx | T. truncatus | |
| Intestine | Rectum | |||
| CNS | Trachea | |||
| Blowhole | ||||
| 38 | Lung | T. truncatus | ||
| CNS | ||||
| 39 | Lung | Oropharynx | T. truncatus | |
| Intestine | Rectum | |||
| CNS | Trachea | |||
| Blowhole | ||||
| 40 | Intestine | S. coeruleoalba | ||
| 41 | Lung | Oropharynx | T. truncatus | |
| Intestine | Rectum | |||
| CNS | Trachea | |||
| Blowhole | ||||
| 2022 | 42 | Lung | Oropharynx | T. truncatus |
| Intestine | Rectum | |||
| CNS | Trachea | |||
| Blowhole | ||||
| 43 | Lung | Oropharynx | T. truncatus | |
| Intestine | Rectum | |||
| CNS | Trachea | |||
| Blowhole |
4.5. Immunofluorescence
| Antigen | Target | Antibody/Antiserum | Host | Dilution | Source | Technique(s) |
|---|---|---|---|---|---|---|
| 7B7 | RBD protein | Mono | Mouse | 1:50 | Kansas State University College of Veterinary Medicine | IF, IHC |
| 5A | S protein | Poly | Rabbit | 1:2000 | Kansas State University College of Veterinary Medicine | IHC |
| 3A | N protein | Poly | Rabbit | 1:1000 | Kansas State University College of Veterinary Medicine | IHC |
| ACE 2 | ACE2 receptor | Poly | Rabbit | 1:200/1:2000 | Abcam | IF, IHC |
| CD 68 | Macrophage | Mono | 1:50 | DAKO | IF |
4.6. Western Blot
4.7. ACE-2 Immunohistochemistry
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nabi, G.; Khan, S. Risk of COVID-19 pneumonia in aquatic mammals. Environ. Res. 2020, 188, 109732. [Google Scholar] [CrossRef]
- Johnstone, C.; Báez, J.C. Placing the COVID-19 Pandemic in a Marine Ecological Context: Potential Risks for Conservation of Marine Air-Breathing Animals and Future Zoonotic Outbreaks. Front. Mar. Sci. 2021, 8, 691682. [Google Scholar] [CrossRef]
- Di Guardo, G.; Agrimi, U.; Morelli, L.; Cardeti, G.; Terracciano, G.; Kennedy, S. Post mortem investigations on cetaceans found stranded on the coasts of Italy between 1990 and 1993. Vet. Rec. 1995, 136, 439–442. [Google Scholar] [CrossRef]
- Casalone, C.; Mazzariol, S.; Pautasso, A.; Di Guardo, G.; Di Nocera, F.; Lucifora, G.; Ligios, C.; Franco, A.; Fichi, G.; Cocumelli, C.; et al. Cetacean strandings in Italy: An unusual mortality event along the Tyrrhenian Sea coast in 2013. Dis. Aquat. Organ. 2014, 109, 81–86. [Google Scholar] [CrossRef]
- Gryseels, S.; De Bruyn, L.; Gyselings, R.; Calvignac-Spencer, S.; Leendertz, F.H.; Leirs, H. Risk of human-to-wildlife transmission of SARS-CoV-2. Mam. Rev. 2021, 51, 272–292. [Google Scholar] [CrossRef]
- Audino, T.; Grattarola, C.; Centelleghe, C.; Peletto, S.; Giorda, F.; Florio, C.L.; Caramelli, M.; Bozzetta, E.; Mazzariol, S.; Di Guardo, G.; et al. SARS-CoV-2, a Threat to Marine Mammals? A Study from Italian Seawaters. Animals 2021, 11, 1663. [Google Scholar] [CrossRef]
- Sun, Z.P.; Yang, S.Y.; Cai, X.; Han, W.D.; Hu, G.W.; Qian, Y.; Wang, Y.Y.; Zhang, R.; Xie, Y.H.; Qu, D. Survival of SARS-CoV-2 in artificial seawater and on the surface of inanimate materials. J. Med. Virol. 2022, 94, 3982–3987. [Google Scholar] [CrossRef]
- Mathavarajah, S.; Dellaire, G. Lions, tigers and kittens too: ACE2 and susceptibility to COVID-19. Evol. Med. Public Health 2020, 2020, 109–113. [Google Scholar] [CrossRef]
- Bogomolni, A.L.; Gast, R.J.; Ellis, J.C.; Dennett, M.; Pugliares, K.R.; Lentell, B.J.; Moore, M.J. Victims or vectors: A survey of marine vertebrate zoonoses from coastal waters of the Northwest Atlantic. Dis. Aquat. Organ. 2008, 81, 13–38. [Google Scholar] [CrossRef]
- Mordecai, G.J.; Hewson, I. Coronaviruses in the sea. Front. Microbiol. 2020, 11, 1795. [Google Scholar] [CrossRef]
- Schütze, H. Coronaviruses in Aquatic Organisms. Aquac. Virol. 2016, 327–335. [Google Scholar] [CrossRef]
- Hasnat, F.; Noman, F.; Moben, A.L.; Sarker, A.S.; Morshed, J.; Mutanabbi, M. Difference in Clinical Patterns between COVID-19 Affected Children and Adults. Mymensingh Med. J. 2021, 30, 1093–1099. [Google Scholar]
- Berni Canani, R.; Comegna, M.; Paparo, L.; Cernera, G.; Bruno, C.; Strisciuglio, C.; Zollo, I.; Gravina, A.G.; Miele, E.; Cantone, E.; et al. Age-Related Differences in the Expression of Most Relevant Mediators of SARS-CoV-2 Infection in Human Respiratory and Gastrointestinal Tract. Front. Pediatr. 2021, 9, 697390. [Google Scholar] [CrossRef]
- Devaux, C.A.; Rolain, J.M.; Raoult, D. ACE2 receptor polymorphism: Susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J. Microbiol. Immunol. Infect. 2020, 53, 425–435. [Google Scholar] [CrossRef]
- Abassi, Z.; Knaney, Y.; Karram, T.; Heyman, S.N. The lung macrophage in SARS-CoV-2 infection: A friend or a foe? Front. Immunol. 2020, 11, 1312. [Google Scholar] [CrossRef]
- Lye, P.; Dunk, C.E.; Zhang, J.; Wei, Y.; Nakpu, J.; Hamada, H.; Imperio, G.E.; Bloise, E.; Matthews, S.G.; Lye, S.J. ACE2 Is Expressed in Immune Cells That Infiltrate the Placenta in Infection-Associated Preterm Birth. Cells 2021, 10, 1724. [Google Scholar] [CrossRef]
- Sefik, E.; Qu, R.; Junqueira, C.; Kaffe, E.; Mirza, H.; Zhao, J.; Brewer, J.R.; Han, A.; Steach, H.R.; Israelow, B.; et al. Inflammasome activation in infected macrophages drives COVID-19 pathology. Nature 2022, 606, 585–593. [Google Scholar] [CrossRef]
- Epstein, J.H.; Price, J.T. The significant but understudied impact of pathogen transmission from humans to animals. Mt. Sinai J. Med. 2022, 76, 448–455. [Google Scholar] [CrossRef]
- Messenger, A.M.; Barnes, A.N.; Gray, G.C. Reverse zoonotic disease transmission (zooanthroponosis): A systematic review of seldom-documented human biological threats to animals. PLoS ONE 2014, 9, e89055. [Google Scholar] [CrossRef]
- La Rosa, G.; Iaconelli, M.; Mancini, P.; Ferraro, G.B.; Veneri, C.; Bonadonna, L.; Lucentini, L.; Suffredini, E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020, 736, 139652. [Google Scholar] [CrossRef]
- Rimoldi, S.G.; Stefani, F.; Gigantiello, A.; Polesello, S.; Comandatore, F.; Mileto, D.; Maresca, M.; Longobardi, C.; Mancon, A.; Romeri, F.; et al. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020, 744, 140911. [Google Scholar] [CrossRef]
- Yeo, C.; Kaushal, S.; Yeo, D. Enteric involvement of coronaviruses: Is faecal–oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020, 5, 335–337. [Google Scholar] [CrossRef]
- Langone, M.; Petta, L.; Cellamare, C.M.; Ferraris, M.; Guzzinati, R.; Mattioli, D.; Sabia, G. SARS-CoV-2 in water services: Presence and impacts. Environ. Pollut. 2021, 268, 115806. [Google Scholar] [CrossRef]
- Fossi, M.C.; Marsili, L.; Baini, M.; Giannetti, M.; Coppola, D.; Guerranti, C.; Caliani, I.; Minutoli, R.; Lauriano, G.; Finoia, M.G.; et al. Fin whales and microplastics: The Mediterranean Sea and the Sea of Cortez scenarios. Environ. Pollut. 2016, 209, 68–78. [Google Scholar] [CrossRef]
- Zhang, E.; Kim, M.; Rueda, L.; Rochman, C.; VanWormer, E.; Moore, J.; Shapiro, K. Association of zoonotic protozoan parasites with microplastics in seawater and im-plications for human and wildlife health. Sci. Rep. 2022, 12, 6532. [Google Scholar] [CrossRef]
- Food and Agriculture Organization (FAO); World Organisation for Animal Health (OIE); World Health Organization (WHO). Joint Statement on the Prioritization of Monitoring SARS-CoV-2 Infection in Wildlife and Preventing the Formation of Animal Reservoirs; Ginevra, Switzerland, 7 March 2022. [Google Scholar]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 2020, 368, 1016–1020. [Google Scholar] [CrossRef]
- Mykytyn, A.Z.; Lamers, M.M.; Okba, N.M.A.; Breugem, T.I.; Schipper, D.; van den Doel, P.B.; van Run, P.; van Amerongen, G.; de Waal, L.; Koopmans, M.P.G.; et al. Susceptibility of rabbits to SARS-CoV-2. Emerg. Microbes. Infect. 2021, 10, 1–7. [Google Scholar] [CrossRef]
- Ulrich, L.; Wernike, K.; Hoffmann, D.; Mettenleiter, T.C.; Beer, M. Experimental Infection of Cattle with SARS-CoV-2. Emerg. Infect. Dis. 2020, 26, 2979–2981. [Google Scholar] [CrossRef]
- Oreshkova, N.; Molenaar, R.J.; Vreman, S.; Harders, F.; Oude Munnink, B.B.; Hakze-van der Honing, R.W.; Gerhards, N.; Tolsma, P.; Bouwstra, R.; Sikkema, R.S.; et al. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Euro Surveill. 2020, 25, 2001005. [Google Scholar] [CrossRef]
- Rabalski, L.; Kosinski, M.; Smura, T.; Aaltonen, K.; Kant, R.; Sironen, T.; Szewczyk, B.; Grzybek, M. Severe Acute Respiratory Syndrome Coronavirus 2 in Farmed Mink (Neovison vison), Poland. Emerg. Infect. Dis. 2021, 27, 2333–2339. [Google Scholar] [CrossRef]
- Damas, J.; Hughes, G.M.; Keough, K.C.; Painter, C.A.; Persky, N.S.; Corbo, M.; Hiller, M.; Koepfli, K.-P.; Pfenning, A.R.; Zhao, H.; et al. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc. Natl. Acad. Sci. USA 2020, 117, 22311–22322. [Google Scholar] [CrossRef]
- Casalone, C.; Di Guardo, G. CoViD-19 and mad cow disease: So different yet so similar. Science 2020. [Google Scholar] [CrossRef]
- Proietto, U.; Fernandez, A.; Manolo, A.; Di Guardo, G. Caratterizzazione istomorfologica ed immunoistochimica dei granulociti neutrofili e dei macrofagi nel polmone di cetacei spiaggiati. Ittiopatologia 2008, 5, 5–17. [Google Scholar]
- Song, X.; Hu, W.; Yu, H.; Zhao, L.; Zhao, Y.; Zhao, X. Little to no expression of angiotensin-converting enzyme-2 on most human peripheral blood immune cells but highly expressed on tissue macrophages. Cytom. A, 2020; Epub ahead of print. [Google Scholar] [CrossRef]
- Di Guardo, G. SARS-CoV-2 susceptibility of domestic and wild mammals: Is it all about the similarity of their ACE2 receptor to the human one? Proc. R. Soc. B 2022, 289, 2021256020212560. [Google Scholar] [CrossRef]
- Geraci, J.R.; Lounsbury, V.J. Marine Mammals Ashore: A Field Guide for Strandings, 2nd ed.; National Aquarium in Baltimore; Mystic Aquarium & Institute for Exploration, Mystic, CT and Program in Comparative Medicine, University of Maryland School of Medicine: Baltimore, MD, USA, 2006. [Google Scholar]
- Zinchuk, V.; Zinchuk, O.; Okada, T. Quantitative colocalization analysis of multicolor confocal immunofluorescence microscopy images: Pushing pixels to explore biological phenomena. Acta Histochem. Cytochem. 2007, 40, 101–111. [Google Scholar] [CrossRef]

| Species | n° | Sex | Age | Origin | IHC ACE2 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Female | Male | Juvenile | Calf | Adult | Captivity | Wild | − | + | ++ | ||
| T. truncatus | 17 | 9 | 8 | 2 | 6 | 9 | 10 | 7 | 3 | 5 | 9 |
| S. coeruleoalba | 15 | 8 | 7 | 6 | 2 | 7 | 0 | 15 | 8 | 2 | 5 |
| Chi-square Test for ACE-2 by IHC | Regression Analyses for ACE2 by IHC | ||
|---|---|---|---|
| p–value | Odds ratio | 95% Conf. Interval | |
| Species | 0.082 | 0.525 ¹ | 0.091–3.034 |
| Origin | 0.054 | 4.8 ² | 0.385–59.895 |
| Age | 1 | / | / |
| Sex | 0.647 | / | / |
| ID | Species | Sex | Age | Origin | Cause of Death | Infections | Pathology (EE LUNG) | IF | Note | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ACE2 | CD68 | Colocalization (Person’s Coefficient/%) | |||||||||
| 123 | T. truncatus | F | Calf | C | Birth hypoxia with meconium aspiration syndrome (MAS) | Burkholderia cepacia, Aeromonas hydrophila | meconium in 46% of 50 fields observed at 40X; diffuse interstitial lympho-plasma cell infiltrates with edema, hemorrhage and hyperemia; hypertrophic and multifocally hyperplastic bronchial and bronchiolar mucosa; macrophages. | +++ | +++ | 0.8580 98.35% | presence of active macrophages due to infection |
| 133 | T. truncatus | F | Adult | C | ND | ND | large areas of severe mixed interstitial infiltrates (lymphocytes, plasma cells, macrophagesand neutrophilic granulocytes); abundant macrophage exudation with rare neutrophilic granulocytes; both alveolar and bronchial with diffuse tissue edema; diffuse anthracosis. | ++ | +++ | 0.4137 5.19 % | Absence of active macrophages (M1) due to lack of infection |
| 142 | T. truncatus | F | Adult | W | Sepsis resulting from systemic mycosis and Toxoplasma gondii encephalitis | Toxoplasma gondii | in bronchial lumen macrophage and lymphocytic inflammation; severe exudation in some bronchioles of active macrophages and neutrophils; necrosis and fungal hyphae. peripheral vascular structures with edema and fibrinoid necrosis associated with thrombosis with fungal hyphae.; diffuse mild fibrosis of septa with macrophage activation and edema. | + | + | 0.8644 89.52 % | presence of active macrophages due to infection, in this case parasitic. |
| 343 | T. truncatus | F | Calf | C | meconium-induced colic constipation. | ND | areas of emphysema, mainly in the sub-pleural area; large portions of parenchyma characterized by neonatal atelectasis and scarce keratin scales in the alveolar spaces; prominent intravascular pulmonary macrophages; occasional circulating neutrophilic granulocytes (neutrophilic margination); flaking erythrocytes and epithelial cells; exogenous materials are noted. minimal macrophage exudation | + (+++ in epithelial tissue) | +++ | 0.5870 25.01 % | Absence of active macrophages (M1) due to lack of infection |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Audino, T.; Berrone, E.; Grattarola, C.; Giorda, F.; Mattioda, V.; Martelli, W.; Pintore, A.; Terracciano, G.; Cocumelli, C.; Lucifora, G.; et al. Potential SARS-CoV-2 Susceptibility of Cetaceans Stranded along the Italian Coastline. Pathogens 2022, 11, 1096. https://doi.org/10.3390/pathogens11101096
Audino T, Berrone E, Grattarola C, Giorda F, Mattioda V, Martelli W, Pintore A, Terracciano G, Cocumelli C, Lucifora G, et al. Potential SARS-CoV-2 Susceptibility of Cetaceans Stranded along the Italian Coastline. Pathogens. 2022; 11(10):1096. https://doi.org/10.3390/pathogens11101096
Chicago/Turabian StyleAudino, Tania, Elena Berrone, Carla Grattarola, Federica Giorda, Virginia Mattioda, Walter Martelli, Antonio Pintore, Giuliana Terracciano, Cristiano Cocumelli, Giuseppe Lucifora, and et al. 2022. "Potential SARS-CoV-2 Susceptibility of Cetaceans Stranded along the Italian Coastline" Pathogens 11, no. 10: 1096. https://doi.org/10.3390/pathogens11101096
APA StyleAudino, T., Berrone, E., Grattarola, C., Giorda, F., Mattioda, V., Martelli, W., Pintore, A., Terracciano, G., Cocumelli, C., Lucifora, G., Nocera, F. D., Di Francesco, G., Di Renzo, L., Rubini, S., Gavaudan, S., Toffan, A., Puleio, R., Bold, D., Brunelli, F., ... Casalone, C. (2022). Potential SARS-CoV-2 Susceptibility of Cetaceans Stranded along the Italian Coastline. Pathogens, 11(10), 1096. https://doi.org/10.3390/pathogens11101096








