Rotavirus Infection in Swine: Genotypic Diversity, Immune Responses, and Role of Gut Microbiome in Rotavirus Immunity
Abstract
1. Rotavirus Genome, Classification and Host Range
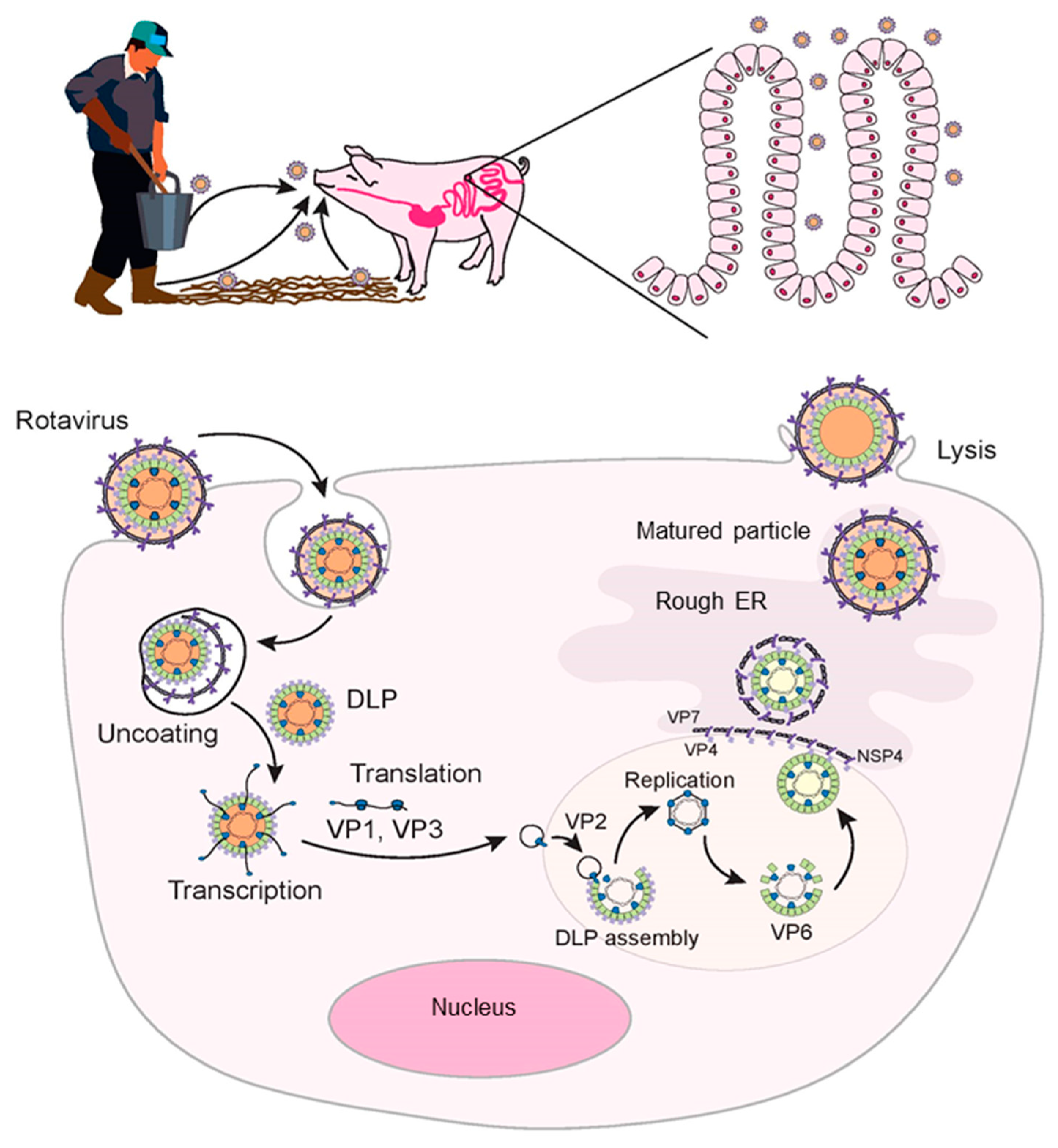
2. Rotavirus Entry and Replication
3. Distribution and Genotypic Diversity of Porcine Rotaviruses
4. Immune Responses to Rotavirus Infection
4.1. Innate Immune Response
4.1.1. Role of RIG-I-like Receptors
4.1.2. Role of Toll-like Receptors (TLRs)
4.1.3. Other Mediators of Innate Immune Response
4.2. Adaptive Immune Response
5. Maternal Immunity and Protection of Piglets
6. Gut Microbiome and Rotavirus Immunity
6.1. Composition of Swine Gut Microbiome
6.2. Evidence from Human Rotavirus Studies
6.3. Role of Probiotics and Vitamin A in Immunity against Rotaviruses
6.4. Gut Microbiome Modulation and Response to Rotavirus Infection
7. Overall Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Desselberger, U. Rotaviruses. Virus Res. 2014, 190, 75–96. [Google Scholar] [CrossRef] [PubMed]
- Shaw, R.D.; Vo, P.T.; Offit, P.A.; Coulsont, B.S.; Greenberg, H.B. Antigenic Mapping of the Surface Proteins of Rhesus Rotavirus. Virology 1986, 155, 434–451. [Google Scholar] [CrossRef]
- Ludert, J.E.; Ruiz, M.C.; Hidalgo, C.; Liprandi, F. Antibodies to Rotavirus Outer Capsid Glycoprotein VP7 Neutralize Infectivity by Inhibiting Virion Decapsidation. J. Virol. 2002, 76, 6643–6651. [Google Scholar] [CrossRef]
- Ramani, S.; Hu, L.; Venkataram Prasad, B.V.; Estes, M.K. Diversity in Rotavirus–Host Glycan Interactions: A “Sweet” Spectrum. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Nejmeddine, M.; Trugnan, G.; Sapin, C.; Kohli, E.; Svensson, L.; Lopez, S.; Cohen, J. Rotavirus Spike Protein VP4 Is Present at the Plasma Membrane and Is Associated with Microtubules in Infected Cells. J. Virol. 2000, 74, 3313–3320. [Google Scholar] [CrossRef]
- Lorrot, M.; Vasseur, M. How Do the Rotavirus NSP4 and Bacterial Enterotoxins Lead Differently to Diarrhea? Virol. J. 2007, 4, 31. [Google Scholar] [CrossRef]
- Matthijnssens, J.; Ciarlet, M.; Rahman, M.; Attoui, H.; Bányai, K.; Estes, M.K.; Gentsch, J.R.; Iturriza-Gómara, M.; Kirkwood, C.D.; Martella, V.; et al. Recommendations for the Classification of Group A Rotaviruses Using All 11 Genomic RNA Segments. Arch. Virol. 2008, 153, 1621–1629. [Google Scholar] [CrossRef]
- Matthijnssens, J.; Otto, P.H.; Ciarlet, M.; Desselberger, U.; van Ranst, M.; Johne, R. VP6-Sequence-Based Cutoff Values as a Criterion for Rotavirus Species Demarcation. Arch. Virol. 2012, 157, 1177–1182. [Google Scholar] [CrossRef] [PubMed]
- Vlasova, A.; Amimo, J.; Saif, L. Porcine Rotaviruses: Epidemiology, Immune Responses and Control Strategies. Viruses 2017, 9, 48. [Google Scholar] [CrossRef] [PubMed]
- Alekseev, K.; Penin, A.; Mukhin, A.; Khametova, K.; Grebennikova, T.; Yuzhakov, A.; Moskvina, A.; Musienko, M.; Raev, S.; Mishin, A.; et al. Genome Characterization of a Pathogenic Porcine Rotavirus B Strain Identified in Buryat Republic, Russia in 2015. Pathogens 2018, 7, 46. [Google Scholar] [CrossRef] [PubMed]
- Chasey, D.; Bridger, J.C.; McCrae, M.A. A New Type of Atypical Rotavirus in Pigs. Arch. Virol. 1986, 89, 235–243. [Google Scholar] [CrossRef]
- Homwong, N.; Diaz, A.; Rossow, S.; Ciarlet, M.; Marthaler, D. Three-Level Mixed-Effects Logistic Regression Analysis Reveals Complex Epidemiology of Swine Rotaviruses in Diagnostic Samples from North America. PLoS ONE 2016, 11, e0154734. [Google Scholar] [CrossRef] [PubMed]
- Marthaler, D.; Rossow, K.; Culhane, M.; Goyal, S.; Collins, J.; Matthijnssens, J.; Nelson, M.; Ciarlet, M. Widespread Rotavirus H in Commercially Raised Pigs, United States. Emerg. Infect. Dis. 2014, 20, 1203–1206. [Google Scholar] [CrossRef] [PubMed]
- Giri, S.; Nair, N.P.; Mathew, A.; Manohar, B.; Simon, A.; Singh, T.; Suresh Kumar, S.; Mathew, M.A.; Babji, S.; Arora, R.; et al. Rotavirus Gastroenteritis in Indian Children < 5 Years Hospitalized for Diarrhoea, 2012 to 2016. BMC Public Health 2019, 19, 69. [Google Scholar] [CrossRef]
- Sadiq, A.; Bostan, N.; Bokhari, H.; Matthijnssens, J.; Yinda, K.C.; Raza, S.; Nawaz, T. Molecular Characterization of Human Group A Rotavirus Genotypes Circulating in Rawalpindi, Islamabad, Pakistan during 2015-2016. PLoS ONE 2019, 14, e0220387. [Google Scholar] [CrossRef]
- Basera, S.S.; Singh, R.; Vaid, N.; Sharma, K.; Chakravarti, S.; Malik, Y.P.S. Detection of Rotavirus Infection in Bovine Calves by RNA-PAGE and RT-PCR. Indian J. Virol. 2010, 21, 144–147. [Google Scholar] [CrossRef][Green Version]
- Kaminjolo, J.S.; Adesiyun, A.A. Rotavirus Infection in Calves, Piglets, Lambs and Goat Kids in Trinidad. Br. Vet. J. 1994, 150, 293–299. [Google Scholar] [CrossRef]
- Legrottaglie, R.; Volpe, A.; Rizzi, V.; Agrimi, P. Isolation and Identification of Rotaviruses as Aetiological Agents of Neonatal Diarrhoea in Kids. Electrophoretical Characterization by PAGE. New Microbiol. 1993, 16, 227–235. [Google Scholar]
- Abe, M.; Yamasaki, A.; Ito, N.; Mizoguchi, T.; Asano, M.; Okano, T.; Sugiyama, M. Molecular Characterization of Rotaviruses in a Japanese Raccoon Dog (Nyctereutes Procyonoides) and a Masked Palm Civet (Paguma Larvata) in Japan. Vet. Microbiol. 2010, 146, 253–259. [Google Scholar] [CrossRef]
- Silva, L.C.; Sanches, A.A.; Gregori, F.; Brandão, P.E.; Alfieri, A.A.; Headley, S.A.; Jerez, J.A. First Description of Group A Rotavirus from Fecal Samples of Ostriches (Struthio Camelus). Res. Vet. Sci. 2012, 93, 1066–1069. [Google Scholar] [CrossRef] [PubMed]
- Pauly, M.; Oni, O.O.; Sausy, A.; Owoade, A.A.; Adeyefa, C.A.O.; Muller, C.P.; Hübschen, J.M.; Snoeck, C.J. Molecular Epidemiology of Avian Rotaviruses Group A and D Shed by Different Bird Species in Nigeria. Virol. J. 2017, 14, 111. [Google Scholar] [CrossRef]
- ter Veen, C.; de Bruijn, N.D.; Dijkman, R.; de Wit, J.J. Prevalence of Histopathological Intestinal Lesions and Enteric Pathogens in Dutch Commercial Broilers with Time. Avian Pathol. 2017, 46, 95–105. [Google Scholar] [CrossRef]
- Ortega, A.F.; Martínez-Castañeda, J.S.; Bautista-Gómez, L.G.; Muñoz, R.F.; Hernández, I.Q. Identification of Co-Infection by Rotavirus and Parvovirus in Dogs with Gastroenteritis in Mexico. Braz. J. Microbiol. 2017, 48, 769–773. [Google Scholar] [CrossRef]
- Nemoto, M.; Ryan, E.; Lyons, P.; Cullinane, A. Molecular Characterisation of Equine Group A Rotaviruses in Ireland (2011–2015). Vet. J. 2017, 226, 12–14. [Google Scholar] [CrossRef]
- Nemoto, M.; Niwa, H.; Murakami, S.; Miki, R.; Higuchi, T.; Bannai, H.; Tsujimura, K.; Kokado, H. Molecular Analyses of G3A/G3B and G14 Equine Group A Rotaviruses Detected between 2012 and 2018 in Japan. J. Gen. Virol. 2019, 100, 913–931. [Google Scholar] [CrossRef]
- Marthaler, D.; Rossow, K.; Gramer, M.; Collins, J.; Goyal, S.; Tsunemitsu, H.; Kuga, K.; Suzuki, T.; Ciarlet, M.; Matthijnssens, J. Detection of Substantial Porcine Group B Rotavirus Genetic Diversity in the United States, Resulting in a Modified Classification Proposal for G Genotypes. Virology 2012, 433, 85–96. [Google Scholar] [CrossRef]
- Medici, K.; Barry, A.; Alfieri, A.; Alfieri, A. Porcine Rotavirus Groups A, B, and C Identified by Polymerase Chain Reaction in a Fecal Sample Collection with Inconclusive Results by Polyacrylamide Gel Electrophoresis. J. Swine Health Prod. 2011, 19, 146–150. [Google Scholar]
- Barman, P.; Ghosh, S.; Das, S.; Varghese, V.; Chaudhuri, S.; Sarkar, S.; Krishnan, T.; Bhattacharya, S.K.; Chakrabarti, A.; Kobayashi, N.; et al. Sequencing and Sequence Analysis of VP7 and NSP5 Genes Reveal Emergence of a New Genotype of Bovine Group B Rotaviruses in India. J. Clin. Microbiol. 2004, 42, 2816–2818. [Google Scholar] [CrossRef]
- Chang, K.O.; Parwani, A.V.; Smith, D.; Saif, L.J. Detection of Group B Rotaviruses in Fecal Samples from Diarrheic Calves and Adult Cows and Characterization of Their VP7 Genes. J. Clin. Microbiol. 1997, 35, 2107–2110. [Google Scholar] [CrossRef]
- Alam, M.M.; Pun, S.B.; Gauchan, P.; Yokoo, M.; Doan, Y.H.; Tran, T.N.H.; Nakagomi, T.; Nakagomi, O.; Pandey, B.D. The First Identification of Rotavirus B from Children and Adults with Acute Diarrhoea in Kathmandu, Nepal. Trop. Med. Health 2013, 41, 129–134. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sanekata, T.; Ahmed, M.U.; Kader, A.; Taniguchi, K.; Kobayashi, N. Human Group B Rotavirus Infections Cause Severe Diarrhea in Children and Adults in Bangladesh. J. Clin. Microbiol. 2003, 41, 2187–2190. [Google Scholar] [CrossRef]
- Theil, K.W.; Saif, L.J.; Moorhead, P.D.; Whitmoyer, R.E. Porcine Rotavirus-like Virus (Group B Rotavirus): Characterization and Pathogenicity for Gnotobiotic Pigs. J. Clin. Microbiol. 1985, 21, 340–345. [Google Scholar] [CrossRef]
- Kattoor, J.J.; Saurabh, S.; Malik, Y.S.; Sircar, S.; Dhama, K.; Ghosh, S.; Bányai, K.; Kobayashi, N.; Singh, R.K. Unexpected Detection of Porcine Rotavirus C Strains Carrying Human Origin VP6 Gene. Vet. Q. 2017, 37, 252–261. [Google Scholar] [CrossRef]
- Marthaler, D.; Homwong, N.; Rossow, K.; Culhane, M.; Goyal, S.; Collins, J.; Matthijnssens, J.; Ciarlet, M. Rapid Detection and High Occurrence of Porcine Rotavirus A, B, and C by RT-QPCR in Diagnostic Samples. J. Virol. Methods. 2014, 209, 30–34. [Google Scholar] [CrossRef]
- Bhat, S.; Kattoor, J.J.; Malik, Y.S.; Sircar, S.; Deol, P.; Rawat, V.; Rakholia, R.; Ghosh, S.; Vlasova, A.N.; Nadia, T.; et al. Species C Rotaviruses in Children with Diarrhea in India, 2010–2013: A Potentially Neglected Cause of Acute Gastroenteritis. Pathogens 2018, 7, 23. [Google Scholar] [CrossRef]
- Kumazaki, M.; Usuku, S. Epidemiological and Genetic Analysis of Human Group C Rotaviruses Isolated from Outbreaks of Acute Gastroenteritis in Yokohama, Japan, between 2006 and 2012. Arch. Virol. 2014, 159, 761–771. [Google Scholar] [CrossRef]
- Tiku, V.R.; Jiang, B.; Kumar, P.; Aneja, S.; Bagga, A.; Bhan, M.K.; Ray, P. First Study Conducted in Northern India That Identifies Group C Rotavirus as the Etiological Agent of Severe Diarrhea in Children in Delhi. Virol. J. 2017, 14, 100. [Google Scholar] [CrossRef]
- Soma, J.; Tsunemitsu, H.; Miyamoto, T.; Suzuki, G.; Sasaki, T.; Suzuki, T. Whole-Genome Analysis of Two Bovine Rotavirus C Strains: Shintoku and Toyama. J. Gen. Virol. 2013, 94, 128–135. [Google Scholar] [CrossRef]
- Otto, P.H.; Rosenhain, S.; Elschner, M.C.; Hotzel, H.; Machnowska, P.; Trojnar, E.; Hoffmann, K.; Johne, R. Detection of Rotavirus Species A, B and C in Domestic Mammalian Animals with Diarrhoea and Genotyping of Bovine Species A Rotavirus Strains. Vet. Microbiol. 2015, 179, 168–176. [Google Scholar] [CrossRef]
- Marton, S.; Mihalov-Kovács, E.; Dóró, R.; Csata, T.; Fehér, E.; Oldal, M.; Jakab, F.; Matthijnssens, J.; Martella, V.; Bányai, K. Canine Rotavirus C Strain Detected in Hungary Shows Marked Genotype Diversity. J. Gen. Virol. 2015, 96, 3059–3071. [Google Scholar] [CrossRef] [PubMed]
- Crawford, S.E.; Ramani, S.; Tate, J.E.; Parashar, U.D.; Svensson, L.; Hagbom, M.; Franco, M.A.; Greenberg, H.B.; O’Ryan, M.; Kang, G.; et al. Rotavirus Infection. Nat. Rev. Dis. Primers 2017, 3, 17083. [Google Scholar] [CrossRef] [PubMed]
- Shaw, D.P.; Morehouse, L.G.; Solorzano, R.F. Experimental Rotavirus Infection in Three-Week-Old Pigs. Am. J. Vet. Res. 1989, 50, 1961–1965. [Google Scholar]
- Dewey, C.; Carman, S.; Pasma, T.; Josephson, G.; McEwen, B. Relationship between Group a Porcine Rotavirus and Management Practices in Swine Herds in Ontario. Can. Vet. J. 2003, 44, 649–653. [Google Scholar]
- Haselhorst, T.; Fleming, F.E.; Dyason, J.C.; Hartnell, R.D.; Yu, X.; Holloway, G.; Santegoets, K.; Kiefel, M.J.; Blanchard, H.; Coulson, B.S.; et al. Sialic Acid Dependence in Rotavirus Host Cell Invasion. Nat. Chem. Biol. 2009, 5, 91–93. [Google Scholar] [CrossRef]
- Hu, L.; Crawford, S.E.; Hyser, J.M.; Estes, M.K.; Prasad, B.V. Rotavirus Non-Structural Proteins: Structure and Function. Curr. Opin. Virol. 2012, 2, 380–388. [Google Scholar] [CrossRef]
- Lopez, S.; Arias, C.F. Early Steps in Rotavirus Cell Entry. Curr. Top. Microbiol. Immunol. 2006, 309, 39–66. [Google Scholar] [CrossRef]
- Ciarlet, M.; Ludert, J.E.; Iturriza-Gómara, M.; Liprandi, F.; Gray, J.J.; Desselberger, U.; Estes, M.K. Initial Interaction of Rotavirus Strains with N -Acetylneuraminic (Sialic) Acid Residues on the Cell Surface Correlates with VP4 Genotype, Not Species of Origin. J. Virol. 2002, 76, 4087–4095. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Martínez, M.A.; López, S.; Arias, C.F.; Isa, P. Gangliosides Have a Functional Role during Rotavirus Cell Entry. J. Virol. 2013, 87, 1115–1122. [Google Scholar] [CrossRef]
- Rolsma, M.D.; Kuhlenschmidt, T.B.; Gelberg, H.B.; Kuhlenschmidt, M.S. Structure and Function of a Ganglioside Receptor for Porcine Rotavirus. J. Virol. 1998, 72, 9079–9091. [Google Scholar] [CrossRef]
- Arias, C.F.; Silva-Ayala, D.; López, S. Rotavirus Entry: A Deep Journey into the Cell with Several Exits. J. Virol. 2015, 89, 890–893. [Google Scholar] [CrossRef]
- Hyser, J.M.; Collinson-Pautz, M.R.; Utama, B.; Estes, M.K. Rotavirus Disrupts Calcium Homeostasis by NSP4 Viroporin Activity. MBio 2010, 1, e00265-10. [Google Scholar] [CrossRef] [PubMed]
- Halaigel, N.; Masia, R.M.; Fernandez-Jimenez, M.; Ribes, J.M.; Montava, R.; de Blas, I.; Girones, O.; Alonso, J.L.; Buesa, J. Enteric Calicivirus and Rotavirus Infections in Domestic Pigs. Epidemiol. Infect. 2010, 138, 542–548. [Google Scholar] [CrossRef]
- Tuanthap, S.; Vongpunsawad, S.; Luengyosluechakul, S.; Sakkaew, P.; Theamboonlers, A.; Amonsin, A.; Poovorawan, Y. Genome Constellations of 24 Porcine Rotavirus Group A Strains Circulating on Commercial Thai Swine Farms between 2011 and 2016. PLoS ONE 2019, 14, e0211002. [Google Scholar] [CrossRef]
- Amimo, J.O.; Vlasova, A.N.; Saif, L.J. Detection and Genetic Diversity of Porcine Group A Rotaviruses in Historic (2004) and Recent (2011 and 2012) Swine Fecal Samples in Ohio: Predominance of the G9P [13] Genotype in Nursing Piglets. J. Clin. Microbiol. 2013, 51, 1142–1151. [Google Scholar] [CrossRef]
- Janke, B.H.; Nelson, J.K.; Benfield, D.A.; Nelson, E.A. Relative Prevalence of Typical and Atypical Strains among Rotaviruses from Diarrheic Pigs in Conventional Swine Herds. J. Vet. Diagn. Investig. 1990, 2, 308–311. [Google Scholar] [CrossRef]
- Collins, P.J.; Martella, V.; Sleator, R.D.; Fanning, S.; O’Shea, H. Detection and Characterisation of Group A Rotavirus in Asymptomatic Piglets in Southern Ireland. Arch. Virol. 2010, 155, 1247–1259. [Google Scholar] [CrossRef]
- Martella, V.; Ciarlet, M.; Bányai, K.; Lorusso, E.; Arista, S.; Lavazza, A.; Pezzotti, G.; Decaro, N.; Cavalli, A.; Lucente, M.S.; et al. Identification of Group a Porcine Rotavirus Strains Bearing a Novel VP4 (P) Genotype in Italian Swine Herds. J. Clin. Microbiol. 2007, 45, 577–580. [Google Scholar] [CrossRef]
- Papp, H.; László, B.; Jakab, F.; Ganesh, B.; de Grazia, S.; Matthijnssens, J.; Ciarlet, M.; Martella, V.; Bányai, K. Review of Group A Rotavirus Strains Reported in Swine and Cattle. Vet. Microbiol. 2013, 165, 190–199. [Google Scholar] [CrossRef]
- Theuns, S.; Conceição-Neto, N.; Zeller, M.; Heylen, E.; Roukaerts, I.D.M.; Desmarets, L.M.B.; van Ranst, M.; Nauwynck, H.J.; Matthijnssens, J. Characterization of a Genetically Heterogeneous Porcine Rotavirus C, and Other Viruses Present in the Fecal Virome of a Non-Diarrheic Belgian Piglet. Infect. Genet. Evol. 2016, 43, 135–145. [Google Scholar] [CrossRef]
- Saif, L.J.; Bohl, E.H.; Theil, K.W.; Cross, R.F.; House, J.A. Rotavirus-Like, Calicivirus-Like, and 23-Nm Virus-Like Particles Associated with Diarrhea in Young Pigs. J. Clin. Microbiol. 1980, 12, 105–111. [Google Scholar] [CrossRef]
- Marthaler, D.; Rossow, K.; Culhane, M.; Collins, J.; Goyal, S.; Ciarlet, M.; Matthijnssens, J. Identification, Phylogenetic Analysis and Classification of Porcine Group C Rotavirus VP7 Sequences from the United States and Canada. Virology 2013, 446, 189–198. [Google Scholar] [CrossRef]
- Amimo, J.O.; Vlasova, A.N.; Saif, L.J. Prevalence and Genetic Heterogeneity of Porcine Group C Rotaviruses in Nursing and Weaned Piglets in Ohio, USA and Identification of a Potential New VP4 Genotype. Vet. Microbiol. 2013, 164, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Niira, K.; Ito, M.; Masuda, T.; Saitou, T.; Abe, T.; Komoto, S.; Sato, M.; Yamasato, H.; Kishimoto, M.; Naoi, Y.; et al. Whole Genome Sequences of Japanese Porcine Species C Rotaviruses Reveal a High Diversity of Genotypes of Individual Genes and Will Contribute to a Comprehensive, Generally Accepted Classification System. Infect. Genet. Evol. 2016, 44, 106–113. [Google Scholar] [CrossRef]
- Suzuki, T.; Inoue, D. Full Genome-Based Genotyping System for Rotavirus H and Detection of Potential Gene Recombination in Nonstructural Protein 3 between Porcine Rotavirus H and Rotavirus C. J. Gen. Virol. 2018, 99, 1582–1589. [Google Scholar] [CrossRef] [PubMed]
- Chepngeno, J.; Diaz, A.; Paim, F.C.; Saif, L.J.; Vlasova, A.N. Rotavirus C: Prevalence in Suckling Piglets and Development of Virus-like Particles to Assess the Influence of Maternal Immunity on the Disease Development. Vet. Res. 2019, 50, 84. [Google Scholar] [CrossRef] [PubMed]
- Roczo-Farkas, S.; Dunlop, R.H.; Donato, C.M.; Kirkwood, C.D.; McOrist, S. Rotavirus Group C Infections in Neonatal and Grower Pigs in Australia. Vet. Rec. 2021, 188, e296. [Google Scholar] [CrossRef] [PubMed]
- Kuga, K.; Miyazaki, A.; Suzuki, T.; Takagi, M.; Hattori, N.; Katsuda, K.; Mase, M.; Sugiyama, M.; Tsunemitsu, H. Genetic Diversity and Classification of the Outer Capsid Glycoprotein VP7 of Porcine Group B Rotaviruses. Arch. Virol. 2009, 154, 1785–1795. [Google Scholar] [CrossRef]
- Shepherd, F.; Murtaugh, M.; Chen, F.; Culhane, M.; Marthaler, D. Longitudinal Surveillance of Porcine Rotavirus B Strains from the United States and Canada and In Silico Identification of Antigenically Important Sites. Pathogens 2017, 6, 64. [Google Scholar] [CrossRef]
- Alam, M.M.; Kobayashi, N.; Ishino, M.; Ahmed, M.S.; Ahmed, M.U.; Paul, S.K.; Muzumdar, B.K.; Hussain, Z.; Wang, Y.-H.; Naik, T.N. Genetic Analysis of an ADRV-N-like Novel Rotavirus Strain B219 Detected in a Sporadic Case of Adult Diarrhea in Bangladesh. Arch. Virol. 2007, 152, 199–208. [Google Scholar] [CrossRef]
- Jiang, S.; Ji, S.; Tang, Q.; Cui, X.; Yang, H.; Kan, B.; Gao, S. Molecular Characterization of a Novel Adult Diarrhoea Rotavirus Strain J19 Isolated in China and Its Significance for the Evolution and Origin of Group B Rotaviruses. J. Gen. Virol. 2008, 89, 2622–2629. [Google Scholar] [CrossRef]
- Nagashima, S.; Kobayashi, N.; Ishino, M.; Alam, M.M.; Ahmed, M.U.; Paul, S.K.; Ganesh, B.; Chawla-Sarkar, M.; Krishnan, T.; Naik, T.N.; et al. Whole Genomic Characterization of a Human Rotavirus Strain B219 Belonging to a Novel Group of the Genus Rotavirus. J. Med. Virol. 2008, 80, 2023–2033. [Google Scholar] [CrossRef]
- Wakuda, M. Porcine Rotavirus Closely Related to Novel Group of Human Rotaviruses. Emerg. Infect. Dis. 2011, 17, 1491–1493. [Google Scholar] [CrossRef]
- Yang, H.; Makeyev, E.V.; Kang, Z.; Ji, S.; Bamford, D.H.; van Dijk, A.A. Cloning and Sequence Analysis of DsRNA Segments 5, 6 and 7 of a Novel Non-Group A, B, C Adult Rotavirus That Caused an Outbreak of Gastroenteritis in China. Virus Res. 2004, 106, 15–26. [Google Scholar] [CrossRef]
- Molinari, B.L.D.; Lorenzetti, E.; Otonel, R.A.A.; Alfieri, A.F.; Alfieri, A.A. Species H Rotavirus Detected in Piglets with Diarrhea, Brazil, 2012. Emerg. Infect. Dis. 2014, 20, 1019–1022. [Google Scholar] [CrossRef]
- Hakim, M.S.; Ding, S.; Chen, S.; Yin, Y.; Su, J.; van der Woude, C.J.; Fuhler, G.M.; Peppelenbosch, M.P.; Pan, Q.; Wang, W. TNF-α Exerts Potent Anti-Rotavirus Effects via the Activation of Classical NF-ΚB Pathway. Virus Res. 2018, 253, 28–37. [Google Scholar] [CrossRef]
- Brisse, M.; Ly, H. Comparative Structure and Function Analysis of the RIG-I-Like Receptors: RIG-I and MDA5. Front. Immunol. 2019, 10, 1586. [Google Scholar] [CrossRef]
- Marshall, J.S.; Warrington, R.; Watson, W.; Kim, H.L. An Introduction to Immunology and Immunopathology. Allergy Asthma Clin. Immunol. 2018, 14, 49. [Google Scholar] [CrossRef]
- Turvey, S.E.; Broide, D.H. Innate Immunity. J. Allergy Clin. Immunol. 2010, 125, S24–S32. [Google Scholar] [CrossRef]
- Amarante-Mendes, G.P.; Adjemian, S.; Branco, L.M.; Zanetti, L.C.; Weinlich, R.; Bortoluci, K.R. Pattern Recognition Receptors and the Host Cell Death Molecular Machinery. Front. Immunol. 2018, 9, 2379. [Google Scholar] [CrossRef]
- Mogensen, T.H. Pathogen Recognition and Inflammatory Signaling in Innate Immune Defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef]
- Broquet, A.H.; Hirata, Y.; McAllister, C.S.; Kagnoff, M.F. RIG-I/MDA5/MAVS Are Required to Signal a Protective IFN Response in Rotavirus-Infected Intestinal Epithelium. J. Immunol. 2011, 186, 1618–1626. [Google Scholar] [CrossRef]
- Sen, A.; Pruijssers, A.J.; Dermody, T.S.; García-Sastre, A.; Greenberg, H.B. The Early Interferon Response to Rotavirus Is Regulated by PKR and Depends on MAVS/IPS-1, RIG-I, MDA-5, and IRF3. J. Virol. 2011, 85, 3717–3732. [Google Scholar] [CrossRef] [PubMed]
- Wen, K.; Azevedo, M.S.P.; Gonzalez, A.; Zhang, W.; Saif, L.J.; Li, G.; Yousef, A.; Yuan, L. Toll-like Receptor and Innate Cytokine Responses Induced by Lactobacilli Colonization and Human Rotavirus Infection in Gnotobiotic Pigs. Vet. Immunol. Immunopathol. 2009, 127, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Chassaing, B.; Shi, Z.; Uchiyama, R.; Zhang, Z.; Denning, T.L.; Crawford, S.E.; Pruijssers, A.J.; Iskarpatyoti, J.A.; Estes, M.K.; et al. Prevention and Cure of Rotavirus Infection via TLR5/NLRC4–Mediated Production of IL-22 and IL-18. Science 2014, 346, 861–865. [Google Scholar] [CrossRef]
- Sanchez David, R.Y.; Combredet, C.; Sismeiro, O.; Dillies, M.-A.; Jagla, B.; Coppée, J.-Y.; Mura, M.; Guerbois Galla, M.; Despres, P.; Tangy, F.; et al. Comparative Analysis of Viral RNA Signatures on Different RIG-I-like Receptors. eLife 2016, 5, e11275. [Google Scholar] [CrossRef]
- Tan, P.; He, L.; Cui, J.; Qian, C.; Cao, X.; Lin, M.; Zhu, Q.; Li, Y.; Xing, C.; Yu, X.; et al. Assembly of the WHIP-TRIM14-PPP6C Mitochondrial Complex Promotes RIG-I-Mediated Antiviral Signaling. Mol. Cell 2017, 68, 293–307.e5. [Google Scholar] [CrossRef]
- Ishizuka, T.; Kanmani, P.; Kobayashi, H.; Miyazaki, A.; Soma, J.; Suda, Y.; Aso, H.; Nochi, T.; Iwabuchi, N.; Xiao, J.; et al. Immunobiotic Bifidobacteria Strains Modulate Rotavirus Immune Response in Porcine Intestinal Epitheliocytes via Pattern Recognition Receptor Signaling. PLoS ONE 2016, 11, e0152416. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.-D.; Feng, N.; Sen, A.; Balan, M.; Tseng, H.-C.; McElrath, C.; Smirnov, S.V.; Peng, J.; Yasukawa, L.L.; Durbin, R.K.; et al. Distinct Roles of Type I and Type III Interferons in Intestinal Immunity to Homologous and Heterologous Rotavirus Infections. PLoS Pathog. 2016, 12, e1005600. [Google Scholar] [CrossRef]
- Li, S.; Shao, Q.; Zhu, Y.; Ji, X.; Luo, J.; Xu, Y.; Liu, X.; Zheng, W.; Chen, N.; Meurens, F.; et al. Porcine RIG-I and MDA5 Signaling CARD Domains Exert Similar Antiviral Function Against Different Viruses. Front. Microbiol. 2021, 12, 677634. [Google Scholar] [CrossRef]
- Reikine, S.; Nguyen, J.B.; Modis, Y. Pattern Recognition and Signaling Mechanisms of RIG-I and MDA5. Front. Immunol. 2014, 5, 342. [Google Scholar] [CrossRef]
- Otte, J.-M.; Cario, E.; Podolsky, D.K. Mechanisms of Cross Hyporesponsiveness to Toll-like Receptor Bacterial Ligands in Intestinal Epithelial Cells. Gastroenterology 2004, 126, 1054–1070. [Google Scholar] [CrossRef]
- Zhou, R.; Wei, H.; Sun, R.; Tian, Z. Recognition of Double-Stranded RNA by TLR3 Induces Severe Small Intestinal Injury in Mice. J. Immunol. 2007, 178, 4548–4556. [Google Scholar] [CrossRef] [PubMed]
- Konkel, J.E.; Maruyama, T.; Carpenter, A.C.; Xiong, Y.; Zamarron, B.F.; Hall, B.E.; Kulkarni, A.B.; Zhang, P.; Bosselut, R.; Chen, W. Control of the Development of CD8αα+ Intestinal Intraepithelial Lymphocytes by TGF-β. Nat. Immunol. 2011, 12, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Iizuka, M.; Nakagomi, O.; Suzuki, M.; Horie, Y.; Konno, S.; Hirasawa, F.; Sasaki, K.; Shindo, K.; Watanabe, S. Rotavirus Double-Stranded RNA Induces Apoptosis and Diminishes Wound Repair in Rat Intestinal Epithelial Cells. J. Gastroenterol. Hepatol. 2006, 21, 521–530. [Google Scholar] [CrossRef]
- Pott, J.; Stockinger, S.; Torow, N.; Smoczek, A.; Lindner, C.; McInerney, G.; Bäckhed, F.; Baumann, U.; Pabst, O.; Bleich, A.; et al. Age-Dependent TLR3 Expression of the Intestinal Epithelium Contributes to Rotavirus Susceptibility. PLoS Pathog. 2012, 8, e1002670. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, R.; Chassaing, B.; Zhang, B.; Gewirtz, A.T. MyD88-Mediated TLR Signaling Protects against Acute Rotavirus Infection While Inflammasome Cytokines Direct Ab Response. Innate Immun. 2015, 21, 416–428. [Google Scholar] [CrossRef]
- Zhu, S.; Ding, S.; Wang, P.; Wei, Z.; Pan, W.; Palm, N.W.; Yang, Y.; Yu, H.; Li, H.-B.; Wang, G.; et al. Nlrp9b Inflammasome Restricts Rotavirus Infection in Intestinal Epithelial Cells. Nature 2017, 546, 667–670. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A. Mucosal Dendritic Cells. Annu. Rev. Immunol. 2007, 25, 381–418. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Guerrero, D.V.; Meza-Perez, S.; Ramirez-Pliego, O.; Santana-Calderon, M.A.; Espino-Solis, P.; Gutierrez-Xicotencatl, L.; Flores-Romo, L.; Esquivel-Guadarrama, F.R. Rotavirus Infection Activates Dendritic Cells from Peyer’s Patches in Adult Mice. J. Virol. 2010, 84, 1856–1866. [Google Scholar] [CrossRef]
- Fasciano, A.C.; Mecsas, J. Eat Your Vitamin A: A Role for Retinoic Acid in the Development of Microfold Cells. Gastroenterology 2020, 159, 34–36. [Google Scholar] [CrossRef]
- di Fiore, I.J.M.; Holloway, G.; Coulson, B.S. Innate Immune Responses to Rotavirus Infection in Macrophages Depend on MAVS but Involve Neither the NLRP3 Inflammasome nor JNK and P38 Signaling Pathways. Virus Res. 2015, 208, 89–97. [Google Scholar] [CrossRef]
- González, A.M.; Azevedo, M.S.P.; Jung, K.; Vlasova, A.; Zhang, W.; Saif, L.J. Innate Immune Responses to Human Rotavirus in the Neonatal Gnotobiotic Piglet Disease Model. Immunology 2010, 131, 242–256. [Google Scholar] [CrossRef]
- Matikainen, S.; Sirén, J.; Tissari, J.; Veckman, V.; Pirhonen, J.; Severa, M.; Sun, Q.; Lin, R.; Meri, S.; Uzé, G.; et al. Tumor Necrosis Factor Alpha Enhances Influenza A Virus-Induced Expression of Antiviral Cytokines by Activating RIG-I Gene Expression. J. Virol. 2006, 80, 3515–3522. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.H.; Webster, R.G. Tumor Necrosis Factor Alpha Exerts Powerful Anti-Influenza Virus Effects in Lung Epithelial Cells. J. Virol. 2002, 76, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xu, L.; Brandsma, J.H.; Wang, Y.; Hakim, M.S.; Zhou, X.; Yin, Y.; Fuhler, G.M.; van der Laan, L.J.W.; van der Woude, C.J.; et al. Convergent Transcription of Interferon-Stimulated Genes by TNF-α and IFN-α Augments Antiviral Activity against HCV and HEV. Sci. Rep. 2016, 6, 25482. [Google Scholar] [CrossRef]
- Gómez del Moral, M.; Ortuño, E.; Fernández-Zapatero, P.; Alonso, F.; Alonso, C.; Ezquerra, A.; Domínguez, J. African Swine Fever Virus Infection Induces Tumor Necrosis Factor Alpha Production: Implications in Pathogenesis. J. Virol. 1999, 73, 2173–2180. [Google Scholar] [CrossRef]
- Jiang, B.; Snipes-Magaldi, L.; Dennehy, P.; Keyserling, H.; Holman, R.C.; Bresee, J.; Gentsch, J.; Glass, R.I. Cytokines as Mediators for or Effectors against Rotavirus Disease in Children. Clin. Vaccine Immunol. 2003, 10, 995–1001. [Google Scholar] [CrossRef]
- Franco, M.A.; Greenberg, H.B. Role of B Cells and Cytotoxic T Lymphocytes in Clearance of and Immunity to Rotavirus Infection in Mice. J. Virol. 1995, 69, 7800–7806. [Google Scholar] [CrossRef]
- MCNEAL, M.M.; BARONE, K.S.; RAE, M.N.; WARD, R.L. Effector Functions of Antibody and CD8+Cells in Resolution of Rotavirus Infection and Protection against Reinfection in Mice. Virology 1995, 214, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.O.; Vandal, O.H.; Yuan, L.; Hodgins, D.C.; Saif, L.J. Antibody-Secreting Cell Responses to Rotavirus Proteins in Gnotobiotic Pigs Inoculated with Attenuated or Virulent Human Rotavirus. J. Clin. Microbiol. 2001, 39, 2807–2813. [Google Scholar] [CrossRef]
- Caddy, S.L.; Vaysburd, M.; Wing, M.; Foss, S.; Andersen, J.T.; O’Connell, K.; Mayes, K.; Higginson, K.; Iturriza-Gómara, M.; Desselberger, U.; et al. Intracellular Neutralisation of Rotavirus by VP6-Specific IgG. PLoS Pathog. 2020, 16, e1008732. [Google Scholar] [CrossRef]
- Lappalainen, S.; Pastor, A.R.; Tamminen, K.; López-Guerrero, V.; Esquivel-Guadarrama, F.; Palomares, L.A.; Vesikari, T.; Blazevic, V. Immune Responses Elicited against Rotavirus Middle Layer Protein VP6 Inhibit Viral Replication in Vitro and in Vivo. Hum. Vaccin. Immunother. 2014, 10, 2039–2047. [Google Scholar] [CrossRef] [PubMed]
- Nair, N.; Feng, N.; Blum, L.K.; Sanyal, M.; Ding, S.; Jiang, B.; Sen, A.; Morton, J.M.; He, X.-S.; Robinson, W.H.; et al. VP4- and VP7-Specific Antibodies Mediate Heterotypic Immunity to Rotavirus in Humans. Sci. Transl. Med. 2017, 9, eaam5434. [Google Scholar] [CrossRef]
- Qiao, X.; Li, G.; Wang, X.; Li, X.; Liu, M.; Li, Y. Recombinant Porcine Rotavirus VP4 and VP4-LTB Expressed in Lactobacillus Casei Induced Mucosal and Systemic Antibody Responses in Mice. BMC Microbiol. 2009, 9, 249. [Google Scholar] [CrossRef]
- Vizzi, E.; Calviño, E.; González, R.; Pérez-Schael, I.; Ciarlet, M.; Kang, G.; Estes, M.K.; Liprandi, F.; Ludert, J.E. Evaluation of Serum Antibody Responses against the Rotavirus Nonstructural Protein NSP4 in Children after Rhesus Rotavirus Tetravalent Vaccination or Natural Infection. Clin. Vaccine Immunol. 2005, 12, 1157–1163. [Google Scholar] [CrossRef]
- Azevedo, M.S.P.; Yuan, L.; Iosef, C.; Chang, K.-O.; Kim, Y.; van Nguyen, T.; Saif, L.J. Magnitude of Serum and Intestinal Antibody Responses Induced by Sequential Replicating and Nonreplicating Rotavirus Vaccines in Gnotobiotic Pigs and Correlation with Protection. Clin. Vaccine Immunol. 2004, 11, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Velázquez, F.R.; Matson, D.O.; Guerrero, M.L.; Shults, J.; Calva, J.J.; Morrow, A.L.; Glass, R.I.; Pickering, L.K.; Ruiz-Palacios, G.M. Serum Antibody as a Marker of Protection against Natural Rotavirus Infection and Disease. J. Infect. Dis. 2000, 182, 1602–1609. [Google Scholar] [CrossRef]
- Blutt, S.E.; Warfield, K.L.; Lewis, D.E.; Conner, M.E. Early Response to Rotavirus Infection Involves Massive B Cell Activation. J. Immunol. 2002, 168, 5716–5721. [Google Scholar] [CrossRef] [PubMed]
- Twitchell, E.L.; Tin, C.; Wen, K.; Zhang, H.; Becker-Dreps, S.; Azcarate-Peril, M.A.; Vilchez, S.; Li, G.; Ramesh, A.; Weiss, M.; et al. Modeling Human Enteric Dysbiosis and Rotavirus Immunity in Gnotobiotic Pigs. Gut Pathog. 2016, 8, 51. [Google Scholar] [CrossRef]
- Wen, K.; Bui, T.; Weiss, M.; Li, G.; Kocher, J.; Yang, X.; Jobst, P.M.; Vaught, T.; Ramsoondar, J.; Ball, S.; et al. B-Cell-Deficient and CD8 T-Cell-Depleted Gnotobiotic Pigs for the Study of Human Rotavirus Vaccine-Induced Protective Immune Responses. Viral Immunol. 2016, 29, 112–127. [Google Scholar] [CrossRef]
- Angel, J.; Franco, M.A.; Greenberg, H.B. Rotavirus Immune Responses and Correlates of Protection. Curr. Opin. Virol. 2012, 2, 419–425. [Google Scholar] [CrossRef]
- de Vos, B.; Han, H.H.; Bouckenooghe, A.; Debrus, S.; Gillard, P.; Ward, R.; Cheuvart, B. Live Attenuated Human Rotavirus Vaccine, RIX4414, Provides Clinical Protection in Infants Against Rotavirus Strains with and Without Shared G and P Genotypes. Pediatric Infect. Dis. J. 2009, 28, 261–266. [Google Scholar] [CrossRef]
- Ruiz-Palacios, G.M.; Pérez-Schael, I.; Velázquez, F.R.; Abate, H.; Breuer, T.; Clemens, S.C.; Cheuvart, B.; Espinoza, F.; Gillard, P.; Innis, B.L.; et al. Safety and Efficacy of an Attenuated Vaccine against Severe Rotavirus Gastroenteritis. N. Engl. J. Med. 2006, 354, 11–22. [Google Scholar] [CrossRef]
- Clarke, E.; Desselberger, U. Correlates of Protection against Human Rotavirus Disease and the Factors Influencing Protection in Low-Income Settings. Mucosal Immunol. 2015, 8, 1–17. [Google Scholar] [CrossRef]
- Green, K.Y.; Taniguchi, K.; Mackow, E.R.; Kapikian, A.Z. Homotypic and Heterotypic Epitope-Specific Antibody Responses in Adult and Infant Rotavirus Vaccinees: Implications for Vaccine Development. J. Infect. Dis. 1990, 161, 667–679. [Google Scholar] [CrossRef]
- Hoshino, Y.; Jones, R.W.; Ross, J.; Honma, S.; Santos, N.; Gentsch, J.R.; Kapikian, A.Z. Rotavirus Serotype G9 Strains Belonging to VP7 Gene Phylogenetic Sequence Lineage 1 May Be More Suitable for Serotype G9 Vaccine Candidates than Those Belonging to Lineage 2 or 3. J. Virol. 2004, 78, 7795–7802. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Fischer, D.D.; Kandasamy, S.; Rauf, A.; Langel, S.N.; Wentworth, D.E.; Stucker, K.M.; Halpin, R.A.; Lam, H.C.; Marthaler, D.; et al. Comparative In Vitro and In Vivo Studies of Porcine Rotavirus G9P [13] and Human Rotavirus Wa G1P [8]. J. Virol. 2016, 90, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Feng, N.; Hu, L.; Ding, S.; Sanyal, M.; Zhao, B.; Sankaran, B.; Ramani, S.; McNeal, M.; Yasukawa, L.L.; Song, Y.; et al. Human VP8* MAbs Neutralize Rotavirus Selectively in Human Intestinal Epithelial Cells. J. Clin. Investig. 2019, 129, 3839–3851. [Google Scholar] [CrossRef]
- Burns, J.W.; Siadat-Pajouh, M.; Krishnaney, A.A.; Greenberg, H.B. Protective Effect of Rotavirus VP6-Specific IgA Monoclonal Antibodies That Lack Neutralizing Activity. Science 1996, 272, 104–107. [Google Scholar] [CrossRef]
- McNeal, M.M.; VanCott, J.L.; Choi, A.H.C.; Basu, M.; Flint, J.A.; Stone, S.C.; Clements, J.D.; Ward, R.L. CD4 T Cells Are the Only Lymphocytes Needed to Protect Mice against Rotavirus Shedding after Intranasal Immunization with a Chimeric VP6 Protein and the Adjuvant LT(R192G). J. Virol. 2002, 76, 560–568. [Google Scholar] [CrossRef]
- Langel, S.N.; Wang, Q.; Vlasova, A.N.; Saif, L.J. Host Factors Affecting Generation of Immunity Against Porcine Epidemic Diarrhea Virus in Pregnant and Lactating Swine and Passive Protection of Neonates. Pathogens 2020, 9, 130. [Google Scholar] [CrossRef]
- Bianchi, A.T.J.; Scholten, J.-W.; Moonen Leusen, B.H.W.M.; Boersma, W.J.A. Development of the Natural Response of Immunoglobulin Secreting Cells in the Pig as a Function of Organ, Age and Housing. Dev. Comp. Immunol. 1999, 23, 511–520. [Google Scholar] [CrossRef]
- Hodgins, D.C.; Kang, S.Y.; deArriba, L.; Parreño, V.; Ward, L.A.; Yuan, L.; To, T.; Saif, L.J. Effects of Maternal Antibodies on Protection and Development of Antibody Responses to Human Rotavirus in Gnotobiotic Pigs. J. Virol. 1999, 73, 186–197. [Google Scholar] [CrossRef]
- Song, D.; Moon, H.; Kang, B. Porcine Epidemic Diarrhea: A Review of Current Epidemiology and Available Vaccines. Clin. Exp. Vaccine Res. 2015, 4, 166–176. [Google Scholar] [CrossRef]
- Fu, Z.F.; Hampson, D.J.; Wilks, C.R. Transfer of Maternal Antibody against Group A Rotavirus from Sows to Piglets and Serological Responses Following Natural Infection. Res. Vet. Sci. 1990, 48, 365–373. [Google Scholar] [CrossRef]
- Tzipori, S.; Chandler, D.; Makin, T.; Smith, M. Escherichia coli and rotavirus infections in four-week-old gnotobiotic piglets fed milk or dry food. Aust. Vet. J. 1980, 56, 279–284. [Google Scholar] [CrossRef]
- Lecce, J.G.; King, M.W. Role of Rotavirus (Reo-like) in Weanling Diarrhea of Pigs. J. Clin. Microbiol. 1978, 8, 454–458. [Google Scholar] [CrossRef]
- Bohl, E.H.; Gupta, R.K.P.; Olquin, M.V.F.; Saif, L.J. Antibody Responses in Serum, Colostrum, and Milk of Swine After Infection or Vaccination with Transmissible Gastroenteritis Virus. Infect. Immun. 1972, 6, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Boyd, W.; Pittman, J.; Wolfe, Z. Comparison of Prefarrow Rotavirus Vaccine and Natural Planned Exposure on Suckling Pig Performance. In Proceedings of the American Association of Swine Veterinarians Annual Meeting, Atlanta, Georgia, 7 March 2020. [Google Scholar]
- Shepherd, F. Enhancing Control of Porcine Rotavirus Through the Identification of Candidate B Cell Epitopes. Ph.D. Thesis, University of Minnesota, Minneapolis, MN, USA, August 2020. [Google Scholar]
- Niu, Q.; Li, P.; Hao, S.; Zhang, Y.; Kim, S.W.; Li, H.; Ma, X.; Gao, S.; He, L.; Wu, W.; et al. Dynamic Distribution of the Gut Microbiota and the Relationship with Apparent Crude Fiber Digestibility and Growth Stages in Pigs. Sci. Rep. 2015, 5, 9938. [Google Scholar] [CrossRef]
- Liu, S.; Zhao, L.; Zhai, Z.; Zhao, W.; Ding, J.; Dai, R.; Sun, T.; Meng, H. Porcine Epidemic Diarrhea Virus Infection Induced the Unbalance of Gut Microbiota in Piglets. Curr. Microbiol. 2015, 71, 643–649. [Google Scholar] [CrossRef]
- Huang, M.-Z.; Wang, S.-Y.; Wang, H.; Cui, D.-A.; Yang, Y.-J.; Liu, X.-W.; Kong, X.-J.; Li, J.-Y. Differences in the Intestinal Microbiota between Uninfected Piglets and Piglets Infected with Porcine Epidemic Diarrhea Virus. PLoS ONE 2018, 13, e0192992. [Google Scholar] [CrossRef]
- Huang, A.; Cai, R.; Wang, Q.; Shi, L.; Li, C.; Yan, H. Dynamic Change of Gut Microbiota During Porcine Epidemic Diarrhea Virus Infection in Suckling Piglets. Front. Microbiol. 2019, 10, 322. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Peng, Q.; Chen, Y.; Zhou, X.; Zhang, F.; Li, A.; Huang, D.; Wu, Q.; Ye, Y.; He, H.; et al. Altered Gut Microbiota Profiles in Sows and Neonatal Piglets Associated with Porcine Epidemic Diarrhea Virus Infection. Sci. Rep. 2017, 7, 17439. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Dong, W.; Ding, Y.; Ding, X.; Zhang, Q.; Jiang, L. Changes in Cecal Microbiota Community of Suckling Piglets Infected with Porcine Epidemic Diarrhea Virus. PLoS ONE 2019, 14, e0219868. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tsai, T.; Deng, F.; Wei, X.; Chai, J.; Knapp, J.; Apple, J.; Maxwell, C.V.; Lee, J.A.; Li, Y.; et al. Longitudinal Investigation of the Swine Gut Microbiome from Birth to Market Reveals Stage and Growth Performance Associated Bacteria. Microbiome 2019, 7, 109. [Google Scholar] [CrossRef]
- Harris, V.; Ali, A.; Fuentes, S.; Korpela, K.; Kazi, M.; Tate, J.; Parashar, U.; Wiersinga, W.J.; Giaquinto, C.; de Weerth, C.; et al. Rotavirus Vaccine Response Correlates with the Infant Gut Microbiota Composition in Pakistan. Gut Microbes 2018, 9, 93–101. [Google Scholar] [CrossRef]
- Shi, Z.; Zou, J.; Zhang, Z.; Zhao, X.; Noriega, J.; Zhang, B.; Zhao, C.; Ingle, H.; Bittinger, K.; Mattei, L.M.; et al. Segmented Filamentous Bacteria Prevent and Cure Rotavirus Infection. Cell 2019, 179, 644–658.e13. [Google Scholar] [CrossRef]
- Niederwerder, M.C.; Constance, L.A.; Rowland, R.R.R.; Abbas, W.; Fernando, S.C.; Potter, M.L.; Sheahan, M.A.; Burkey, T.E.; Hesse, R.A.; Cino-Ozuna, A.G. Fecal Microbiota Transplantation Is Associated with Reduced Morbidity and Mortality in Porcine Circovirus Associated Disease. Front. Microbiol. 2018, 9, 1631. [Google Scholar] [CrossRef]
- Harris, V.C.; Haak, B.W.; Handley, S.A.; Jiang, B.; Velasquez, D.E.; Hykes, B.L.; Droit, L.; Berbers, G.A.M.; Kemper, E.M.; van Leeuwen, E.M.M.; et al. Effect of Antibiotic-Mediated Microbiome Modulation on Rotavirus Vaccine Immunogenicity: A Human, Randomized-Control Proof-of-Concept Trial. Cell Host Microbe. 2018, 24, 197–207.e4. [Google Scholar] [CrossRef]
- Harris, V.C.; Armah, G.; Fuentes, S.; Korpela, K.E.; Parashar, U.; Victor, J.C.; Tate, J.; de Weerth, C.; Giaquinto, C.; Wiersinga, W.J.; et al. Significant Correlation Between the Infant Gut Microbiome and Rotavirus Vaccine Response in Rural Ghana. J. Infect. Dis. 2017, 215, 34–41. [Google Scholar] [CrossRef]
- Parker, E.P.K.; Praharaj, I.; Zekavati, A.; Lazarus, R.P.; Giri, S.; Operario, D.J.; Liu, J.; Houpt, E.; Iturriza-Gómara, M.; Kampmann, B.; et al. Influence of the Intestinal Microbiota on the Immunogenicity of Oral Rotavirus Vaccine given to Infants in South India. Vaccine 2018, 36, 264–272. [Google Scholar] [CrossRef]
- Park, M.S.; Kwon, B.; Ku, S.; Ji, G.E. The Efficacy of Bifidobacterium Longum BORI and Lactobacillus Acidophilus AD031 Probiotic Treatment in Infants with Rotavirus Infection. Nutrients 2017, 9, 887. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Twitchell, E.; Li, G.; Wen, K.; Weiss, M.; Kocher, J.; Lei, S.; Ramesh, A.; Ryan, E.P.; Yuan, L. High Protective Efficacy of Rice Bran against Human Rotavirus Diarrhea via Enhancing Probiotic Growth, Gut Barrier Function and Innate Immunity. Sci. Rep. 2015, 5, 15004. [Google Scholar] [CrossRef] [PubMed]
- Vlasova, A.N.; Shao, L.; Kandasamy, S.; Fischer, D.D.; Rauf, A.; Langel, S.N.; Chattha, K.S.; Kumar, A.; Huang, H.-C.; Rajashekara, G.; et al. Escherichia Coli Nissle 1917 Protects Gnotobiotic Pigs against Human Rotavirus by Modulating PDC and NK-Cell Responses. Eur. J. Immunol. 2016, 46, 2426–2437. [Google Scholar] [CrossRef]
- Pant, N.; Marcotte, H.; Bruessow, H.; Svensson, L.; Hammarstrom, L. Effective Prophylaxis against Rotavirus Diarrhea Using a Combination of Lactobacillus Rhamnosus GG and Antibodies. BMC Microbiol. 2007, 7, 86. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, R.P.; John, J.; Shanmugasundaram, E.; Rajan, A.K.; Thiagarajan, S.; Giri, S.; Babji, S.; Sarkar, R.; Kaliappan, P.S.; Venugopal, S.; et al. The Effect of Probiotics and Zinc Supplementation on the Immune Response to Oral Rotavirus Vaccine: A Randomized, Factorial Design, Placebo-Controlled Study among Indian Infants. Vaccine 2018, 36, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Mao, M.; Yu, T.; Xiong, Y.; Wang, Z.; Liu, H.; Gotteland, M.; Brunser, O. Effect of a Lactose-Free Milk Formula Supplemented with Bifidobacteria and Streptococci on the Recovery from Acute Diarrhoea. Asia Pac. J. Clin. Nutr. 2008, 17, 30–34. [Google Scholar]
- Chattha, K.S.; Kandasamy, S.; Vlasova, A.N.; Saif, L.J. Vitamin A Deficiency Impairs Adaptive B and T Cell Responses to a Prototype Monovalent Attenuated Human Rotavirus Vaccine and Virulent Human Rotavirus Challenge in a Gnotobiotic Piglet Model. PLoS ONE 2013, 8, e82966. [Google Scholar] [CrossRef]
- Kandasamy, S.; Chattha, K.S.; Vlasova, A.N.; Saif, L.J. Prenatal Vitamin A Deficiency Impairs Adaptive Immune Responses to Pentavalent Rotavirus Vaccine (RotaTeq®) in a Neonatal Gnotobiotic Pig Model. Vaccine 2014, 32, 816–824. [Google Scholar] [CrossRef]
- Vlasova, A.N.; Chattha, K.S.; Kandasamy, S.; Siegismund, C.S.; Saif, L.J. Prenatally Acquired Vitamin A Deficiency Alters Innate Immune Responses to Human Rotavirus in a Gnotobiotic Pig Model. J. Immunol. 2013, 190, 4742–4753. [Google Scholar] [CrossRef]
- Michael, H.; Langel, S.N.; Miyazaki, A.; Paim, F.C.; Chepngeno, J.; Alhamo, M.A.; Fischer, D.D.; Srivastava, V.; Kathayat, D.; Deblais, L.; et al. Malnutrition Decreases Antibody Secreting Cell Numbers Induced by an Oral Attenuated Human Rotavirus Vaccine in a Human Infant Fecal Microbiota Transplanted Gnotobiotic Pig Model. Front. Immunol. 2020, 11, 296. [Google Scholar] [CrossRef]
- Langel, S.N.; Paim, F.C.; Alhamo, M.A.; Lager, K.M.; Vlasova, A.N.; Saif, L.J. Oral Vitamin A Supplementation of Porcine Epidemic Diarrhea Virus Infected Gilts Enhances IgA and Lactogenic Immune Protection of Nursing Piglets. Vet. Res. 2019, 50, 101. [Google Scholar] [CrossRef] [PubMed]
- Zijlstra, R.T.; McCracken, B.A.; Odle, J.; Donovan, S.M.; Gelberg, H.B.; Petschow, B.W.; Zuckermann, F.A.; Gaskins, H.R. Malnutrition Modifies Pig Small Intestinal Inflammatory Responses to Rotavirus. J. Nutr. 1999, 129, 838–843. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Vlasova, A.N.; Deblais, L.; Huang, H.-C.; Wijeratne, A.; Kandasamy, S.; Fischer, D.D.; Langel, S.N.; Paim, F.C.; Alhamo, M.A.; et al. Impact of Nutrition and Rotavirus Infection on the Infant Gut Microbiota in a Humanized Pig Model. BMC Gastroenterol. 2018, 18, 93. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, A.; Kandasamy, S.; Michael, H.; Langel, S.N.; Paim, F.C.; Chepngeno, J.; Alhamo, M.A.; Fischer, D.D.; Huang, H.-C.; Srivastava, V.; et al. Protein Deficiency Reduces Efficacy of Oral Attenuated Human Rotavirus Vaccine in a Human Infant Fecal Microbiota Transplanted Gnotobiotic Pig Model. Vaccine 2018, 36, 6270–6281. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, D.; Shepherd, F.K.; Springer, N.L.; Mwangi, W.; Marthaler, D.G. Rotavirus Infection in Swine: Genotypic Diversity, Immune Responses, and Role of Gut Microbiome in Rotavirus Immunity. Pathogens 2022, 11, 1078. https://doi.org/10.3390/pathogens11101078
Kumar D, Shepherd FK, Springer NL, Mwangi W, Marthaler DG. Rotavirus Infection in Swine: Genotypic Diversity, Immune Responses, and Role of Gut Microbiome in Rotavirus Immunity. Pathogens. 2022; 11(10):1078. https://doi.org/10.3390/pathogens11101078
Chicago/Turabian StyleKumar, Deepak, Frances K Shepherd, Nora L. Springer, Waithaka Mwangi, and Douglas G. Marthaler. 2022. "Rotavirus Infection in Swine: Genotypic Diversity, Immune Responses, and Role of Gut Microbiome in Rotavirus Immunity" Pathogens 11, no. 10: 1078. https://doi.org/10.3390/pathogens11101078
APA StyleKumar, D., Shepherd, F. K., Springer, N. L., Mwangi, W., & Marthaler, D. G. (2022). Rotavirus Infection in Swine: Genotypic Diversity, Immune Responses, and Role of Gut Microbiome in Rotavirus Immunity. Pathogens, 11(10), 1078. https://doi.org/10.3390/pathogens11101078





