Virulence Determinants and Genetic Diversity of Yersinia Species Isolated from Retail Meat
Abstract
:1. Introduction
2. Results
2.1. Prevalence of Yersinia spp. and Pathogenic Yersinia Enterocolitica Bioserovars in Meats
2.2. Antimicrobial Resistance in Yersinia enterocolitica 2/O:9 and 4/O:3 Isolates
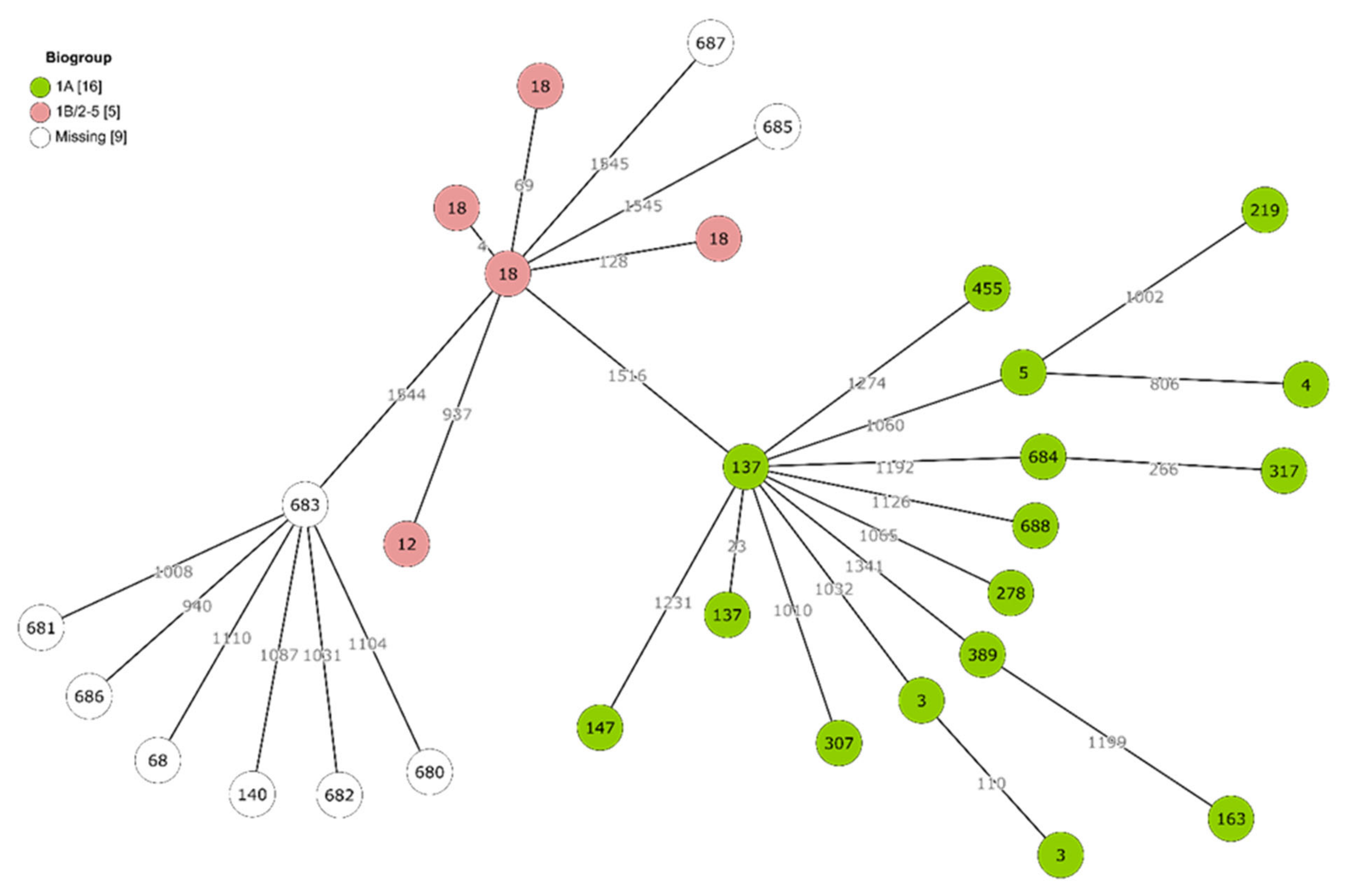
2.3. Genetic Diversity and Virulence of Yersinia Isolates
3. Discussion
4. Materials and Methods
4.1. Sampling
4.2. Microbiological Testing of Samples
4.3. Detection of Biotypes and Serogroups of Yersinia Enterocolitica
4.4. Detection of Antimicrobial Resistance of Pathogenic Yersinia Enterocolitica
4.5. Screening of Pathogenicity of Yersinia Enterocolitica with qPCR
4.6. Genome Sequencing and Analysis
4.7. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bottone, E.J. Yersinia enterocolitica: Revisitation of enduring human pathogen. Clin. Microbiol. Newsl. 2015, 37, 1–8. [Google Scholar] [CrossRef]
- Hammerl, J.A.; Barac, A.; Erben, P.; Fuhrmann, J.; Gadicherla, A.; Kumsteller, F.; Lauckner, A.; Müller, F.; Hertwig, S. Properties of two broad host range phages of Yersinia enterocolitica isolated from wild animals. Int. J. Mol. Sci. 2021, 22, 11381. [Google Scholar] [CrossRef] [PubMed]
- Rosner, B.M.; Stark, K.; Werber, D. Epidemiology of reported Yersinia enterocolitica infections in Germany, 2001–2008. BMC Public Health 2010, 10, 337. [Google Scholar] [CrossRef] [Green Version]
- Le Guern, A.S.; Martin, L.; Savin, C.; Carniel, E. Yersiniosis in France: Overview and potential sources of infection. Int. J. Infect. Dis. 2016, 46, 1–7. [Google Scholar] [CrossRef] [Green Version]
- European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and CDC). The European Union One Health 2019 Zoonoses Report. EFSA J. 2021, 19, e06406. [Google Scholar] [CrossRef]
- Cornelis, G.R.; Boland, A.P.; Boyn, A.P.; Geuijen, C.; Iriarte, M.; Neyt, C.; Sory, M.P.; Stainier, I. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 1998, 62, 1315–1352. [Google Scholar] [CrossRef] [Green Version]
- Howard, S.L.; Gaunt, M.W.; Hinds, J.; Witney, A.A.; Stabler, R.; Wren, B.W. Application of comparative phylogenomics to study the evolution of Yesinia enterocolitica and to identify genetic differences relating to pathogenicity. J. Bacteriol. 2006, 188, 3645–3653. [Google Scholar] [CrossRef] [Green Version]
- Tennant, S.M.; Grant, T.H.; Robins-Browne, R.M. Pathogenicity of Yersinia enterocolitica biotype 1A. FEMS Immunol. Med. Microbiol. 2003, 38, 127–137. [Google Scholar] [CrossRef] [Green Version]
- Murros, A.; Säde, E.; Johansson, P.; Korkeala, H.; Fredriksson-Ahomaa, M.; Björkroth, J. Characterization of European Yersinia enterocolitica 1A strains using restriction fragment length polymorphism and multilocus sequence analysis. Lett. Appl. Microbiol. 2016, 63, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Laukkanen-Ninious, R.; Fredriksson-Ahomaa, M.; Korkeala, H. Enteropathogenic Yersinia in the pork production chain: Challenges for control. Compr. Rev. Food Sci. Food Saf. 2014, 13, 1165–1191. [Google Scholar] [CrossRef]
- Bari, M.L.; Hossain, H.A.; Isshiki, K.; Ukuku, D. Behavior of Yersinia enterocolitica in foods. J. Pathog. 2011, 420732. [Google Scholar] [CrossRef] [Green Version]
- Van Damme, I.; Berkvens, D.; Vanantwerpen, G.; Baré, J.; Houf, K.; Wauters, G.; De Zutter, L. Contamination of freshly slaughtered pig carcasses with enteropathogenic Yersinia spp.: Distribution, quantification and identification of risk factors. Int. J. Food Microbiol. 2015, 204, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Bonardi, S.; Paris, A.; Bassi, L.; Salmi, F.; Bacci, C.; Riboldi, E.; Boni, E.; D’Incau, M.; Tagliabue, S.; Brindani, F. Detection, semiquantitative enumeration, and antimicrobial susceptibility of Yersinia enterocolitica in pork and chicken meats in Italy. J. Food Prot. 2010, 73, 1785–1792. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson-Ahomaa, M.; Koch, U.; Bucher, M.; Stolle, A. Different genotypes of Yersinia enterocolitica 4/O:3 strains widely distributed in butchers shops in the Munich area. Int. J. Food Microbiol. 2004, 95, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Lorencova, A.; Slany, M. Prevalence of pathogenic Yersinia enterocolitica in minced meat, pig tongues and hearts at the retail level in the Czech Republic detected by real time PCR. Potravinarstvo 2016, 10, 282–286. [Google Scholar] [CrossRef]
- Laukkanen-Ninios, R.; Fredriksson-Ahomaa, M.; Maijala, R.; Korkeala, H. High prevalence of pathogenic Yersinia enterocolitica in pig cheeks. Food Microbiol. 2014, 43, 5–52. [Google Scholar] [CrossRef] [PubMed]
- Lucero-Estrada, C.S.M.; Favier, G.I.; Escudero, M.E. An overview of Yersinia enterocolitica and related species in samples of different origin from San Luis, Argentina. Food Microbiol. 2020, 86, 103345. [Google Scholar] [CrossRef]
- Guillier, L.; Fravalo, P.; Leclercq, A.; Thébault, A.; Kooh, P.; Cadavez, V.; Gonzales-Barron, U. Risk factors for sporadic Yersinia enterocolitica infections: A systematic review and meta-analysis. Microb. Risk Anal. 2021, 17, 100141. [Google Scholar] [CrossRef]
- Morka, K.; Wałecka-Zacharska, E.; Schubert, J.; Dudek, B.; Woźniak-Biel, A.; Kuczkowski, M.; Wieliczko, A.; Bystrón, J.; Bania, J.; Bugla-Płoskońska, G. Genetic diversity and distribution of virulence-associated genes in Y. enterocolitica and Y. enterocolitica-like isolates from humans and animals in Poland. Pathogens 2021, 10, 65. [Google Scholar] [CrossRef]
- Bancerz-Kisiel, A.; Pieczywek, M.; Łada, P.; Szweda, W. The most important virulence markers of Yersinia enterocolitica and their role during infection. Genes 2018, 9, 235. [Google Scholar] [CrossRef] [Green Version]
- ISO 18867:2015; Microbiology of the Food Chain–Polymerase Chain Reaction–Detection of Pathogenic Yersinia enterocolitica and Yersinia pseudotuberculosis; International Organization of Standartization (ISO): Geneva, Switzerland, 2015.
- Imori, P.F.M.; Passaglia, J.; Souza, R.A.; Rocha, L.B.; Falcao, J.P. Virulence-related genes, adhesion and invasion of some Yersinia enterocolitica-like strains suggests its pathogenic potential. Microb. Pathog. 2017, 104, 72–77. [Google Scholar] [CrossRef]
- Kraushaar, B.; Dieckmann, R.; Wittwer, M.; Knabner, D.; Konietzny, A.; Mäde, D.; Strauch, E. Characterization of a Yersinia enterocolitica biotype 1A strain harbouring an ail gene. J. Appl. Microb. 2011, 111, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Ashton, P.M.; Nair, S.; Peters, T.M.; Bale, J.A.; Powell, D.G.; Painset, A.; Tewolde, R.; Schaefer, U.; Jenkins, C.; Dallman, T.J.; et al. Identification of Salmonella for public health surveillance using whole genome sequencing. PeerJ 2016, 4, e1752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inns, T.; Flanagan, S.; Greig, D.R.; Jenkins, C.; Seddon, K.; Chin, T.; Cartwright, J. First use of whole-genome sequencing to investigate a cluster of Yersinia enterocolitica, Liverpool, United Kingdom, 2017. J. Med. Microbiol. 2018, 67, 1747–1752. [Google Scholar] [CrossRef]
- Hunter, E.; Greig, D.R.; Schaefer, U.; Wright, M.J.; Dallman, T.J.; McNally, A.; Jenkins, C. Identification and typing of Yersinia enterocolitica and Yersinia pseudotuberculosis isolated from human clinical specimens in England between 2004 and 2018. J. Med. Microbiol. 2018, 68, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Özdemir, F.; Arslan, S. Genotypic and phenotypic virulence characteristics and antimicrobial resistance of Yersinia spp. isolated from meat and milk products. J. Food Sci. 2015, 80, 1306–1313. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.K.; Ooi, P.T.; Thong, K.L. Prevalence of Yersinia enterocolitica from food and pigs in selected states of Malaysia. Food Control 2014, 35, 94–100. [Google Scholar] [CrossRef]
- Zadernowska, A.; Chajęcka-Wierzchowska, W. Prevalence, biofilm formation and virulence markers of Salmonella sp. and Yersinia enterocolitica in food of animal origin in Poland. LWT-Food Sci. Technol. 2017, 75, 552–556. [Google Scholar] [CrossRef]
- Younis, G.; Mady, M.; Awad, A. Yersinia enterocolitica: Prevalence, virulence, and antimicrobial resistance from retail and processed meat in Egypt. Vet. World 2019, 12, 1078–1084. [Google Scholar] [CrossRef]
- Syczyło, K.; Platt-Samoraj, A.; Bancerz-Kisiel, A.; Szczerba-Turek, A.; Pajdak-Czaus, J.; Łabuć, S.; Procajło, Z.; Socha, P.; Chuzhebayeva, G.; Szweda, W. The prevalence of Yersinia enterocolitica in game animals in Poland. PLoS ONE 2018, 13, e0195136. [Google Scholar] [CrossRef]
- McNally, A.; Cheasty, T.; Fearnley, C.; Dalziel, R.W.; Paiba, G.A.; Manning, G.; Newell, D.G. Comparison of the biotypes of Yersinia enterocolitica isolated from pigs, cattle and sheep at slaughter and from humans with yersiniosis in Great Britain during 1999–2000. Lett. Appl. Microbiol. 2004, 39, 103–108. [Google Scholar] [CrossRef]
- Bonardi, S.; Paris, A.; Bacci, C.; D’Incau, M.; Ferroni, L.; Brindani, F. Detection and characterization of Yersinia enterocolitica from pigs and cattle. Vet. Res. Commun. 2007, 31, 347–350. [Google Scholar] [CrossRef]
- Longenberger, A.H.; Gronostaj, M.P.; Yee, G.Y.; Johnson, L.M.; Lando, J.F.; Voorhees, R.E.; Waller, K.; Weltman, A.C.; Moll, M.; Lyss, S.B.; et al. Yersinia enterocolitica infections associated with improperly pasteurized milk products: Southwest Pennsylvania, March-August, 2011. Epidemiol. Infect. 2014, 142, 1640–1650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonardi, S.; Le Guern, A.S.; Savin, C.; Pupillo, G.; Bolzoni, L.; Cavalca, M.; Pongolini, S. Detection, virulence and antimicrobial resistance of Yersinia enterocolitica in pulk tank milk in Italy. Int. Dairy J. 2018, 84, 46–53. [Google Scholar] [CrossRef]
- Liang, J.; Duan, R.; Xia, S.; Hao, Q.; Yang, J.; Xiao, Y.; Qui, H.; Shi, G.; Wand, S.; Gu, W.; et al. Ecology and geographic distribution of Yersinia enterocolitica among livestock and wildlife in China. Vet. Microbiol. 2015, 178, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson-Ahomaa, M.; Stolle, A.; Korkeala, H. Molecular epidemiology of Yersinia enterocolitica infections. FEMS Immunol. Med. Microbiol. 2006, 47, 315–329. [Google Scholar] [CrossRef] [Green Version]
- Grahek-Ogden, D.; Schimmer, B.; Cudjoe, K.S.; Nygård, K.; Kapperud, G. Outbreak of Yersinia enterocolitica serogroup O:9 infection and processed pork, Norway. Emerg. Infect. Dis. 2007, 13, 754–756. [Google Scholar] [CrossRef]
- Van Damme, I.; Berkvens, D.; Botteldoorn, N.; Dierick, K.; Wits, J.; Pochet, B.; De Zutter, L. Evaluation of the ISO 10273:2003 method for the isolation of human pathogenic Yersinia enterocolitica from pig carcasses and minced meat. Food Microbiol. 2013, 36, 170–175. [Google Scholar] [CrossRef]
- Peruzy, M.F.; Aponte, M.; Proroga, Y.T.R.; Capuano, F.; Cristiano, D.; Delibato, E.; Houf, K.; Murru, N. Yersinia enterocolitica detection in pork products: Evaluation of isolation protocols. Food Microbiol. 2020, 92, 103593. [Google Scholar] [CrossRef] [PubMed]
- Messelhäusser, U.; Kämpf, P.; Colditz, J.; Bauer, H.; Schreiner, H.; Höller, C.; Busch, U. Qualitative and quantitative detection of human pathogenic Yersinia enterocolitica in different food matrices at retail level in Bavaria. Foodborne Pathog. Dis. 2011, 8, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Terentjeva, M.; Bērziņš, A. Prevalence of Yersinia enterocolitica 4/O:3 in raw pork at retail market in Latvia. Arch. Lebensmittelhyg 2013, 64, 125–156. [Google Scholar] [CrossRef]
- Shoaib, M.; Shehzad, A.; Raza, H.; Niazi, S.; Khan, I.M.; Akhtar, W.; Safdar, W.; Wand, Z. A comprehensive review of the prevalence, pathogenesis and detection of Yersinia enterocolitica. RSC Adv. 2019, 70, 41010–41021. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Liu, M.; Wang, H.; Wu, Q.; Xu, T.; Ma, G.; Zhong, Y.; Zhang, J.; Chen, M.; Zue, L.; et al. Occurrence, molecular characterization, and antimicrobial susceptibility of Yersinia enterocolitica isolated from retail food samples in China. LWT 2021, 150, 111876. [Google Scholar] [CrossRef]
- Felek, S.; Krukonis, E.S. The Yersinia pestis Ail protein mediates binding and Yop delivery to host cells required for plague virulence. Infect. Immun. 2009, 77, 825–836. [Google Scholar] [CrossRef] [Green Version]
- Platt-Samoraj, A.; Kończyk-Kmiecik, K.; Bakuła, T. Occurrence and genetic correlations of Yersinia spp. isolated from commensal rodents in Northeastern Poland. Pathogens 2021, 10, 1247. [Google Scholar] [CrossRef] [PubMed]
- Novoslavskij, A.; Kudirkiené, E.; Marcinkuté, A.; Bajoriūniené, A.; Korkeala, H.; Malakauskas, M. Genetic diversity and antimicrobial resistance of Yersinia enterocolitica isolated from pigs and humans in Lithuania. J. Sci. Food Agric. 2013, 93, 1858–1862. [Google Scholar] [CrossRef] [PubMed]
- Bonardi, S.; Bruini, I.; D’Incau, M.; VanDamme, I.; Carniel, E.; Brémont, S.; Cavallini, P.; Tagliabue, S.; Brindani, F. Detection, seroprevalence and antimicrobial resistance of Yersinia enterocolitica and Yersinia pseudotuberculosis in pig tonsils in Northern Italy. Int. J. Food Microbiol. 2016, 235, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Wu, Q.; Hu, H.; Zhang, J.; Huang, H. Prevalence and characterization of Yersinia enterocolitica isolated from retail foods in China. Food Control 2016, 61, 20–27. [Google Scholar] [CrossRef]
- Bonke, R.; Wacheck, S.; Stüber, E.; Meyer, C.; Märlbauer, E.; Fredriksson-Ahomaa, M. Antimicrobial susceptibility and distribution of β-lactamase A(blaA) and β-lactamase B (blaB) genes in enteropathogenic Yersinia species. Microb. Drug Resist. 2011, 17, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Seoane, A.; García Lobo, J.M. Identification of a streptogramin a acetyltransferase gene in the chromosome of Yersinia enterocolitica. Antimicrob. Agents Chemother. 2000, 44, 905–999. [Google Scholar] [CrossRef] [Green Version]
- Bolton, D.J.; Ivory, C.; McDowell, D. A small study of Yersinia enterocolitica in pigs from birth to carcass and characterization of porcine and human strains. Food Control 2013, 33, 521–524. [Google Scholar] [CrossRef]
- Karlsson, P.A.; Tano, E.; Jernberg, C.; Hickman, R.A.; Guy, L.; Järhult, J.D.; Wang, H. Molecular characterization of multidrug-resistant Yersinia enterocolitica from foodborne outbreaks in Sweden. Front. Microbiol. 2021, 12, 664665. [Google Scholar] [CrossRef]
- Bhagat, N.; Virgi, J.S. Yersinia enterocolitica 1A correlates with clonal groups and not the source of isolation. FEMS Microbiol. Lett. 2007, 266, 177–183. [Google Scholar] [CrossRef] [Green Version]
- Thong, K.L.; Tan, L.K.; Ooi, P.T. Genetic diversity, virulotyping and antimicrobial resistance susceptibility of Yersinia enterocolitica isolated from pigs and porcine products in Malaysia. J. Sci. Food Agric. 2018, 98, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Rusak, L.A.; Junqueira, R.M.; Hofer, E.; Vallim, D.C.; Asensi, M.D. Next-generation sequencing virulome analysis of a Yersinia enterecolitica subsp. palearctica bioserotype 4/O:3 ST18 isolated from human blood in Brazil. Braz J. Infect. Dis. 2017, 21, 550–553. [Google Scholar] [CrossRef]
- Singh, I.; Virdi, J.S. Production of Yersinia stable toxin (YST) and distribution of yst genes in biotype 1A strains of Yersinia enterocolitica. J. Med. Microb. 2004, 53, 1065–1068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, H.; Sun, Y.; Mao, Z.; Jiang, B. Investigation of virulence genes in clinical isolates of Yersinia enterocolitica. FEMS Immunol. Med. Microbiol. 2008, 53, 368–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garzetti, D.; Susen, R.; Fruth, A.; Tietze, E.; Heesemann, J.; Rakin, A. A molecular scheme for Yersinia enterocolitica patho-serotyping derived from genome-wide analysis. Int. J. Med. Microbiol. 2014, 304, 275–283. [Google Scholar] [CrossRef]
- Bren, A.; Eisenbach, M. How signals are heard during bacterial chemotaxis: Protein-protein interactions in sensory signal propagation. J. Bacteriol. 2000, 182, 6865–6873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.J.; Young, B.M.; Young, G.M. Effect of flagellar mutations on Yersinia enterocolitica biofilm formation. Appl. Environ. Microbiol. 2008, 74, 5466–5474. [Google Scholar] [CrossRef] [Green Version]
- Strydom, H.; Wand, J.; Paine, S.; Dyet, K.; Cullen, K.; Wright, J. Evaluating sub-typing methods for pathogenic Yersinia enterocolitica to support outbreak investigations in New Zealand. Epidemiol. Infect. 2019, 147, e186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ISO 10273:2017; Microbiology of Food and Animal Feed. Horizontal Method for the Detection of Presumably Pathogenic Yersinia enterocolitica; International Organization of Standartization (ISO): Geneva, Switzerland, 2017.
- Wauters, G.; Kandolo, K.; Janssens, M. Revised biogrouping scheme of Yersinia enterocolitica. Contrib. Microbiol. Immunol. 1987, 9, 14–21. [Google Scholar] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoints Tables for Interpretation of MICs and Zones Diameters. Version 11.0. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_11.0_Breakpoint_Tables.pdf (accessed on 8 October 2021).
- Mäde, D.; Reiting, R.; Strauch, E.; Ketteritzsch, K.; Wicke, A. A real-time PCR for detection of pathogenic Yersinia enterocolitica in food combined with an universal internal amplification control system. J. Verbr. Lebensm 2008, 3, 141–151. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prjibelski, A.D.; Puglia, G.D.; Antipov, D.; Bushmanova, E.; Giordano, D.; Mikheenko, A.; Vitale, D.; Lapidus, A. Extending rnaSPAdes functionality for hybrid transcriptome assembly. BMC Bioinform. 2020, 21, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 302. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Alikhan, N.F.; Mohamed, K.; the Agama Study Group; Achtman, M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny and Escherichia core genomic diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, M.; Chattaway, M.A.; Reuter, S.; Savin, C.; Strauch, E.; Carniel, E.; Connor, T.; Van Damme, I.; Rajakaruna, L.; Rajendram, D.; et al. Use of whole-genus genome sequence data to develop a multilocus sequence typing tool that accurately identifies Yersinia isolates to the species and subspecies levels. J. Clin. Microbiol. 2015, 53, 35–42. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Alikhan, N.F.; Sergeant, M.J.; Luhmann, N.; Vaz, C.; Francisco, A.P.; Carrico, J.A.; Achtman, M. GrapeTree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018, 28, 1395–1404. [Google Scholar] [CrossRef] [Green Version]
| Meat Category | Sample Category | No. of Sample | Yersinia spp. | Y. enterocolitica | Y. intermedia | Y. kristensenii | Y. massiliensis | Y. frederiksenii | Y. molaretti |
|---|---|---|---|---|---|---|---|---|---|
| No. of Positive Samples (%) | |||||||||
| Pork | Pork cuts | 160 | 42 (26) | 36 (23) | 4 (3) | 1 (1) | 1 (1) | 2 (1) | 0 (0) |
| Minced pork | 9 | 7 (78) | 3 (33) | 3 (33) | 2 (22) | 1 (11) | 0 (0) | 0 (0) | |
| Offal | 11 | 6 (55) | 5 (45) | 3 (28) | 1 (9) | 0 (0) | 1 (9) | 1 (9) | |
| Beef | Beef cuts | 150 | 24 (16) | 19 (13) | 6 (4) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Total | 330 | 79 (24) | 63 (19) b | 16 (5) | 4 (1) | 2 (1) | 3 (1) | 1 (1) | |
| Agent | MIC Resistance Breakpoint (mg/L) | Identified MIC (mg/L) Range | No. of Resistant Isolates (%) |
|---|---|---|---|
| Ampicillin | 8 | 16–64 | 6 (100) |
| Azithromycin | NA | <2–4 | NA |
| Cefotaxime | 2 | <0.25 | 0 (0) |
| Ceftazidime | 4 | <0.5 | 0 (0) |
| Ciprofloxacin | 0.5 | <0.015 | 0 (0) |
| Chloramphenicol | 8 | <8 | 0 (0) |
| Colistin | 2 | <1 | 0 (0) |
| Gentamicin | 2 | <0.5 | 0 (0) |
| Meropenem | 8 | <0.03 | 0 (0) |
| Nalidixic acid | NA | <4 | NA |
| Tetracycline | 4 | <2 | 0 (0) |
| Tigecycline | 0.5 | <0.25 | 0 (0) |
| Trimetoprim | 4 | 0.5–2 | 0 (0) |
| Sulfametoxazole | NA | <8–16 | NA |
| Y. enterocolitica | Y. frederiksenii | Y. intermedia | Y. kristensenii | |
|---|---|---|---|---|
| 1A | 1B/2-5 | |||
| ST (No. of Isolates) | ||||
| 3 (2) | 12 (1) | 685 (1) a | 68 (1) | 687 (1) a |
| 4 (1) | 18 (4) | 140 (1) | ||
| 137 (2) | 680 (1) a | |||
| 147 (1) | 681 (1) a | |||
| 163 (1) | 682 (1) a | |||
| 219 (1) | 683 (1) a | |||
| 278 (1) | 686 (1) a | |||
| 307 (1) | ||||
| 317 (1) | ||||
| 389 (1) | ||||
| 455 (1) | ||||
| 684 (1) | ||||
| 688 (1) | ||||
| Yersinia Species | ST | Virulence Genes | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ail | fepD | fes | hreP | inv | myfA | myfB | myfC | sat | virF | yadA | ymoA | ystA | ystB | blaA | blaB | ||
| Y. enterocolitica | 12 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 |
| 18 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | |
| 3 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | |
| 4 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | |
| 137 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | |
| 147 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | |
| 163 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | |
| 219 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | |
| 278 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | |
| 307 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | |
| 317 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | |
| 389 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | |
| 455 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | |
| 684 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | |
| 688 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | |
| Y. frederiksenii | 685 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 |
| Y. intermedia | All STs | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 |
| Y. kristensenii | 687 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Terentjeva, M.; Ķibilds, J.; Meistere, I.; Gradovska, S.; Alksne, L.; Streikiša, M.; Ošmjana, J.; Valciņa, O. Virulence Determinants and Genetic Diversity of Yersinia Species Isolated from Retail Meat. Pathogens 2022, 11, 37. https://doi.org/10.3390/pathogens11010037
Terentjeva M, Ķibilds J, Meistere I, Gradovska S, Alksne L, Streikiša M, Ošmjana J, Valciņa O. Virulence Determinants and Genetic Diversity of Yersinia Species Isolated from Retail Meat. Pathogens. 2022; 11(1):37. https://doi.org/10.3390/pathogens11010037
Chicago/Turabian StyleTerentjeva, Margarita, Juris Ķibilds, Irēna Meistere, Silva Gradovska, Laura Alksne, Madara Streikiša, Jevgēnija Ošmjana, and Olga Valciņa. 2022. "Virulence Determinants and Genetic Diversity of Yersinia Species Isolated from Retail Meat" Pathogens 11, no. 1: 37. https://doi.org/10.3390/pathogens11010037
APA StyleTerentjeva, M., Ķibilds, J., Meistere, I., Gradovska, S., Alksne, L., Streikiša, M., Ošmjana, J., & Valciņa, O. (2022). Virulence Determinants and Genetic Diversity of Yersinia Species Isolated from Retail Meat. Pathogens, 11(1), 37. https://doi.org/10.3390/pathogens11010037







