Epstein–Barr Virus Detection in the Central Nervous System of HIV-Infected Patients
Abstract
1. Introduction
2. Results
2.1. Selection of Articles
2.2. Quality Assessment
2.3. Characteristics of Studies Included in the Review
2.4. Participant Characteristics
2.5. Laboratory Diagnosis of EBV
2.6. Empirical Treatments
2.7. Prevalence
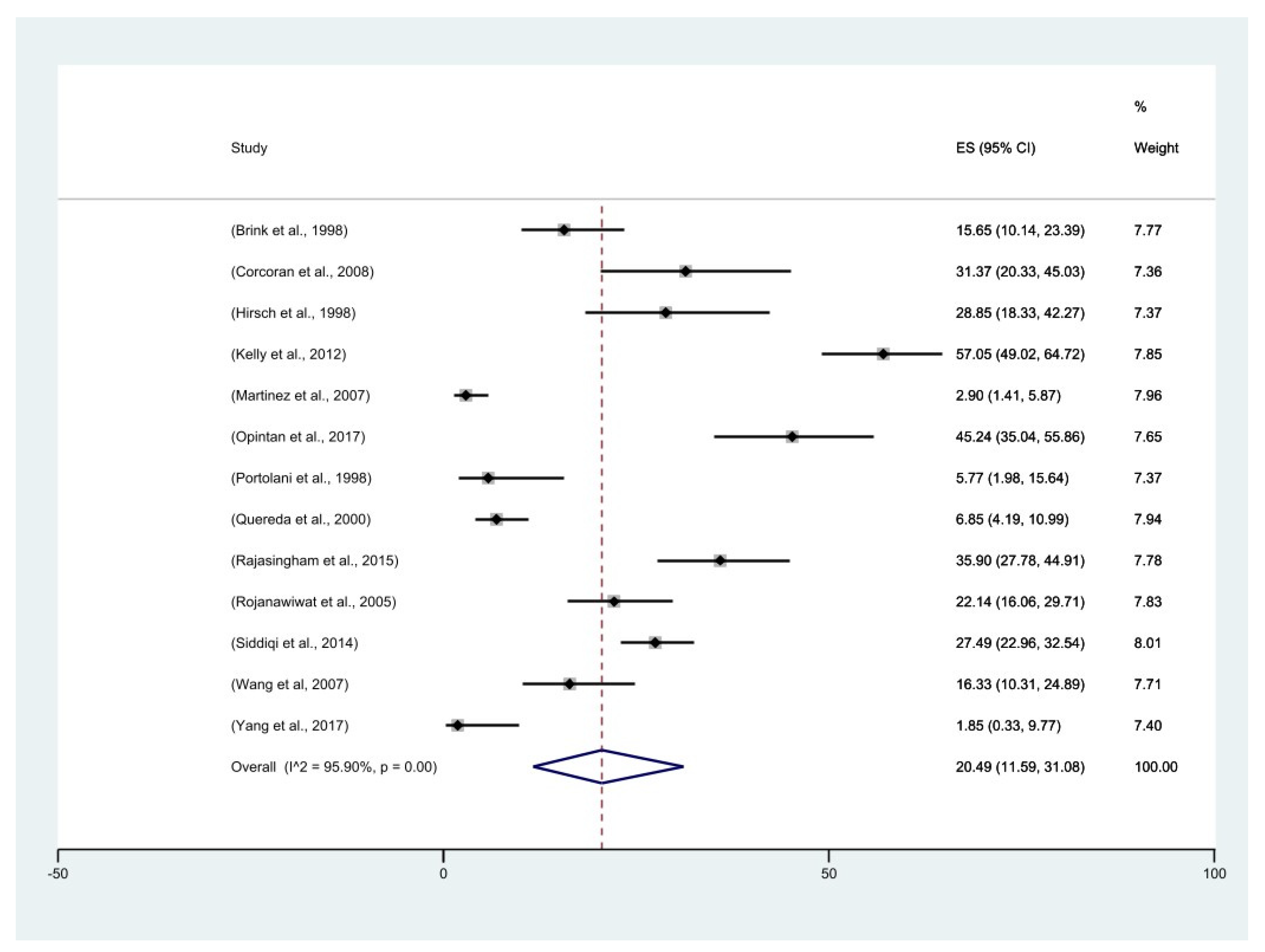
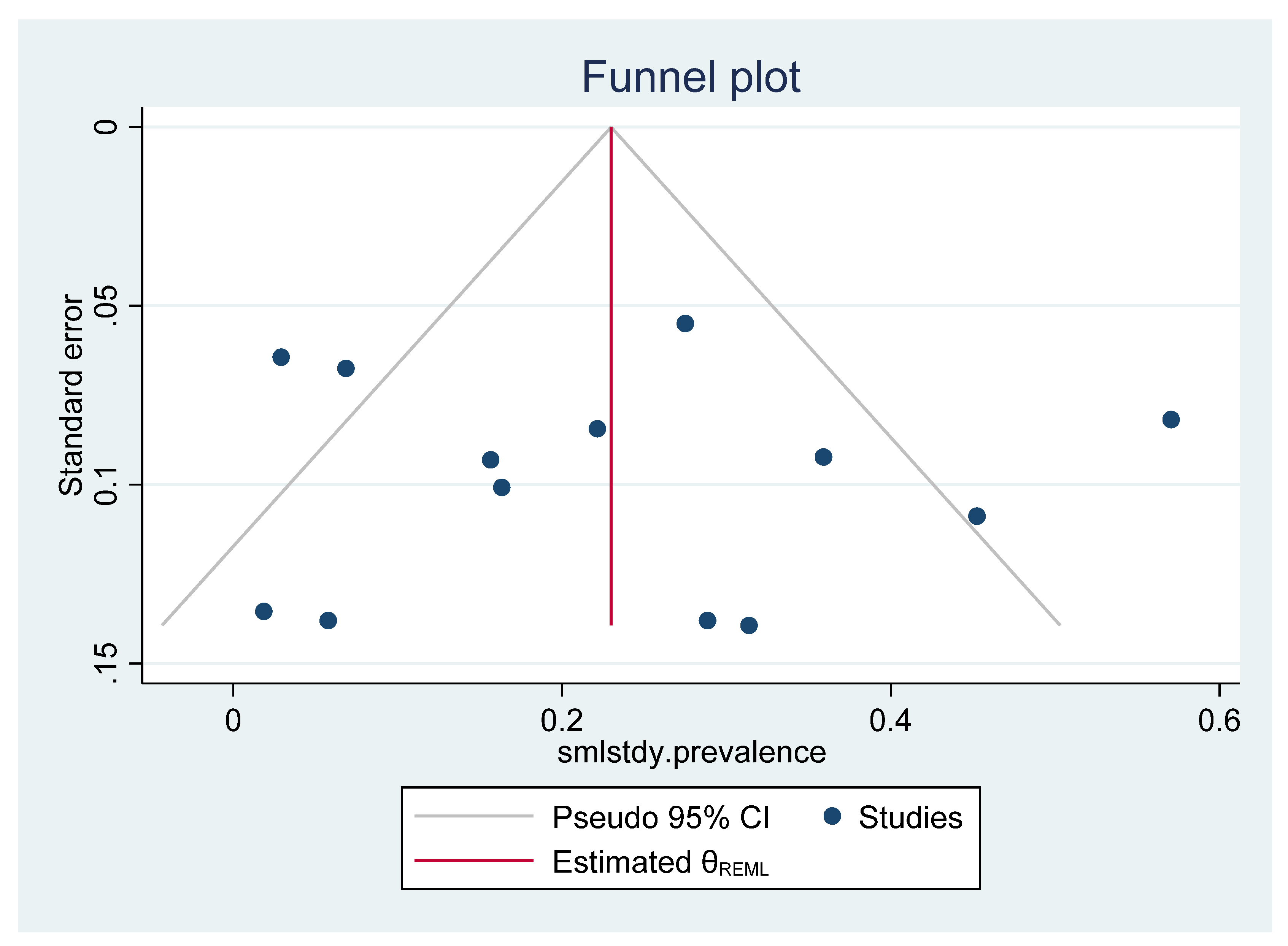
2.8. Meta-Analysis
3. Discussion
4. Materials and Methods
4.1. Literature Search
4.2. Data Management
4.3. Eligibility Criteria
4.4. Selection Process
4.5. Data Collection
4.6. Quality Assessment
4.7. Systematic Review and Meta-Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chiaretti, A.; Capozzi, D.; Mariotti, P.; Valentini, P.; Manni, L.; Buonsenso, D.; Fantacci, C.; Ferrara, P. Increased levels of neurotrophins in the cerebrospinal fluid of children with Epstein-Barr virus meningoencephalitis. Int. J. Infect. Dis. 2014, 20, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Martelius, T.; Lappalainen, M.; Palomaki, M.; Anttila, V.J. Clinical characteristics of patients with Epstein Barr virus in cerebrospinal fluid. BMC Infect. Dis. 2011, 11, 281. [Google Scholar] [CrossRef]
- Gequelin, L.C.; Riediger, I.N.; Nakatani, S.M.; Biondo, A.W.; Bonfim, C.M. Epstein-Barr virus: General factors, virus-related diseases and measurement of viral load after transplant. Rev. Bras. Hematol. Hemoter. 2011, 33, 383–388. [Google Scholar] [CrossRef]
- Zhang, N.; Zuo, Y.; Jiang, L.; Peng, Y.; Huang, X.; Zuo, L. Epstein-Barr Virus and Neurological Diseases. Front. Mol. Biosci. 2022, 8, 816098. [Google Scholar] [CrossRef]
- Slyker, J.A.; Casper, C.; Tapia, K.; Richardson, B.; Bunts, L.; Huang, M.L.; Maleche-Obimbo, E.; Nduati, R.; John-Stewart, G. Clinical and virologic manifestations of primary Epstein-Barr virus (EBV) infection in Kenyan infants born to HIV-infected women. J. Infect. Dis. 2013, 207, 1798–1806. [Google Scholar] [CrossRef]
- Doris, K.; Trevor, K.; Evans, M.; Rabecca, T.; Ephraim, Z.; Chris, C.; Annie, K.; Mulemba, S.; Pascal, P.; Geoffrey, K. Evidence of EBV infection in lymphomas diagnosed in Lusaka, Zambia. PAMJ 2018, 29, 11847. [Google Scholar] [CrossRef]
- Houldcroft, C.J.; Kellam, P. Host genetics of Epstein-Barr virus infection, latency and disease. Rev. Med. Virol. 2015, 25, 71–84. [Google Scholar] [CrossRef]
- Abbott, R.J.; Pachnio, A.; Pedroza-Pacheco, I.; Leese, A.M.; Begum, J.; Long, H.M.; Croom-Carter, D.; Stacey, A.; Moss, P.A.H.; Hislop, A.D.; et al. Asymptomatic Primary Infection with Epstein-Barr Virus: Observations on Young Adult Cases. J. Virol. 2017, 91, e00382-17. [Google Scholar] [CrossRef] [PubMed]
- Chaganti, S.; Heath, E.M.; Bergler, W.; Kuo, M.; Buettner, M.; Niedobitek, G.; Rickinson, A.B.; Bell, A.I. Epstein-Barr virus colonization of tonsillar and peripheral blood B-cell subsets in primary infection and persistence. Blood 2009, 113, 6372–6381. [Google Scholar] [CrossRef]
- Thorley-Lawson, D.A.; Gross, A. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N. Engl. J. Med. 2004, 350, 1328–1337. [Google Scholar] [CrossRef]
- Shindiapina, P.; Ahmed, E.H.; Mozhenkova, A.; Abebe, T.; Baiocchi, R.A. Immunology of EBV-Related Lymphoproliferative Disease in HIV-Positive Individuals. Front. Oncol. 2020, 10, 1723. [Google Scholar] [CrossRef] [PubMed]
- Bjornevik, K.; Cortese, M.; Healy, B.C.; Kuhle, J.; Mina, M.J.; Leng, Y.; Elledge, S.J.; Niebuhr, D.W.; Scher, A.I.; Munger, K.L.; et al. Longitudinal analysis reveals high prevalence of Epstein-Barr virus associated with multiple sclerosis. Science 2022, 375, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Piriou, E.; van Dort, K.; Nanlohy, N.M.; van Oers, M.H.; Miedema, F.; van Baarle, D. Loss of EBNA1-specific memory CD4+ and CD8+ T cells in HIV-infected patients progressing to AIDS-related non-Hodgkin lymphoma. Blood 2005, 106, 3166–3174. [Google Scholar] [CrossRef]
- Dávila-Collado, R.; Jarquín-Durán, O.; Dong, L.T.; Espinoza, J.L. Epstein-Barr Virus and Helicobacter Pylori Co-Infection in Non-Malignant Gastroduodenal Disorders. Pathogens 2020, 9, 104. [Google Scholar] [CrossRef] [PubMed]
- Kelly, M.J.; Benjamin, L.A.; Cartwright, K.; Ajdukiewicz, K.M.; Cohen, D.B.; Menyere, M.; Galbraith, S.; Guiver, M.; Neuhann, F.; Solomon, T.; et al. Epstein-barr virus coinfection in cerebrospinal fluid is associated with increased mortality in Malawian adults with bacterial meningitis. J. Infect. Dis. 2012, 205, 106–110. [Google Scholar] [CrossRef]
- Macedo, C.; Webber, S.A.; Donnenberg, A.D.; Popescu, I.; Hua, Y.; Green, M.; Rowe, D.; Smith, L.; Brooks, M.M.; Metes, D. EBV-specific CD8+ T cells from asymptomatic pediatric thoracic transplant patients carrying chronic high EBV loads display contrasting features: Activated phenotype and exhausted function. J. Immunol. 2011, 186, 5854–5862. [Google Scholar] [CrossRef]
- Njie, R.; Bell, A.I.; Jia, H.; Croom-Carter, D.; Chaganti, S.; Hislop, A.D.; Whittle, H.; Rickinson, A.B. The effects of acute malaria on Epstein-Barr virus (EBV) load and EBV-specific T cell immunity in Gambian children. J. Infect. Dis. 2009, 199, 31–38. [Google Scholar] [CrossRef]
- Hernández, D.M.; Valderrama, S.; Gualtero, S.; Hernández, C.; López, M.; Herrera, M.V.; Solano, J.; Fiorentino, S.; Quijano, S. Loss of T-Cell Multifunctionality and TCR-Vβ Repertoire Against Epstein-Barr Virus Is Associated With Worse Prognosis and Clinical Parameters in HIV(+) Patients. Front. Immunol. 2018, 9, 2291. [Google Scholar] [CrossRef]
- Vella, S.; Schwartländer, B.; Sow, S.P.; Eholie, S.P.; Murphy, R.L. The history of antiretroviral therapy and of its implementation in resource-limited areas of the world. AIDS 2012, 26, 1231–1241. [Google Scholar] [CrossRef]
- Friis, A.M.; Gyllensten, K.; Aleman, A.; Ernberg, I.; Akerlund, B. The Effect of Antiretroviral Combination Treatment on Epstein-Barr Virus (EBV) Genome Load in HIV-Infected Patients. Viruses 2010, 2, 867–879. [Google Scholar] [CrossRef]
- Stevens, S.J.C.; Blank, B.S.N.; Smits, P.H.M.; Meenhorst, P.L.; Middeldorp, J.M. High Epstein–Barr virus (EBV) DNA loads in HIV-infected patients: Correlation with antiretroviral therapy and quantitative EBV serology. AIDS 2002, 16, 993–1001. [Google Scholar] [CrossRef]
- Prado, J.G.; Frater, J. Editorial: Immune Surveillance of the HIV Reservoir: Mechanisms, Therapeutic Targeting and New Avenues for HIV Cure. Front. Immunol. 2020, 11, 70. [Google Scholar] [CrossRef]
- Meyding-Lamadé, U.; Strank, C. Herpesvirus infections of the central nervous system in immunocompromised patients. Ther. Adv. Neurol. Disord. 2012, 5, 279–296. [Google Scholar] [CrossRef]
- Kleines, M.; Scheithauer, S.; Schiefer, J.; Hausler, M. Clinical application of viral cerebrospinal fluid PCR testing for diagnosis of central nervous system disorders: A retrospective 11-year experience. Diagn. Microbiol. Infect. Dis. 2014, 80, 207–215. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, J.; Wen, Y. Lessons from Epstein-Barr virus DNA detection in cerebrospinal fluid as a diagnostic tool for EBV-induced central nervous system dysfunction among HIV-positive patients. Biomed. Pharm. 2022, 145, 112392. [Google Scholar] [CrossRef]
- Benjamin, L.; Kelly, M.; Cohen, D.; Neuhann, F.; Galbraith, S.; Mallewa, M.; Hopkins, M.; Hart, I.; Guiver, M.; Lalloo, D. Detection of herpes viruses in the cerebrospinal fluid of adults with suspected viral meningitis in Malawi. Infection 2013, 41, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Kerr, J.R. Epstein-Barr virus (EBV) reactivation and therapeutic inhibitors. J. Clin. Pathol. 2019, 72, 651. [Google Scholar] [CrossRef]
- Pagano, J.S.; Whitehurst, C.B.; Andrei, G. Antiviral Drugs for EBV. Cancers 2018, 10, 197. [Google Scholar] [CrossRef]
- Brooks, J.M.; Long, H.M.; Tierney, R.J.; Shannon-Lowe, C.; Leese, A.M.; Fitzpatrick, M.; Taylor, G.S.; Rickinson, A.B. Early T Cell Recognition of B Cells following Epstein-Barr Virus Infection: Identifying Potential Targets for Prophylactic Vaccination. PLoS Pathog. 2016, 12, e1005549. [Google Scholar] [CrossRef] [PubMed]
- Ivers, L.C.; Kim, A.Y.; Sax, P.E. Predictive value of polymerase chain reaction of cerebrospinal fluid for detection of Epstein-Barr virus to establish the diagnosis of HIV-related primary central nervous system lymphoma. Clin. Infect Dis. 2004, 38, 1629–1632. [Google Scholar] [CrossRef]
- Tselis, A.; Duman, R.; Storch, G.A.; Lisak, R.P. Epstein-Barr virus encephalomyelitis diagnosed by polymerase chain reaction: Detection of the genome in the CSF. Neurology 1997, 48, 1351–1355. [Google Scholar] [CrossRef]
- He, T.; Kaplan, S.; Kamboj, M.; Tang, Y.W. Laboratory Diagnosis of Central Nervous System Infection. Curr. Infect. Dis. Rep. 2016, 18, 35. [Google Scholar] [CrossRef]
- Cinque, P.; Vago, L.; Dahl, H.; Brytting, M.; Terreni, M.R.; Fornara, C.; Racca, S.; Castagna, A.; Monforte, A.D.; Wahren, B.; et al. Polymerase chain reaction on cerebrospinal fluid for diagnosis of virus-associated opportunistic diseases of the central nervous system in HIV-infected patients. AIDS 1996, 10, 951–958. [Google Scholar] [CrossRef]
- Roddie, C.; Peggs, K.S. Immunotherapy for transplantation-associated viral infections. J. Clin. Investig. 2017, 127, 2513–2522. [Google Scholar] [CrossRef]
- Portolani, M.; Pietrosemoli, P.; Meacci, M.; Sabbatini, A.M.; Pecorari, M.; Mantovani, G.; Cermelli, C. Detection of Epstein-Barr virus DNA in cerebrospinal fluid from immunocompetent individuals with brain disorders. New Microbiol. 1998, 21, 77–79. [Google Scholar]
- Opintan, J.A.; Awadzi, B.K.; Biney, I.J.K.; Ganu, V.; Doe, R.; Kenu, E.; Adu, R.F.; Osei, M.M.; Akumwena, A.; Grigg, M.E.; et al. High rates of cerebral toxoplasmosis in HIV patients presenting with meningitis in Accra, Ghana. Trans. R. Soc. Trop. Med. Hyg. 2017, 111, 464–471. [Google Scholar] [CrossRef]
- Rajasingham, R.; Rhein, J.; Klammer, K.; Musubire, A.; Nabeta, H.; Akampurira, A.; Mossel, E.C.; Williams, D.A.; Boxrud, D.J.; Crabtree, M.B.; et al. Epidemiology of meningitis in an HIV-infected Ugandan cohort. Am. J. Trop. Med. Hyg. 2015, 92, 274–279. [Google Scholar] [CrossRef]
- Yang, R.; Zhang, H.; Xiong, Y.; Gui, X.; Zhang, Y.; Deng, L.; Gao, S.; Luo, M.; Hou, W.; Guo, D. Molecular diagnosis of central nervous system opportunistic infections and mortality in HIV-infected adults in Central China. AIDS Res. 2017, 14, 24. [Google Scholar] [CrossRef]
- Hirsch, H.H.; Meylan, P.R.; Zimmerli, W.; Iten, A.; Battegay, M.; Erb, P.; Swiss HIV Cohort Study. HIV-1-infected patients with focal neurologic signs: Diagnostic role of PCR for Toxoplasma gondii, Epstein-Barr virus, and JC virus. Clin. Microbiol. Infect. 1998, 4, 577–584. [Google Scholar] [CrossRef]
- Brink, N.S.; Sharvell, Y.; Howard, M.R.; Fox, J.D.; Harrison, M.J.; Miller, R.F. Detection of Epstein-Barr virus and Kaposi’s sarcoma-associated herpesvirus DNA in CSF from persons infected with HIV who had neurological disease. J. Neurol. Neurosurg. Psychiatry 1998, 65, 191–195. [Google Scholar] [CrossRef][Green Version]
- Wang, J.; Ozzard, A.; Nathan, M.; Atkins, M.; Nelson, M.; Gazzard, B.; Bower, M. The significance of Epstein–Barr virus detected in the cerebrospinal fluid of people with HIV infection. HIV Med. 2007, 8, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Quereda, C.; Corral, I.; Laguna, F.; Valencia, M.E.; Tenorio, A.; Echeverria, J.E.; Navas, E.; Martin-Davila, P.; Moreno, A.; Moreno, V.; et al. Diagnostic utility of a multiplex herpesvirus PCR assay performed with cerebrospinal fluid from human immunodeficiency virus-infected patients with neurological disorders. J. Clin. Microbiol. 2000, 38, 3061–3067. [Google Scholar] [CrossRef]
- Rojanawiwat, A.; Miura, T.; Thaisri, H.; Pathipvanich, P.; Umnajsirisuk, S.; Koibuchi, T.; Vongsheree, S.; Iwamoto, A.; Ariyoshi, K.; Sawanpanyalert, P. Frequent detection of Epstein-Barr Virus and cytomegalovirus but not JC virus DNA in cerebrospinal fluid samples from human immunodeficiency virus-infected patients in northern Thailand. J. Clin. Microbiol. 2005, 43, 3484–3486. [Google Scholar] [CrossRef]
- Martínez, P.A.; Díaz, R.; González, D.; Oropesa, L.; González, R.; Pérez, L.; Viera, J.; Kourí, V. The effect of highly active antiretroviral therapy on outcome of central nervous system herpesviruses infection in Cuban human immunodeficiency virus-infected individuals. J. NeuroVirol. 2007, 13, 446–451. [Google Scholar] [CrossRef]
- Corcoran, C.; Rebe, K.; van der Plas, H.; Myer, L.; Hardie, D.R. The predictive value of cerebrospinal fluid Epstein-Barr viral load as a marker of primary central nervous system lymphoma in HIV-infected persons. J. Clin. Virol. 2008, 42, 433–436. [Google Scholar] [CrossRef]
- Siddiqi, O.K.; Ghebremichael, M.; Dang, X.; Atadzhanov, M.; Kaonga, P.; Khoury, M.N.; Koralnik, I.J. Molecular diagnosis of central nervous system opportunistic infections in HIV-infected Zambian adults. Clin. Infect. Dis. 2014, 58, 1771–1777. [Google Scholar] [CrossRef]
- Kwok, H.; Chan, K.W.; Chan, K.H.; Chiang, A.K. Distribution, persistence and interchange of Epstein-Barr virus strains among PBMC, plasma and saliva of primary infection subjects. PLoS ONE 2015, 10, e0120710. [Google Scholar] [CrossRef][Green Version]
- Zanella, L.; Riquelme, I.; Buchegger, K.; Abanto, M.; Ili, C.; Brebi, P. A reliable Epstein-Barr Virus classification based on phylogenomic and population analyses. Sci. Rep. 2019, 9, 9829. [Google Scholar] [CrossRef]
- Smatti, M.K.; Al-Sadeq, D.W.; Ali, N.H.; Pintus, G.; Abou-Saleh, H.; Nasrallah, G.K. Epstein-Barr Virus Epidemiology, Serology, and Genetic Variability of LMP-1 Oncogene Among Healthy Population: An Update. Front. Oncol. 2018, 8, 211. [Google Scholar] [CrossRef] [PubMed]
- Peh, S.C.; Kim, L.H.; Poppema, S. Frequent presence of subtype A virus in Epstein-Barr virus-associated malignancies. Pathology 2002, 34, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Aitken, C.; Sengupta, S.K.; Aedes, C.; Moss, D.J.; Sculley, T.B. Heterogeneity within the Epstein-Barr virus nuclear antigen 2 gene in different strains of Epstein-Barr virus. J. Gen. Virol. 1994, 75 Pt 1, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Ayee, R.; Ofori, M.E.O.; Tagoe, E.A.; Languon, S.; Searyoh, K.; Armooh, L.; Bilson-Amoah, E.; Baidoo, K.; Kitcher, E.; Wright, E.; et al. Genotypic Characterization of Epstein Barr Virus in Blood of Patients with Suspected Nasopharyngeal Carcinoma in Ghana. Viruses 2020, 12, 766. [Google Scholar] [CrossRef]
- Neves, M.; Marinho-Dias, J.; Ribeiro, J.; Sousa, H. Epstein-Barr virus strains and variations: Geographic or disease-specific variants? J. Med. Virol. 2017, 89, 373–387. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Hamid, M.; Chen, J.J.; Constantine, N.; Massoud, M.; Raab-Traub, N. EBV strain variation: Geographical distribution and relation to disease state. Virology 1992, 190, 168–175. [Google Scholar] [CrossRef]
- Petrara, M.R.; Penazzato, M.; Massavon, W.; Nabachwa, S.; Nannyonga, M.; Mazza, A.; Gianesin, K.; Del Bianco, P.; Lundin, R.; Sumpter, C.; et al. Epstein-Barr virus load in children infected with human immunodeficiency virus type 1 in Uganda. J. Infect. Dis. 2014, 210, 392–399. [Google Scholar] [CrossRef]
- Bouvard, V.; Baan, R.; Straif, K.; Grosse, Y.; Secretan, B.; El Ghissassi, F.; Benbrahim-Tallaa, L.; Guha, N.; Freeman, C.; Galichet, L.; et al. A review of human carcinogens--Part B: Biological agents. Lancet Oncol. 2009, 10, 321–322. [Google Scholar] [CrossRef]
- Wong, Y.; Meehan, M.T.; Burrows, S.R.; Doolan, D.L.; Miles, J.J. Estimating the global burden of Epstein–Barr virus-related cancers. J. Cancer Res. Clin. Oncol. 2022, 148, 31–46. [Google Scholar] [CrossRef]
- Bossolasco, S.; Cinque, P.; Ponzoni, M.; Vigano, M.G.; Lazzarin, A.; Linde, A.; Falk, K.I. Epstein-Barr virus DNA load in cerebrospinal fluid and plasma of patients with AIDS-related lymphoma. J. Neuro Virol. 2002, 8, 432–438. [Google Scholar] [CrossRef]
- Gaeta, A.; Verzaro, S.; Cristina, L.M.; Mancini, C.; Nazzari, C. Diagnosis of neurological herpesvirus infections: Real time PCR in cerebral spinal fluid analysis. New Microbiol. 2009, 32, 333–340. [Google Scholar]
- Ryan, J.L.; Fan, H.; Glaser, S.L.; Schichman, S.A.; Raab-Traub, N.; Gulley, M.L. Epstein-Barr virus quantitation by real-time PCR targeting multiple gene segments: A novel approach to screen for the virus in paraffin-embedded tissue and plasma. J. Mol. Diagn. 2004, 6, 378–385. [Google Scholar] [CrossRef]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan-a web and mobile app for systematic reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef]
- Higgins, J.P.; Thompson, S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002, 21, 1539–1558. [Google Scholar] [CrossRef]


| Author | Was the Study on the Detection of Epstein–Barr Virus DNA in the CSF? | Was the Research Problem Clearly Described? | Was the Hypothesis/Aim/Objective of the Study Clearly Described? | Was the Study Design Clearly Reported? | Was the Study Population Clearly Described? | Were the Study Population Inclusion and Exclusion Criteria Clearly Stated? | Was the EBV Screening Procedure Well Described? | Was the EBV Target Gene Clearly Stated? | Was the Sample Size Cleared Stated? | Were the Characteristics of the Patients Included in the Study Clearly Described? | Were The Statistical Tests Used To Assess The Primary Outcomes Well Described? | Were the Main Outcomes to be Measured Clearly Described in the Methods Section? | Were the Main Findings of the Study Clearly Described? | Were the Main Findings Linked to the Study Aims or Objectives? | Were the Study Results Appropriately Interpreted, e.g., in Terms of the Strength of the Evidence, Its Application/Implications, and Causality? | Was a Summary of the Key Outcomes Clearly Stated? | Were the Findings Compared to Other Relevant Studies? | Total | % Score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Brink et al., 1998 | Yes | Yes | Yes | Yes | Yes | P * | Yes | Yes | Yes | P * | P * | Yes | Yes | Yes | Yes | Yes | Yes | 14 | 82 |
| Corcoran et al., 2008 | Yes | Yes | Yes | Yes | Yes | No | No | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | 14 | 82 |
| Hirsh et al., 1998 | Yes | Yes | Yes | Yes | Yes | P * | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | 15 | 88 |
| Kelly et al., 2012 | Yes | Yes | Yes | Yes | Yes | P * | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 16 | 94 |
| Martinez et al., 2007 | Yes | Yes | Yes | Yes | Yes | P * | Yes | ! | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | 14 | 82 |
| Opintan et al., 2017 | Yes | Yes | Yes | Yes | Yes | Yes | P * | ! | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 15 | 88 |
| Portolani et al., 1998 | Yes | P * | Yes | No | No | No | Yes | Yes | Yes | P * | No | Yes | Yes | Yes | Yes | Yes | Yes | 11 | 65 |
| Quereda et al., 2000 | Yes | Yes | Yes | Yes | Yes | P * | Yes | ! | Yes | Yes | P * | Yes | Yes | Yes | Yes | Yes | Yes | 14 | 82 |
| Rajasingham et al., 2015 | Yes | Yes | Yes | Yes | Yes | Yes | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 15 | 88 |
| Rojanawiwat et al., 2005 | Yes | Yes | Yes | P* | Yes | P * | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes | Yes | No | Yes | 13 | 76 |
| Siddiqi et al., 2014 | Yes | Yes | Yes | Yes | Yes | P * | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 16 | 94 |
| Wang et al., 2007 | Yes | Yes | Yes | Yes | Yes | P * | P * | Yes | Yes | Yes | P * | Yes | Yes | Yes | Yes | Yes | Yes | 14 | 82 |
| Yang et al., 2017 | Yes | Yes | P * | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 16 | 94 |
| Author, Year [Ref] | Country | Study Design | Sample Size | EBV-Positive via Qualitative PCR % (n/n) | EBV-Positive via Quantitative PCR % (n/n) | Conclusions Drawn from Study | |
|---|---|---|---|---|---|---|---|
| Europe | |||||||
| 1 | Hirsh H et al. (1998) [39] | Switzerland | Prospective study and retrospective study | 53 | 29 (15/52) | N/A |
|
| 2 | Brink et al. (1998) [40] | United Kingdom | Prospective study | 115 | N/A | 16 (18/115) |
|
| 3 | Wang et al. (2007) [41] | United Kingdom | Retrospective analysis study | 98 | 16 (16/98) | N/A |
|
| 4 | Portolani et al. (1998) [35] | Italy | Retrospective analysis study | 52 | 6 (3/52) | N/A |
|
| 5 | Quereda et al. (2000) [42] | Spain | Retrospective analysis study | 219 | 7 (15/219) | N/A |
|
| Asia | |||||||
| 6 | Rojanawiwat et al. (2005) [43] | Thailand | Prospective study | 140 | 22 (31/140) | N/A |
|
| 7 | Yang et al. (2017) [38] | China | Hospital-based | 54 | 2 (1/54) | N/A |
|
| South America | |||||||
| 8 | Martinez et al. (2007) [44] | Cuba | Cross-sectional | 241 | 3 (7/241) | N/A |
|
| Africa | |||||||
| 9 | (Corcoran et al. (2008) [45] | South Africa | Retrospective study | 55 | N/A | 36 (20/55) |
|
| 10 | Opintan et al. (2017) [36] | Ghana | Prospective cohort study | 84 | 45 (38/84) | N/A |
|
| 11 | Rajasingham et al. (2015) [37] | Uganda | Prospective cohort study | 314 | 36 (42/117) | N/A |
|
| 12 | Kelly et al. (2011) [15] | Malawi | Retrospective analysis study | 188 | N/A | 45 (85/188) |
|
| 13 | Siddiqi et al. (2014) [46] | Zambia | Cross-sectional | 331 | 27 (91/331) | N/A |
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Musukuma-Chifulo, K.; Siddiqi, O.K.; Chilyabanyama, O.N.; Bates, M.; Chisenga, C.C.; Simuyandi, M.; Sinkala, E.; Dang, X.; Koralnik, I.J.; Chilengi, R.; et al. Epstein–Barr Virus Detection in the Central Nervous System of HIV-Infected Patients. Pathogens 2022, 11, 1080. https://doi.org/10.3390/pathogens11101080
Musukuma-Chifulo K, Siddiqi OK, Chilyabanyama ON, Bates M, Chisenga CC, Simuyandi M, Sinkala E, Dang X, Koralnik IJ, Chilengi R, et al. Epstein–Barr Virus Detection in the Central Nervous System of HIV-Infected Patients. Pathogens. 2022; 11(10):1080. https://doi.org/10.3390/pathogens11101080
Chicago/Turabian StyleMusukuma-Chifulo, Kalo, Omar Khalik Siddiqi, Obvious Nchimunya Chilyabanyama, Matthew Bates, Caroline Cleopatra Chisenga, Michelo Simuyandi, Edford Sinkala, Xin Dang, Igor Jerome Koralnik, Roma Chilengi, and et al. 2022. "Epstein–Barr Virus Detection in the Central Nervous System of HIV-Infected Patients" Pathogens 11, no. 10: 1080. https://doi.org/10.3390/pathogens11101080
APA StyleMusukuma-Chifulo, K., Siddiqi, O. K., Chilyabanyama, O. N., Bates, M., Chisenga, C. C., Simuyandi, M., Sinkala, E., Dang, X., Koralnik, I. J., Chilengi, R., & Munsaka, S. (2022). Epstein–Barr Virus Detection in the Central Nervous System of HIV-Infected Patients. Pathogens, 11(10), 1080. https://doi.org/10.3390/pathogens11101080









