Abstract
Yersinia enterocolitica is an important foodborne pathogen, and the determination of its virulence factors and genetic diversity within the food chain could help understand the epidemiology of yersiniosis. The aim of the present study was to detect the prevalence, and characterize the virulence determinants and genetic diversity, of Yersinia species isolated from meat. A total of 330 samples of retailed beef (n = 150) and pork (n = 180) in Latvia were investigated with culture and molecular methods. Whole genome sequencing (WGS) was applied for the detection of virulence and genetic diversity. The antimicrobial resistance of pathogenic Y. enterocolitica isolates was detected in accordance with EUCAST. Yersinia species were isolated from 24% (79/330) of meats, and the prevalence of Y. enterocolitica in pork (24%, 44/180) was significantly higher (p < 0.05) than in beef (13%, 19/150). Y. enterocolitica pathogenic bioserovars 2/O:9 and 4/O:3 were isolated from pork samples (3%, 6/180). Only resistance to ampicillin was confirmed in Y. enterocolitica 4/O:3 and 2/O:9 isolates, but not in other antimicrobials. Major virulence determinants, including ail, inv, virF, ystA and myfA, were confirmed with WGS in Y. enterocolitica 2/O:9 and 4/O:3. MLST typing revealed 15 STs (sequence types) of Y. enterocolitica with ST12 and ST18, which were associated with pathogenic bioserovars. For Y. enterocolitica 1A, Y. kristensenii, Y. intermedia and Y. frederiksenii, novel STs were registered (ST680-688). The presence of virulence genes and genetic characteristics of certain Y. enterocolitica STs confirm the common knowledge that pork could be an important source of pathogenic Yersinia.
Keywords:
Yersinia enterocolitica; prevalence; antimicrobial resistance; pork; WGS; cgMLST; virulence factors; Latvia 1. Introduction
The Yersinia genus currently consists of 28 species, of which three are human pathogenic, while others are considered as non-pathogenic, Yersinia-like microorganisms [1,2]. Pathogenic Yersinia enterocolitica and Yersinia pseudotuberculosis are reported to cause yersiniosis, which is a zoonotic foodborne infection characterized by gastrointestinal manifestations, and post-infection sequelas, such as reactive arthritis or erythema nodosum [3,4]. Yersiniosis is reported to be the fourth most common bacterial zoonosis within the European Union [5].
Y. enterocolitica is a very heterogeneous species and is divided into six biotypes and various serogroups with different bioserovars showing distinctive virulence properties, hosts and geographical distribution [1]. Y. enterocolitica biotype 1A is non-pathogenic since it lacks classical virulence markers, which are important for the invasion of the human host and survival in the organisms [4,6,7]. Non-pathogenic Yersinia and Y. enterocolitica are widely distributed in the environment, animals and food and were isolated from clinical patients [8,9]. Y. enterocolitica biotypes 1B-5 are pathogenic, and bioserovars of 2/O:5,27, 2/O:9, 3/O:3 and 4/O:3 were recorded in clinical cases in Europe [1].
Pathogenic Y. enterocolitica were reported to be present in animal hosts, although they were rarely associated with meats other than pork [10,11]. Pigs are suspected to be important carriers of pathogenic Yersinia, and the contamination of pork may occur during slaughter as a result of cross-contamination [12]. Pathogenic Yersinia were identified in pig carcasses at the slaughterhouses, meat processing environment and at the retail [10,13]. Pathogenic Y. enterocolitica has been frequently isolated from pork—retail cuts, minced pork, offal and pork sausages—with the majority of isolated strains belonging to the same bioserotypes that were identified in pigs—4/O:3 [13,14,15,16,17]. Undercooked pork meat has been significantly associated with sporadic yersiniosis cases, but the genetic similarity between the human and porcine isolates indicates transmission through the pork production chain [10,18,19]. Thus, studies on the prevalence of pathogenic Yersinia species in meats are important for the recognition of foodborne transmission and the assessment of the distribution within the food chain.
Pathogenic Y. enterocolitica carry both chromosomal (ail, invA and ystA) and plasmid-borne (plasmid of Yersinia virulence, pYV) genes, e.g., yadA and virF, which are required for full virulence [20]. The present, widely recognized methodology to differentiate between non-pathogenic and pathogenic Yersinia species mostly relies on the detection of the ail gene (adhesion and invasion locus) [21]. Notwithstanding, the presence of virulence markers, including the ail gene, was reported in non-pathogenic Yersinia species and Y. enterocolitica 1A isolates [22]. Therefore, the characterization of virulence factors in Yersinia isolates is important for an understanding of the pathogenicity potential of the Yersinia species as different Y. enterocolitica virulotypes and virulence traits could be established in Yersinia species [19,23].
New advances in food safety research show that the application of novel microbial typing methods as whole genome sequencing (WGS) may contribute to the knowledge on the virulence and phylogenetic relationships of the microbial isolates of public health importance [24]. The highly discriminatory approach provided by the WGS is crucial for surveillance, epidemiological investigations of yersiniosis and the virulence assessment of Yersinia species and may provide a new insight into the epidemiology of Yersinia in the food chain [25,26]).
Since there is limited information on the virulence characteristics and genetic diversity of the Yersinia species in meat, the aim of the present study was to investigate the prevalence, characterize virulence factors and describe the genetic diversity of Yersinia isolates recovered from retail meats.
2. Results
2.1. Prevalence of Yersinia spp. and Pathogenic Yersinia Enterocolitica Bioserovars in Meats
The overall prevalence of Yersinia spp. in meats was 24% (79/330). One to three Yersinia spp. were found in one investigated sample. The highest number of Yersinia was found in pork cuts with five isolated species: Y. enterocolitica (23%, 36/160), Y. intermedia (3%, 4/160), Y. kristensenii (1%, 1/160) and Y. frederiksenii (2%, 1/160). The lowest diversity of the Yersinia species was recovered from beef, where 19% (13/150) of Y. enterocolitica- and 4% (6/150) of Y. intermedia-positive samples were identified. The prevalence of Y. enterocolitica in meats was higher than the prevalence of other Yersinia species (p < 0.05) (Table 1).

Table 1.
Prevalence of Yersinia species in meats at the retail market.
A significantly higher prevalence of Y. enterocolitica of 24% (44/180) was identified in pork in comparison to 13% (19/150) (p < 0.05) in beef. In addition to meat categories, the highest prevalence of the Yersinia species of 55% (6/11) and Y. enterocolitica of 45% (5/11) was detected in offal, while the lowest was detected in beef cuts of 16% (24/150) and 19% (13/150), respectively (Table 1).
Out of the 63 Y. enterocolitica-positive samples, five belonged to bioserovar 4/O:3, one to 2/O: 9 and 57 to biotype 1A. The presence of the ail gene was confirmed in all Y. enterocolitica 4/O:3 and 2/O:9 isolates with qPCR (Supplementary Table S3).
2.2. Antimicrobial Resistance in Yersinia enterocolitica 2/O:9 and 4/O:3 Isolates
Antimicrobial resistance against ampicillin was identified in 100% of Y. enterocolitica 4/O:3 and 2/O:9. All Y. enterocolitica 4/O:3 and 2/O:9 isolates were susceptible to cefotaxime, ceftazidime, ciprofloxacin, chloramphenicol, colistin, gentamicin, meropenem, tetracycline and trimethoprim (Table 2). Differences between the antimicrobial resistance pattern of Y. enterocolitica of biotypes 4/O:3 and 2/O:9 were not found.

Table 2.
Antimicrobial resistance in Yersinia enterocolitica 4/O:3 and 2/O:9 isolates.
2.3. Genetic Diversity and Virulence of Yersinia Isolates
MLST sequence types were identified for all sequenced isolates. Among these, nine novel STs were identified and registered in Enterobase (ST680-ST688). Most of the novel STs were from non-enterocolitica species.
Based on WGS data analysis, 15 STs of Y. enterocolitica were identified where all pathogenic 4/O:3 isolates belonged to ST18 but all 2/O:9 isolates belonged to ST12. One Y. enterocolitica 4/O:3 was excluded from WGS analysis due to contamination (Supplementary Table S3). Non-pathogenic Y. enterocolitica belonged to 13 STs, and one to two isolates of each ST were recovered (Table 3). Each isolate of Y. frederiksenii, Y. intermedia and Y. kristensenii represented one ST (Table 3). All but one isolates originated from pork, while ST137 was identified in beef.

Table 3.
Sequence types (STs) of Yersinia isolates found in meat samples.
The genetic structure of the Yersinia population was explored in more detail with the whole-genome multilocus sequence typing (cgMLST) approach, which is based on 1553 loci (Figure 1). On average, >1000 allelic differences separated individual isolates. No dense clusters of genotypes could be observed. Instead, they appeared to be scattered with large distances between them, with exceptions when multiple isolates shared the same ST (e.g., multiple strains representing ST3, ST18 and ST137).
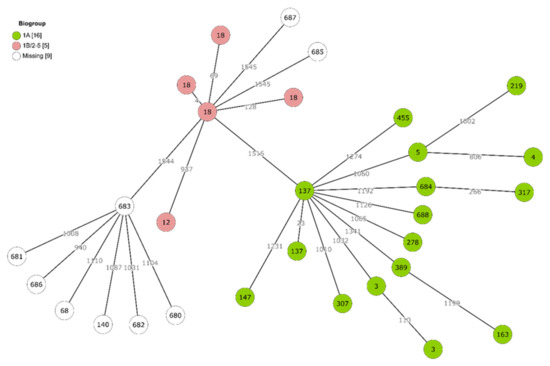
Figure 1.
Minimum spanning tree of Yersinia cgMLST profiles: Branch lengths are drawn in log scale. For each node, MLST sequence type number is indicated. Coloured nodes represent virulent or non-virulent Y. enterocolitica biotypes that were determined based on presence of ail, inv, ystA and ystB genes. Uncoloured nodes represent non-enterocolitica species for which this biotype determination was not applicable.
The most common virulence determinants in all Yersinia species were ymoA (100%) followed by fepD and fes. All Y. enterocolitica harboured hreP, inv, myfB, myfC, sat and ymoA virulence genes. Out of pathogenic Y. enterocolitica, ST18 was the only fepD- and fes-negative ST, but shared ail, hreP, inv, myfA, myfB, myfC, sat, virF, yadA, ymoA and ystA (Table 4, Supplementary Table S1). Y. enterocolitica ST12 contained all virulence factors of ST18, with the exception of yadA, and was fepD and fes positive.

Table 4.
Distribution of major virulence determinants in Yersinia species isolated from meats.
Based on the combined presence or absence of ail, inv, ystA and ystB virulence genes, Y. enterocolitica strains could be classified as virulent or non-virulent biotypes. Only two STs (ST12 and ST18) were represented among the virulent biotypes 1B/2–5. Many more isolates and a wide range of STs were classified as the non-virulent 1A biotype (Figure 1).
Limited diversity in virulence was observed between Y. enterocolitica 1A STs and was related to the presence of myfA in ST 317, 389, 684 and 688, and the absence of fepD and fes in ST688.
All Yersinia species other than Y. enterocolitica shared fepD and ymoA, and all were lacking ail, inv, myfA, myfB, myfC, virF, yadA and ystB. Y. kristensenii isolates harboured the ystA gene. Differences between the distribution of virulence factors among Y. intermedia STs were not observed (Table 4). The virulence determinants of mobility (flgA-flgN, flhA-flhE, fliA-fliT and fliZ), chemotaxis mechanisms (cheA, cheB, cheD, cheR, cheW, cheY and cheZ) and genes that encode flagellar motor proteins (motA and motB) were found in all Y. enterocolitica isolates (Supplementary Table S1).
3. Discussion
The contamination of retailed meats with the Yersinia species (24%) with Y. enterocolitica being predominant was consistent with previous findings [13,27]. The prevalence of Y. enterocolitica in beef and pork in our report was higher than that previously reported in Malaysia, Poland, Italy and Egypt [13,28,29,30]. Yersinia are psychrotrophic microorganisms, and temperate climatic conditions, including a cold winter season, may enhance the survival of Yersinia species in animals and the environment [31]. The unhygienic handling of meat may facilitate the spread of Yersinia species, leading to a higher prevalence at the retail level [14].
Only pork was found to be contaminated by pathogenic Y. enterocolitica 4/O:3 and 2/O:9, while all isolates from beef belonged to non-pathogenic biotype 1A. Pathogenic Y. enterocolitica bioserovars (3/O:5,27 and 3/O:9) were identified in cattle [32,33]. Since pathogenic Y. enterocolitica (2/O:5,27) was found in bulk milk at dairy farms, and improperly treated pasteurized milk, contaminated with Y. enterocolitica, was reported to be the source of yersiniosis outbreak, beef and cattle could be involved in the epidemiology of yersinosis [34,35]. Liang et al. [36] concluded that cattle may act as occasional hosts, while domestic pigs could be the principal reservoir. Pathogenic bioserovars, especially 4/O:3, were often isolated from slaughtered pigs in Europe, being exclusively predominated in pigs from Belgium, Germany and Finland [10,11,12,13,14,16]. The identification of identical genotypes of Y. enterocolitica in pigs and retail pork and human isolates confirms their importance in the epidemiology of human yersiniosis [37]. Y. enterocolitica 4/O:3 was identified as an important source of sporadic yersiniosis, and Y. enterocolitica 2/O:9 was involved in the yersiniosis outbreak in Norway related to undercooked pork meat consumption [18,38]. Since Y. enterocolitica 4/O:3 and O:9 were identified at the retail level, this indicates public health implications as contaminated pork could represent a risk for consumers.
The low recovery of pathogenic Y. enterocolitica from foods was linked to the low sensitivity of the conventional detection methods due to the application of ISO 10273:2017 for food testing and the poor ability of Y. enterocolitica to compete with background microbiota [39]. Y. enterocolitica 4/O:3 counts of 10–102 cfu/g were undetected, while non-pathogenic Y. enterocolitica 1A was accurately identified in experimentally contaminated pork cuts [40]. Reported widespread occurrence of Y. enterocolitica 1A was in agreement with previous studies [13,29].
The ail gene (adhesion and invasion locus) was identified in 3% (6/180) of Y. enterocolitica-positive pork samples with qPCR. All ail-positive Y. enterocolitica isolates belonged to 4/O:3 and 2/O:9 bioserovars. This was in line with previous findings, where the prevalence of pathogenic Y. enterocolitica in pork varied from 0% (0/96) in Poland to 10% (46/446) in Germany, detected using a culture method [29,41]. A higher prevalence of the pathogen was recovered when the combination of ISO 10273 and ail-based qPCR methods was applied [13,28,29,41,42,43,44]. In 11 isolates of Y. enterocolitica 1A, Y. intermedia and Y. kristensenii, Cts > 35 was identified, which was later confirmed as ail negative using WGS. The ail gene is crucial for the adhesion and invasion of the pathogen to the host cell and provides serum resistance, thus making it important for the pathogenesis of yersiniosis [45]. The ail gene is widely targeted to confirm Y. enterocolitica pathogenicity [23]. Previous reports show the sporadic presence of the ail gene in Y. enterocolitica 1A and other Yersinia species in clinical, animal and food samples, raising debates regarding its significance in epidemiology of human yersiniosis [13,23,26,46].
The observed high antimicrobial resistance rates in Y. enterocolitica 4/O:3 and 2/O:9 pork isolates against ampicillin (100%) were in agreement with the 100% reported in Y. enterocolitica 4/O:3 isolates from pigs in Lithuania and Italy [47,48] and Y. enterocolitica 1A from foods in China [49]. Y. enterocolitica was reported to be naturally resistant to ampicillin and other beta-lactam and streptogramin antibiotics due to the presence of vat(F), blaA and blaB genes [50,51]. The presence of blaA and blaB genes in non-pathogenic and pathogenic Y. enterocolitica, as well as in other Yersinia species, was in line with previous reports (49,50). Additionally, resistance to neomycin, streptomycin, tetracycline, chloramphenicol, cephalosporins and carbapenems was reported, which indicates the potential for the development of antimicrobial resistance in Y. enterocolitica. The occurrence of antimicrobial resistance in Y. enterocolitica in meats may be attributed to applications in animals; thus, the antimicrobial resistance in Y. enterocolitica should be monitored [49,52,53].
ymoA, fepD and fes genes were the most common in the Yersinia species, while Y. enterocolitica harbored hreP, inv, myfB, myfC, sat and ymoA virulence genes. ymoA (modulator of the expression of virulence function) was identified in 100% of Y. enterocolitica isolated previously [19]. hreP, fepD, sat and fes genes were mostly associated with Y. enterocolitica 1A [44]. Occasionally, those genes were reported in pathogenic isolates, e.g., fepD (enrochelin ABC transporter) in Y.enterocolitica 1B/O:8 or sat (streptogramin acetyltransferase) in 1B/O:8, 4/O:3 and 3/O:3 bioserovars [19,54,55].
In the present work, the presence of inv, ail, ystA, virF, mufA, myfB, myfC and yop virulon was confirmed in all pathogenic Y. enterocolitica, with the exception of yadA in the 2/O:9 bioserovar. Plasmid and chromosomal virulence genes are important for the full pathogenicity of Y. enterocolitica. yadA and virF are present in pathogenic strains and located at the virulence plasmid, and are crucial for adherence, the transcriptional activity of yop and yadA and invasion into the host cell [20,53]. Strains of pathogenic bioserotype 1B/O:8 from pork were reported to be ail, ystA and virulence plasmid negative due to the apparent loss of pYV [55,56].
Aside from chromosomal virulence factors, inv (invasion), which is responsible for host cell penetration, was present in all Y. enterocolitica [19,49]. yst encodes heat stable endotoxins; however, ystA is usually confirmed in pathogenic Y. enterocolitica and is responsible for diarrhea induction. ystB and ystC are usually expressed in non-pathogenic Y. enterocolitica, but their presence was confirmed in clinical isolates [57,58]. We identified ystA in Y. enterocolitica 2/O:9 and 4/O:3 and ystB in non-pathogenic Y. enterocolitica, which corresponds to previous findings [19,49]. The detection of ail and ystB was proposed for the differentiation of Y. enterocolitica 1A and pathogenic 1B/2-5 biotypes by Garzetti et al., 2014 [59]. Our study confirms the correct identification of pathogenic bioserovars using the WGS approach.
The myfA gene promotes the adhesion of the pathogen to enterocytes and was identified in clinical and animal Y. enterocolitica 4/O:3 isolates and sporadically in Y. enterocolitica 1A isolates. myfB and myfC are encoded by the myf operon and form the fibrillar structure functioning during adhesion, and were associated with pathogenic Y. enterocolitica [19,56]. The main differences between Y. enterocolitica 1A STs in the present study were related to the distribution of myfA, myfB and myfC genes.
fepD, fes, ymoA, ystA and ystB virulence genes were confirmed in Y. kristensenii and ymoA and ystB in Y. intermedia in the present study. Despite lacking classical virulence markers, with the exception of ystA in Y. kristensenii, in the present study, other pathogenicity factors may contribute to Yersinia virulence. ystB of Y. enterocolitica 1A was considered as potentially pathogenic, and high similarity between clinical and rodent isolates of ystB, ail and inv fragments was shown [46]. The presence of virulence genes of clinical importance (ail, myfA and ystA) was identified previously in non-pathogenic Y. enterocolitica and other Yersinia—Y. kristensenii and Y. intermedia [9,39,46].
All characterized Y. enterocolitica, Y. intermedia and Y. kristensenii in the present study shared virulence factors for mobility control, which contribute to invasion, biofilm formation and the secretion system (flg and flh); genes responsible for chemotaxis mechanisms (che); and genes which encode the flagellar motor (mot) protein [60,61]. These genes were described in a clinical isolate of Y. enterocolitica 4/O:3, and authors stated that a variety of virulence factors could contribute to the successful dissemination of Y. enterocolitica 4/O:3 clones globally [56].
Out of the STs associated with pathogenic Y. enterocolitica, ST18 was reported to correspond to 4/O:3 and ST12 to biotype 2-3/O:9 [62]. ST18 was isolated from clinical cases in Sweden, Germany, New Zealand, France, the United Kingdom and Brazil [26,53,56,63]. ST18 was identified in pigs, dogs and bovine sources [53]. In general, the present study confirmed that Y. enterocolitica STs 12 and 18 were associated with pathogenic Y. enterocolitica 4/O:3 and 2/O:9 bioserovars.
Among non-pathogenic Y. enterocolitica 1A and Y. intermedia, a higher degree of diversity was found with fifteen and seven STs identified, respectively. Since ail-negative Y. enterocolitica isolates are usually considered as non-pathogenic and rejected without further analysis, the data on the genetic diversity of Y. enterocolitica 1A are limited. Y. enterocolitica 1A of ST3, ST4, ST137 and ST307 were reported in human cases in England, and ST3 was among the most widespread [26]. This shows that WGS-based techniques may provide new knowledge on the pathogenicity and epidemiology of non-pathogenic and pathogenic Y. enterocolitica isolates since the data on the distribution of the MLST types are more informative for understanding the ecology of Y. enterocolitica in comparison with routine biotyping and serotyping.
In the present study, the WGS methodology facilitated the identification and evaluation of the virulence characteristics of pathogenic Y. enterocolitica strains, and the correct identification of all pathogenic strains of ST18 and ST12 was shown. Additionally, the diversity of Y. enterocolitica 1A and the association of the virulence of pathogenic STs with the presence of key virulence determinants in food isolates were shown.
4. Materials and Methods
4.1. Sampling
A total of 330 samples of raw pork and beef were collected between 2015 and 2021 from 32 retail outlets in Latvia. Raw pork samples (n = 180) included pork cuts, minced pork and offal (tongue, liver and kidney), and for beef (n = 150), beef cuts were selected in supermarkets from the meats available to consumers. From one to three samples from the same producer were purchased at once, aseptically placed in sample transportation containers and immediately delivered on ice to the laboratory. Investigations were started within 2 h after collection.
4.2. Microbiological Testing of Samples
Samples were investigated according to the ISO 10273:2017 [63]. In brief, 25 g of sample was diluted in 225 mL of Peptone Sorbitol Bile (PSB) broth, which was incubated at 25 °C for 44 h. Enriched broth was placed onto Cefsulodin Irgasan Novobiocin (CIN, Biolife, Milan, Italy) agar with and without 0.5% KOH treatment for 20 s; inoculated agars were incubated at 30 °C for 24 h. Suspicious colonies of Yersinia species with red centres and transparent surrounding areas were selected for biochemical confirmation for urea production, sugar fermentation in Triple Sugar Agar (TSI, Biolife) and Decarboxylase Lysine broth (Biolife). After incubation at 30 °C for 24 h, presumed Yersinia species colonies were confirmed via matrix-assisted laser desorption/ionization mass spectrometry (MALDI-TOF, Bruker, Bremen, Germany). Cultures of Yersinia species were stored in 10% glycerol and Brain Heart Infusion (BHI) media at -80 °C until further investigation.
4.3. Detection of Biotypes and Serogroups of Yersinia Enterocolitica
Biotypes were detected according to Wauters [64], and Y. enterocolitica isolates were tested for pyrazimidase and lipase activity, salicine, xylose and trehalose fermentation. Serogroups of Y. enterocolitica were detected with commercially available antisera against O:3, O:5, O:8, O:9 and O:27 according to the manufacturer’s instructions (Sifin, Berlin, Germany).
4.4. Detection of Antimicrobial Resistance of Pathogenic Yersinia Enterocolitica
The antimicrobial resistance of Y. enterocolitica 4/O:3 and 2/O:9 isolates was detected with broth microdilution method using the EUVSEC panel (TREK Diagnostic Systems Ltd., East Grinstead, UK). The bacterial suspension (0.5 McFarland) in 11 mL of cation-adjusted Mueller-Hinton (MH) broth was used for the inoculation of MIC test panels. Inoculated panels were incubated at 30 °C for 24 h. The antimicrobial resistance was tested against ampicillin (1–64 mg/L), cefotaxime (0.25–4 mg/L), ceftazidime (0.5–8 mg/L), meropenem (0.03–16 mg/L), nalidixic acid (4–128 mg/L), ciprofloxacin (0.015–8 mg/L), tetracycline (2–64 mg/L), colistin (1–16 mg/L), gentamicin (0.5–32 mg/L), trimethoprim (0.25–32 mg/L), sulfamethoxazole (8–1024 mg/L), chloramphenicol (8–128 mg/L), azithromycin (2–64 mg/L) and tigecycline (0.25–8 mg/L). The resistance thresholds were interpreted in accordance with EUCAST [65].
4.5. Screening of Pathogenicity of Yersinia Enterocolitica with qPCR
DNA was extracted from fresh cultures using the MagMAX™ Viral/Pathogen II Nucleic Acid Isolation Kit on a KingFisher Flex instrument (ThermoFisher Scientific, Waltham, MA, USA). The ail gene of Yersinia enterocolitica was targeted for the screening of the pathogenicity of Yersinia species. An amount of 2.5 µL was added to 17.5 µL PCR mastermix containing a Luminaris Color Probe qPCR mix (1X) (Thermo Fisher Scientific), 300 nM ail primers (ail-F: 5′-GGT TAT GCA CAA AGC CAT GTA AA-3′, ail-R: 5′-AAA CGA ACC TAT TAC TCC CCA GTT-3′, 93 bp, Bioneer, Daejeon, Korea), 125 nM ail-tmp-probe (5′FAM-AAC CTG AAG TAC CGT TAT GAA CTC GAT GA-BHQ1-3′, 29 bp, Bioneer) and 6.25 µL RNA-free water [66]. The PCR conditions were 50 °C for 2 min, and 95 °C for 10 min, followed by 45 cycles at 95 °C for 10 s and 30 s at 60 °C (QuantStudio 6, ThermoFisher Scientific).
4.6. Genome Sequencing and Analysis
At least one Y. enterocolitica isolate from pork and beef recovered from the same meat sample was chosen for WGS (Supplementary Table S3).
Whole genome sequencing libraries were prepared from the DNA using either a Nextera XT (Illumina, San Diego, CA, USA), Illumina DNA Prep (Illumina) or QIAseq FX (Qiagen, Hilden, Germany) reagent kit. In all library preparation protocols, the final magnetic bead clean-up procedure was modified to select libraries with a longer insert size (approx. 500 bp). The final libraries were sequenced on the MiSeq instrument (Illumina) to yield 2 × 250 or 2 × 300 bp paired-end reads.
The Trimmomatic v0.38 software was used to remove sequencing adapters and low-quality bases from the raw reads [67]. The trimmed reads were then de novo assembled by the SPAdes assembler v3.14.0 [68]. Bacterial species assignment and the presence of contamination were verified by the taxonomic classification of reads against the MiniKraken (v1_8GB_201904) database using Kraken v2.0.8 [69]. Genomes that appeared contaminated, too fragmented (N50 < 10 kb) or were of inappropriate length were excluded from further analysis.
The presence of virulence trait-encoding genes was determined using a BLAST-based approach and gene reference sequences from the Virulence Factor Database [70]. All genes from the Yersina section of VFDB were included, and a few others were added (see Supplementary Table S2). Any gene was considered to be present in the genome if at least 70% of its length was matched with at least 70% nucleotide identity in the contigs (except for ystA and ystB, for which 80% minimum identity was required). Based on the presence of ail, inv, ystA and ystB virulence determinants, Y. enterocolitica strains were grouped into non-virulent or virulent biotypes (1A or 1B/2-5, respectively), as described by Garzetti et al. (2014) [59].
To explore the diversity of Yersinia genomes, an allele-by-allele approach was used. Raw reads were uploaded to Enterobase, where multi-locus sequence typing (MLST) and core genome MLST (cgMLST) were performed [71]. The McNally seven-gene MLST scheme was used [72]. Genomic relationships based on cgMLST profiles were calculated with the MSTree V2 algorithm and visualized in GrapeTree [73].
4.7. Data Analysis
The significance (p < 0.05) of differences in the prevalence of Yersinia spp. and Y. enterocolitica in different meat categories was calculated using the Chi-square test.
5. Conclusions
Higher genetic diversity was observed for Y. enterocolitica 1A and other Yersinia species in comparison to pathogenic Y. enterocolitica 4/O:3 and 2/O:9. Virulence markers may represent the unique virulence properties of each ST, providing important information on the significance of pathogenic Y. enterocolitica and non-pathogenic Yersinia species in the epidemiology of yersiniosis. The WGS analysis of Y. enterocolitica showed the accurate identification of non-pathogenic 1A and pathogenic 1B/2-5 biotypes. Retail pork contaminated with pathogenic Y. enterocolitica represents public health concerns, since pathogenic Y. enterocolitica harbours key virulence factors for the induction of infection in humans.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pathogens11010037/s1, Table S1: Virulence factors of Yersinia species; Table S2: List of virulence gene references used in the analysis; Table S3: Yersinia species isolates selected for whole genome sequencing (WGS) analysis.
Author Contributions
Conceptualization, M.T. and O.V.; methodology, M.T., J.Ķ., I.M., L.A. and O.V.; software, J.Ķ.; validation, M.T., J.Ķ. and S.G.; formal analysis, M.T., J.Ķ., I.M., S.G. and O.V.; investigation, M.T., J.Ķ., I.M., S.G., L.A., M.S., J.O. and O.V.; resources, M.T., I.M., M.S., L.A. and O.V.; data curation, M.T., J.Ķ., I.M., S.G. and L.A.; writing—original draft preparation, M.T.; writing—review and editing, M.T., J.Ķ., S.G. and O.V.; visualization, J.Ķ.; supervision, M.T. and O.V., project administration, M.T. and O.V.; funding acquisition, I.M., M.S. and O.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Latvian Council of Science, grant number lzp-2020/2-0418, “Epidemiology and genetic characterization of Yersinia spp. within the food chain”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Raw sequence reads have been deposited in the European Nucleotide Archive under the study accession number PRJEB49068.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bottone, E.J. Yersinia enterocolitica: Revisitation of enduring human pathogen. Clin. Microbiol. Newsl. 2015, 37, 1–8. [Google Scholar] [CrossRef]
- Hammerl, J.A.; Barac, A.; Erben, P.; Fuhrmann, J.; Gadicherla, A.; Kumsteller, F.; Lauckner, A.; Müller, F.; Hertwig, S. Properties of two broad host range phages of Yersinia enterocolitica isolated from wild animals. Int. J. Mol. Sci. 2021, 22, 11381. [Google Scholar] [CrossRef] [PubMed]
- Rosner, B.M.; Stark, K.; Werber, D. Epidemiology of reported Yersinia enterocolitica infections in Germany, 2001–2008. BMC Public Health 2010, 10, 337. [Google Scholar] [CrossRef] [Green Version]
- Le Guern, A.S.; Martin, L.; Savin, C.; Carniel, E. Yersiniosis in France: Overview and potential sources of infection. Int. J. Infect. Dis. 2016, 46, 1–7. [Google Scholar] [CrossRef] [Green Version]
- European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and CDC). The European Union One Health 2019 Zoonoses Report. EFSA J. 2021, 19, e06406. [Google Scholar] [CrossRef]
- Cornelis, G.R.; Boland, A.P.; Boyn, A.P.; Geuijen, C.; Iriarte, M.; Neyt, C.; Sory, M.P.; Stainier, I. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 1998, 62, 1315–1352. [Google Scholar] [CrossRef] [Green Version]
- Howard, S.L.; Gaunt, M.W.; Hinds, J.; Witney, A.A.; Stabler, R.; Wren, B.W. Application of comparative phylogenomics to study the evolution of Yesinia enterocolitica and to identify genetic differences relating to pathogenicity. J. Bacteriol. 2006, 188, 3645–3653. [Google Scholar] [CrossRef] [Green Version]
- Tennant, S.M.; Grant, T.H.; Robins-Browne, R.M. Pathogenicity of Yersinia enterocolitica biotype 1A. FEMS Immunol. Med. Microbiol. 2003, 38, 127–137. [Google Scholar] [CrossRef] [Green Version]
- Murros, A.; Säde, E.; Johansson, P.; Korkeala, H.; Fredriksson-Ahomaa, M.; Björkroth, J. Characterization of European Yersinia enterocolitica 1A strains using restriction fragment length polymorphism and multilocus sequence analysis. Lett. Appl. Microbiol. 2016, 63, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Laukkanen-Ninious, R.; Fredriksson-Ahomaa, M.; Korkeala, H. Enteropathogenic Yersinia in the pork production chain: Challenges for control. Compr. Rev. Food Sci. Food Saf. 2014, 13, 1165–1191. [Google Scholar] [CrossRef]
- Bari, M.L.; Hossain, H.A.; Isshiki, K.; Ukuku, D. Behavior of Yersinia enterocolitica in foods. J. Pathog. 2011, 420732. [Google Scholar] [CrossRef] [Green Version]
- Van Damme, I.; Berkvens, D.; Vanantwerpen, G.; Baré, J.; Houf, K.; Wauters, G.; De Zutter, L. Contamination of freshly slaughtered pig carcasses with enteropathogenic Yersinia spp.: Distribution, quantification and identification of risk factors. Int. J. Food Microbiol. 2015, 204, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Bonardi, S.; Paris, A.; Bassi, L.; Salmi, F.; Bacci, C.; Riboldi, E.; Boni, E.; D’Incau, M.; Tagliabue, S.; Brindani, F. Detection, semiquantitative enumeration, and antimicrobial susceptibility of Yersinia enterocolitica in pork and chicken meats in Italy. J. Food Prot. 2010, 73, 1785–1792. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson-Ahomaa, M.; Koch, U.; Bucher, M.; Stolle, A. Different genotypes of Yersinia enterocolitica 4/O:3 strains widely distributed in butchers shops in the Munich area. Int. J. Food Microbiol. 2004, 95, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Lorencova, A.; Slany, M. Prevalence of pathogenic Yersinia enterocolitica in minced meat, pig tongues and hearts at the retail level in the Czech Republic detected by real time PCR. Potravinarstvo 2016, 10, 282–286. [Google Scholar] [CrossRef]
- Laukkanen-Ninios, R.; Fredriksson-Ahomaa, M.; Maijala, R.; Korkeala, H. High prevalence of pathogenic Yersinia enterocolitica in pig cheeks. Food Microbiol. 2014, 43, 5–52. [Google Scholar] [CrossRef] [PubMed]
- Lucero-Estrada, C.S.M.; Favier, G.I.; Escudero, M.E. An overview of Yersinia enterocolitica and related species in samples of different origin from San Luis, Argentina. Food Microbiol. 2020, 86, 103345. [Google Scholar] [CrossRef]
- Guillier, L.; Fravalo, P.; Leclercq, A.; Thébault, A.; Kooh, P.; Cadavez, V.; Gonzales-Barron, U. Risk factors for sporadic Yersinia enterocolitica infections: A systematic review and meta-analysis. Microb. Risk Anal. 2021, 17, 100141. [Google Scholar] [CrossRef]
- Morka, K.; Wałecka-Zacharska, E.; Schubert, J.; Dudek, B.; Woźniak-Biel, A.; Kuczkowski, M.; Wieliczko, A.; Bystrón, J.; Bania, J.; Bugla-Płoskońska, G. Genetic diversity and distribution of virulence-associated genes in Y. enterocolitica and Y. enterocolitica-like isolates from humans and animals in Poland. Pathogens 2021, 10, 65. [Google Scholar] [CrossRef]
- Bancerz-Kisiel, A.; Pieczywek, M.; Łada, P.; Szweda, W. The most important virulence markers of Yersinia enterocolitica and their role during infection. Genes 2018, 9, 235. [Google Scholar] [CrossRef] [Green Version]
- ISO 18867:2015; Microbiology of the Food Chain–Polymerase Chain Reaction–Detection of Pathogenic Yersinia enterocolitica and Yersinia pseudotuberculosis; International Organization of Standartization (ISO): Geneva, Switzerland, 2015.
- Imori, P.F.M.; Passaglia, J.; Souza, R.A.; Rocha, L.B.; Falcao, J.P. Virulence-related genes, adhesion and invasion of some Yersinia enterocolitica-like strains suggests its pathogenic potential. Microb. Pathog. 2017, 104, 72–77. [Google Scholar] [CrossRef]
- Kraushaar, B.; Dieckmann, R.; Wittwer, M.; Knabner, D.; Konietzny, A.; Mäde, D.; Strauch, E. Characterization of a Yersinia enterocolitica biotype 1A strain harbouring an ail gene. J. Appl. Microb. 2011, 111, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Ashton, P.M.; Nair, S.; Peters, T.M.; Bale, J.A.; Powell, D.G.; Painset, A.; Tewolde, R.; Schaefer, U.; Jenkins, C.; Dallman, T.J.; et al. Identification of Salmonella for public health surveillance using whole genome sequencing. PeerJ 2016, 4, e1752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inns, T.; Flanagan, S.; Greig, D.R.; Jenkins, C.; Seddon, K.; Chin, T.; Cartwright, J. First use of whole-genome sequencing to investigate a cluster of Yersinia enterocolitica, Liverpool, United Kingdom, 2017. J. Med. Microbiol. 2018, 67, 1747–1752. [Google Scholar] [CrossRef]
- Hunter, E.; Greig, D.R.; Schaefer, U.; Wright, M.J.; Dallman, T.J.; McNally, A.; Jenkins, C. Identification and typing of Yersinia enterocolitica and Yersinia pseudotuberculosis isolated from human clinical specimens in England between 2004 and 2018. J. Med. Microbiol. 2018, 68, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Özdemir, F.; Arslan, S. Genotypic and phenotypic virulence characteristics and antimicrobial resistance of Yersinia spp. isolated from meat and milk products. J. Food Sci. 2015, 80, 1306–1313. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.K.; Ooi, P.T.; Thong, K.L. Prevalence of Yersinia enterocolitica from food and pigs in selected states of Malaysia. Food Control 2014, 35, 94–100. [Google Scholar] [CrossRef]
- Zadernowska, A.; Chajęcka-Wierzchowska, W. Prevalence, biofilm formation and virulence markers of Salmonella sp. and Yersinia enterocolitica in food of animal origin in Poland. LWT-Food Sci. Technol. 2017, 75, 552–556. [Google Scholar] [CrossRef]
- Younis, G.; Mady, M.; Awad, A. Yersinia enterocolitica: Prevalence, virulence, and antimicrobial resistance from retail and processed meat in Egypt. Vet. World 2019, 12, 1078–1084. [Google Scholar] [CrossRef]
- Syczyło, K.; Platt-Samoraj, A.; Bancerz-Kisiel, A.; Szczerba-Turek, A.; Pajdak-Czaus, J.; Łabuć, S.; Procajło, Z.; Socha, P.; Chuzhebayeva, G.; Szweda, W. The prevalence of Yersinia enterocolitica in game animals in Poland. PLoS ONE 2018, 13, e0195136. [Google Scholar] [CrossRef]
- McNally, A.; Cheasty, T.; Fearnley, C.; Dalziel, R.W.; Paiba, G.A.; Manning, G.; Newell, D.G. Comparison of the biotypes of Yersinia enterocolitica isolated from pigs, cattle and sheep at slaughter and from humans with yersiniosis in Great Britain during 1999–2000. Lett. Appl. Microbiol. 2004, 39, 103–108. [Google Scholar] [CrossRef]
- Bonardi, S.; Paris, A.; Bacci, C.; D’Incau, M.; Ferroni, L.; Brindani, F. Detection and characterization of Yersinia enterocolitica from pigs and cattle. Vet. Res. Commun. 2007, 31, 347–350. [Google Scholar] [CrossRef]
- Longenberger, A.H.; Gronostaj, M.P.; Yee, G.Y.; Johnson, L.M.; Lando, J.F.; Voorhees, R.E.; Waller, K.; Weltman, A.C.; Moll, M.; Lyss, S.B.; et al. Yersinia enterocolitica infections associated with improperly pasteurized milk products: Southwest Pennsylvania, March-August, 2011. Epidemiol. Infect. 2014, 142, 1640–1650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonardi, S.; Le Guern, A.S.; Savin, C.; Pupillo, G.; Bolzoni, L.; Cavalca, M.; Pongolini, S. Detection, virulence and antimicrobial resistance of Yersinia enterocolitica in pulk tank milk in Italy. Int. Dairy J. 2018, 84, 46–53. [Google Scholar] [CrossRef]
- Liang, J.; Duan, R.; Xia, S.; Hao, Q.; Yang, J.; Xiao, Y.; Qui, H.; Shi, G.; Wand, S.; Gu, W.; et al. Ecology and geographic distribution of Yersinia enterocolitica among livestock and wildlife in China. Vet. Microbiol. 2015, 178, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson-Ahomaa, M.; Stolle, A.; Korkeala, H. Molecular epidemiology of Yersinia enterocolitica infections. FEMS Immunol. Med. Microbiol. 2006, 47, 315–329. [Google Scholar] [CrossRef] [Green Version]
- Grahek-Ogden, D.; Schimmer, B.; Cudjoe, K.S.; Nygård, K.; Kapperud, G. Outbreak of Yersinia enterocolitica serogroup O:9 infection and processed pork, Norway. Emerg. Infect. Dis. 2007, 13, 754–756. [Google Scholar] [CrossRef]
- Van Damme, I.; Berkvens, D.; Botteldoorn, N.; Dierick, K.; Wits, J.; Pochet, B.; De Zutter, L. Evaluation of the ISO 10273:2003 method for the isolation of human pathogenic Yersinia enterocolitica from pig carcasses and minced meat. Food Microbiol. 2013, 36, 170–175. [Google Scholar] [CrossRef]
- Peruzy, M.F.; Aponte, M.; Proroga, Y.T.R.; Capuano, F.; Cristiano, D.; Delibato, E.; Houf, K.; Murru, N. Yersinia enterocolitica detection in pork products: Evaluation of isolation protocols. Food Microbiol. 2020, 92, 103593. [Google Scholar] [CrossRef] [PubMed]
- Messelhäusser, U.; Kämpf, P.; Colditz, J.; Bauer, H.; Schreiner, H.; Höller, C.; Busch, U. Qualitative and quantitative detection of human pathogenic Yersinia enterocolitica in different food matrices at retail level in Bavaria. Foodborne Pathog. Dis. 2011, 8, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Terentjeva, M.; Bērziņš, A. Prevalence of Yersinia enterocolitica 4/O:3 in raw pork at retail market in Latvia. Arch. Lebensmittelhyg 2013, 64, 125–156. [Google Scholar] [CrossRef]
- Shoaib, M.; Shehzad, A.; Raza, H.; Niazi, S.; Khan, I.M.; Akhtar, W.; Safdar, W.; Wand, Z. A comprehensive review of the prevalence, pathogenesis and detection of Yersinia enterocolitica. RSC Adv. 2019, 70, 41010–41021. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Liu, M.; Wang, H.; Wu, Q.; Xu, T.; Ma, G.; Zhong, Y.; Zhang, J.; Chen, M.; Zue, L.; et al. Occurrence, molecular characterization, and antimicrobial susceptibility of Yersinia enterocolitica isolated from retail food samples in China. LWT 2021, 150, 111876. [Google Scholar] [CrossRef]
- Felek, S.; Krukonis, E.S. The Yersinia pestis Ail protein mediates binding and Yop delivery to host cells required for plague virulence. Infect. Immun. 2009, 77, 825–836. [Google Scholar] [CrossRef] [Green Version]
- Platt-Samoraj, A.; Kończyk-Kmiecik, K.; Bakuła, T. Occurrence and genetic correlations of Yersinia spp. isolated from commensal rodents in Northeastern Poland. Pathogens 2021, 10, 1247. [Google Scholar] [CrossRef] [PubMed]
- Novoslavskij, A.; Kudirkiené, E.; Marcinkuté, A.; Bajoriūniené, A.; Korkeala, H.; Malakauskas, M. Genetic diversity and antimicrobial resistance of Yersinia enterocolitica isolated from pigs and humans in Lithuania. J. Sci. Food Agric. 2013, 93, 1858–1862. [Google Scholar] [CrossRef] [PubMed]
- Bonardi, S.; Bruini, I.; D’Incau, M.; VanDamme, I.; Carniel, E.; Brémont, S.; Cavallini, P.; Tagliabue, S.; Brindani, F. Detection, seroprevalence and antimicrobial resistance of Yersinia enterocolitica and Yersinia pseudotuberculosis in pig tonsils in Northern Italy. Int. J. Food Microbiol. 2016, 235, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Wu, Q.; Hu, H.; Zhang, J.; Huang, H. Prevalence and characterization of Yersinia enterocolitica isolated from retail foods in China. Food Control 2016, 61, 20–27. [Google Scholar] [CrossRef]
- Bonke, R.; Wacheck, S.; Stüber, E.; Meyer, C.; Märlbauer, E.; Fredriksson-Ahomaa, M. Antimicrobial susceptibility and distribution of β-lactamase A(blaA) and β-lactamase B (blaB) genes in enteropathogenic Yersinia species. Microb. Drug Resist. 2011, 17, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Seoane, A.; García Lobo, J.M. Identification of a streptogramin a acetyltransferase gene in the chromosome of Yersinia enterocolitica. Antimicrob. Agents Chemother. 2000, 44, 905–999. [Google Scholar] [CrossRef] [Green Version]
- Bolton, D.J.; Ivory, C.; McDowell, D. A small study of Yersinia enterocolitica in pigs from birth to carcass and characterization of porcine and human strains. Food Control 2013, 33, 521–524. [Google Scholar] [CrossRef]
- Karlsson, P.A.; Tano, E.; Jernberg, C.; Hickman, R.A.; Guy, L.; Järhult, J.D.; Wang, H. Molecular characterization of multidrug-resistant Yersinia enterocolitica from foodborne outbreaks in Sweden. Front. Microbiol. 2021, 12, 664665. [Google Scholar] [CrossRef]
- Bhagat, N.; Virgi, J.S. Yersinia enterocolitica 1A correlates with clonal groups and not the source of isolation. FEMS Microbiol. Lett. 2007, 266, 177–183. [Google Scholar] [CrossRef] [Green Version]
- Thong, K.L.; Tan, L.K.; Ooi, P.T. Genetic diversity, virulotyping and antimicrobial resistance susceptibility of Yersinia enterocolitica isolated from pigs and porcine products in Malaysia. J. Sci. Food Agric. 2018, 98, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Rusak, L.A.; Junqueira, R.M.; Hofer, E.; Vallim, D.C.; Asensi, M.D. Next-generation sequencing virulome analysis of a Yersinia enterecolitica subsp. palearctica bioserotype 4/O:3 ST18 isolated from human blood in Brazil. Braz J. Infect. Dis. 2017, 21, 550–553. [Google Scholar] [CrossRef]
- Singh, I.; Virdi, J.S. Production of Yersinia stable toxin (YST) and distribution of yst genes in biotype 1A strains of Yersinia enterocolitica. J. Med. Microb. 2004, 53, 1065–1068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, H.; Sun, Y.; Mao, Z.; Jiang, B. Investigation of virulence genes in clinical isolates of Yersinia enterocolitica. FEMS Immunol. Med. Microbiol. 2008, 53, 368–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garzetti, D.; Susen, R.; Fruth, A.; Tietze, E.; Heesemann, J.; Rakin, A. A molecular scheme for Yersinia enterocolitica patho-serotyping derived from genome-wide analysis. Int. J. Med. Microbiol. 2014, 304, 275–283. [Google Scholar] [CrossRef]
- Bren, A.; Eisenbach, M. How signals are heard during bacterial chemotaxis: Protein-protein interactions in sensory signal propagation. J. Bacteriol. 2000, 182, 6865–6873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.J.; Young, B.M.; Young, G.M. Effect of flagellar mutations on Yersinia enterocolitica biofilm formation. Appl. Environ. Microbiol. 2008, 74, 5466–5474. [Google Scholar] [CrossRef] [Green Version]
- Strydom, H.; Wand, J.; Paine, S.; Dyet, K.; Cullen, K.; Wright, J. Evaluating sub-typing methods for pathogenic Yersinia enterocolitica to support outbreak investigations in New Zealand. Epidemiol. Infect. 2019, 147, e186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ISO 10273:2017; Microbiology of Food and Animal Feed. Horizontal Method for the Detection of Presumably Pathogenic Yersinia enterocolitica; International Organization of Standartization (ISO): Geneva, Switzerland, 2017.
- Wauters, G.; Kandolo, K.; Janssens, M. Revised biogrouping scheme of Yersinia enterocolitica. Contrib. Microbiol. Immunol. 1987, 9, 14–21. [Google Scholar] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoints Tables for Interpretation of MICs and Zones Diameters. Version 11.0. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_11.0_Breakpoint_Tables.pdf (accessed on 8 October 2021).
- Mäde, D.; Reiting, R.; Strauch, E.; Ketteritzsch, K.; Wicke, A. A real-time PCR for detection of pathogenic Yersinia enterocolitica in food combined with an universal internal amplification control system. J. Verbr. Lebensm 2008, 3, 141–151. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prjibelski, A.D.; Puglia, G.D.; Antipov, D.; Bushmanova, E.; Giordano, D.; Mikheenko, A.; Vitale, D.; Lapidus, A. Extending rnaSPAdes functionality for hybrid transcriptome assembly. BMC Bioinform. 2020, 21, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 302. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Alikhan, N.F.; Mohamed, K.; the Agama Study Group; Achtman, M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny and Escherichia core genomic diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, M.; Chattaway, M.A.; Reuter, S.; Savin, C.; Strauch, E.; Carniel, E.; Connor, T.; Van Damme, I.; Rajakaruna, L.; Rajendram, D.; et al. Use of whole-genus genome sequence data to develop a multilocus sequence typing tool that accurately identifies Yersinia isolates to the species and subspecies levels. J. Clin. Microbiol. 2015, 53, 35–42. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Alikhan, N.F.; Sergeant, M.J.; Luhmann, N.; Vaz, C.; Francisco, A.P.; Carrico, J.A.; Achtman, M. GrapeTree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018, 28, 1395–1404. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).