Challenges of an Emerging Disease: The Evolving Approach to Diagnosing Devil Facial Tumour Disease
Abstract
:1. Introduction
2. Diagnosis of Infectious Diseases and Cancer
3. Devil Facial Tumour Disease
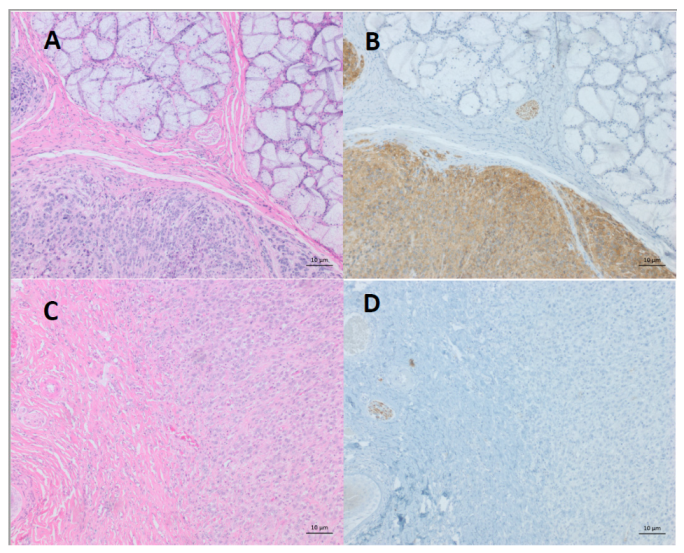
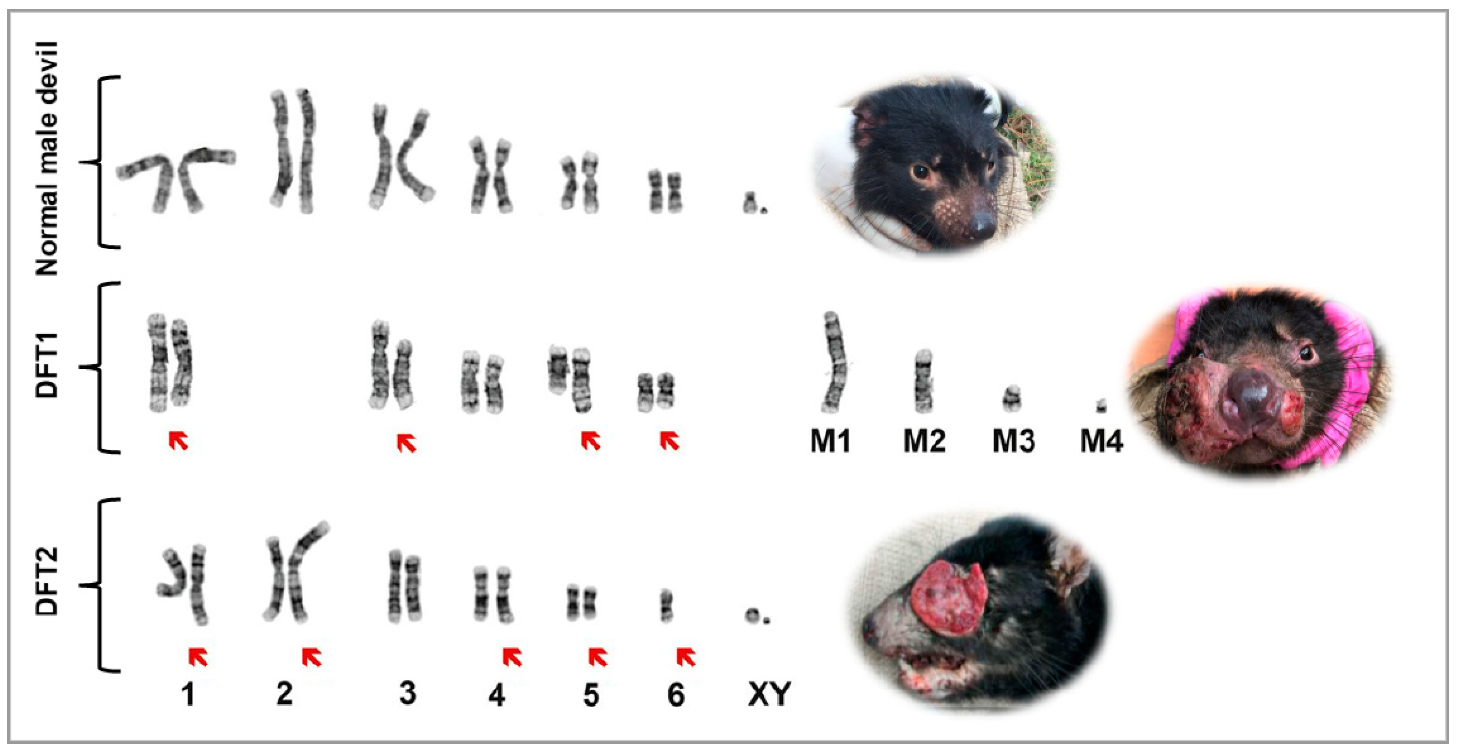
4. Differentiation of DFTDs (DFT1 and DFT2)
5. Liquid Biopsy to Differentiate DFTDs (DFT1 and DFT2)
6. Pre-Diagnostic Approaches
7. Extracellular Vesicles
8. Multi-Omics
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Chibuk, J.; Flory, A.; Kruglyak, K.M.; Leibman, N.; Nahama, A.; Dharajiya, N.; van den Boom, D.; Jensen, T.J.; Friedman, J.S.; Shen, M.R.; et al. Horizons in Veterinary Precision Oncology: Fundamentals of Cancer Genomics and Applications of Liquid Biopsy for the Detection, Characterization, and Management of Cancer in Dogs. Front. Vet. Sci. 2021, 8, 664718. [Google Scholar] [CrossRef] [PubMed]
- Paoloni, M.C.; Khanna, C. Comparative oncology today. Vet. Clin. N. Am. Small Anim. Pract. 2007, 37, 1023–1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hawkins, C.E.; Baars, C.; Hesterman, H.; Hocking, G.J.; Jones, M.E.; Lazenby, B.; Mann, D.; Mooney, N.; Pemberton, D.; Pyecroft, S.; et al. Emerging disease and population decline of an island endemic, the Tasmanian devil Sarcophilus harrisii. Biol. Conserv. 2006, 131, 307–324. [Google Scholar] [CrossRef]
- Loh, R.; Bergfeld, J.; Hayes, D.; O’Hara, A.; Pyecroft, S.; Raidal, S.; Sharpe, R. The pathology of devil facial tumor disease (DFTD) in Tasmanian devils (Sarcophilus harrisii). Vet. Pathol. 2006, 43, 890–895. [Google Scholar] [CrossRef] [PubMed]
- Loh, R.; Hayes, D.; Mahjoor, A.; O’Hara, A.; Pyecroft, S.; Raidal, S. The immunohistochemical characterization of devil facial tumor disease (DFTD) in the Tasmanian Devil (Sarcophilus harrisii). Vet. Pathol. 2006, 43, 896–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murchison, E.P.; Tovar, C.; Hsu, A.; Bender, H.S.; Kheradpour, P.; Rebbeck, C.A.; Obendorf, D.; Conlan, C.; Bahlo, M.; Blizzard, C.A.; et al. The Tasmanian devil transcriptome reveals Schwann cell origins of a clonally transmissible cancer. Science 2010, 327, 84–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tovar, C.; Obendorf, D.; Murchison, E.P.; Papenfuss, A.T.; Kreiss, A.; Woods, G.M. Tumor-specific diagnostic marker for transmissible facial tumors of Tasmanian devils: Immunohistochemistry studies. Vet. Pathol. 2011, 48, 1195–1203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pearse, A.M.; Swift, K. Allograft theory: Transmission of devil facial-tumour disease. Nature 2006, 439, 549. [Google Scholar] [CrossRef] [PubMed]
- Siddle, H.V.; Kreiss, A.; Eldridge, M.D.; Noonan, E.; Clarke, C.J.; Pyecroft, S.; Woods, G.M.; Belov, K. Transmission of a fatal clonal tumor by biting occurs due to depleted MHC diversity in a threatened carnivorous marsupial. Proc. Natl. Acad. Sci. USA 2007, 104, 16221–16226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murchison, E.P.; Schulz-Trieglaff, O.B.; Ning, Z.; Alexandrov, L.B.; Bauer, M.J.; Fu, B.; Hims, M.; Ding, Z.; Ivakhno, S.; Stewart, C.; et al. Genome sequencing and analysis of the Tasmanian devil and its transmissible cancer. Cell 2012, 148, 780–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, W.; Hayes, V.M.; Ratan, A.; Petersen, D.C.; Wittekindt, N.E.; Miller, J.; Walenz, B.; Knight, J.; Qi, J.; Zhao, F. Genetic diversity and population structure of the endangered marsupial Sarcophilus harrisii (Tasmanian devil). Proc. Natl. Acad. Sci. USA 2011, 108, 12348–12353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deakin, J.E.; Bender, H.S.; Pearse, A.M.; Rens, W.; O’Brien, P.C.; Ferguson-Smith, M.A.; Cheng, Y.; Morris, K.; Taylor, R.; Stuart, A.; et al. Genomic restructuring in the Tasmanian devil facial tumour: Chromosome painting and gene mapping provide clues to evolution of a transmissible tumour. PLoS Genet. 2012, 8, e1002483. [Google Scholar] [CrossRef] [PubMed]
- Pye, R.J.; Pemberton, D.; Tovar, C.; Tubio, J.M.; Dun, K.A.; Fox, S.; Darby, J.; Hayes, D.; Knowles, G.W.; Kreiss, A.; et al. A second transmissible cancer in Tasmanian devils. Proc. Natl. Acad. Sci. USA 2016, 113, 374–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hogg, C.; Fox, S.; Pemberton, D.; Belov, K. Saving the Tasmanian Devil: Recovery through Science-Based Management; CSIRO Publishing: Clayton, Australia, 2019. [Google Scholar]
- Kwon, Y.M.; Stammnitz, M.R.; Wang, J.; Swift, K.; Knowles, G.W.; Pye, R.J.; Kreiss, A.; Peck, S.; Fox, S.; Pemberton, D.; et al. Tasman-PCR: A genetic diagnostic assay for Tasmanian devil facial tumour diseases. R. Soc. Open Sci. 2018, 5, 180870. [Google Scholar] [CrossRef] [Green Version]
- Stammnitz, M.R.; Coorens, T.H.H.; Gori, K.C.; Hayes, D.; Fu, B.; Wang, J.; Martin-Herranz, D.E.; Alexandrov, L.B.; Baez-Ortega, A.; Barthorpe, S.; et al. The origins and vulnerabilities of two transmissible cancers in Tasmanian devils. Cancer Cell 2018, 33, 607–619.e615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patchett, A.L.; Coorens, T.H.H.; Darby, J.; Wilson, R.; McKay, M.J.; Kamath, K.S.; Rubin, A.; Wakefield, M.; McIntosh, L.; Mangiola, S.; et al. Two of a kind: Transmissible Schwann cell cancers in the endangered Tasmanian devil (Sarcophilus harrisii). Cell. Mol. Life Sci. 2019, 77, 1847–1858. [Google Scholar] [CrossRef]
- Patchett, A.L.; Flies, A.S.; Lyons, A.B.; Woods, G.M. Curse of the devil: Molecular insights into the emergence of transmissible cancers in the Tasmanian devil (Sarcophilus harrisii). Cell. Mol. Life Sci. 2020, 77, 2507–2525. [Google Scholar] [CrossRef]
- Arthur-Farraj, P.J.; Latouche, M.; Wilton, D.K.; Quintes, S.; Chabrol, E.; Banerjee, A.; Woodhoo, A.; Jenkins, B.; Rahman, M.; Turmaine, M.; et al. c-Jun reprograms Schwann cells of injured nerves to generate a repair cell essential for regeneration. Neuron 2012, 75, 633–647. [Google Scholar] [CrossRef] [Green Version]
- Arthur-Farraj, P.J.; Morgan, C.C.; Adamowicz, M.; Gomez-Sanchez, J.A.; Fazal, S.V.; Beucher, A.; Razzaghi, B.; Mirsky, R.; Jessen, K.R.; Aitman, T.J. Changes in the Coding and Non-coding Transcriptome and DNA Methylome that Define the Schwann Cell Repair Phenotype after Nerve Injury. Cell Rep. 2017, 20, 2719–2734. [Google Scholar] [CrossRef] [Green Version]
- Clements, M.P.; Byrne, E.; Camarillo Guerrero, L.F.; Cattin, A.L.; Zakka, L.; Ashraf, A.; Burden, J.J.; Khadayate, S.; Lloyd, A.C.; Marguerat, S.; et al. The Wound Microenvironment Reprograms Schwann Cells to Invasive Mesenchymal-like Cells to Drive Peripheral Nerve Regeneration. Neuron 2017, 96, 98–114.e117. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Luo, X.; Wang, W.; Wang, Y.; Zhu, F.; Wang, W. Interleukin-1beta Promotes Schwann Cells De-Differentiation in Wallerian Degeneration via the c-JUN/AP-1 Pathway. Front. Cell Neurosci. 2019, 13, 304. [Google Scholar] [CrossRef]
- Caldwell, A.; Coleby, R.; Tovar, C.; Stammnitz, M.R.; Kwon, Y.M.; Owen, R.S.; Tringides, M.; Murchison, E.P.; Skjodt, K.; Thomas, G.J.; et al. The newly-arisen Devil facial tumour disease 2 (DFT2) reveals a mechanism for the emergence of a contagious cancer. eLife 2018, 7, e35314. [Google Scholar] [CrossRef] [PubMed]
- Siddle, H.V.; Kreiss, A.; Tovar, C.; Yuen, C.K.; Cheng, Y.; Belov, K.; Swift, K.; Pearse, A.M.; Hamede, R.; Jones, M.E.; et al. Reversible epigenetic down-regulation of MHC molecules by devil facial tumour disease illustrates immune escape by a contagious cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 5103–5108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patchett, A.L.; Tovar, C.; Blackburn, N.B.; Woods, G.M.; Lyons, A.B. Mesenchymal plasticity of devil facial tumour cells during in vivo vaccine and immunotherapy trials. Immunol. Cell Biol. 2021, 99, 711–723. [Google Scholar] [CrossRef]
- Pastushenko, I.; Blanpain, C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol. 2019, 29, 212–226. [Google Scholar] [CrossRef] [Green Version]
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 12 August 2021).
- Kreth, F.; Muacevic, A.; Medele, R.; Bise, K.; Meyer, T.; Reulen, H.-J. The risk of haemorrhage after image guided stereotactic biopsy of intra-axial brain tumours–a prospective study. Acta Neurochir. 2001, 143, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Robertson, E.; Baxter, G. Tumour seeding following percutaneous needle biopsy: The real story! Clin. Radiol. 2011, 66, 1007–1014. [Google Scholar] [CrossRef]
- Alix-Panabieres, C. The future of liquid biopsy. Nature 2020, 579, S9. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.-C.; Zhou, Q.; Wu, Y.-L. The emerging roles of NGS-based liquid biopsy in non-small cell lung cancer. J. Hematol. Oncol. 2017, 10, 167. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Yi, M.; Dong, B.; Tan, X.; Luo, S.; Wu, K. The role of exosomes in liquid biopsy for cancer diagnosis and prognosis prediction. Int. J. Cancer 2021, 148, 2640–2651. [Google Scholar] [CrossRef]
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience 2015, 65, 783–797. [Google Scholar] [CrossRef] [Green Version]
- Willms, E.; Cabanas, C.; Mager, I.; Wood, M.J.A.; Vader, P. Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front. Immunol. 2018, 9, 738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maas, S.L.; Breakefield, X.O.; Weaver, A.M. Extracellular vesicles: Unique intercellular delivery vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef] [Green Version]
- Rontogianni, S.; Synadaki, E.; Li, B.; Liefaard, M.C.; Lips, E.H.; Wesseling, J.; Wu, W.; Altelaar, M. Proteomic profiling of extracellular vesicles allows for human breast cancer subtyping. Commun. Biol. 2019, 2, 167. [Google Scholar] [CrossRef] [Green Version]
- Zabegina, L.; Nazarova, I.; Knyazeva, M.; Nikiforova, N.; Slyusarenko, M.; Titov, S.; Vasilyev, D.; Sleptsov, I.; Malek, A. MiRNA let-7 from TPO (+) extracellular vesicles is a potential marker for a differential diagnosis of follicular thyroid nodules. Cells 2020, 9, 1917. [Google Scholar] [CrossRef] [PubMed]
- Espejo, C.; Wilson, R.; Willms, E.; Ruiz-Aravena, M.; Pye, R.J.; Jones, M.E.; Hill, A.F.; Woods, G.M.; Lyons, A.B. Extracellular vesicle proteomes of two transmissible cancers of Tasmanian devils reveal tenascin-C as a serum-based differential diagnostic biomarker. Cell. Mol. Life Sci. 2021, 78, 7537–7555. [Google Scholar] [CrossRef]
- Brezgyte, G.; Shah, V.; Jach, D.; Crnogorac-Jurcevic, T. Non-Invasive Biomarkers for Earlier Detection of Pancreatic Cancer-A Comprehensive Review. Cancers 2021, 13, 2722. [Google Scholar] [CrossRef]
- Stamey, T.A.; Yang, N.; Hay, A.R.; McNeal, J.E.; Freiha, F.S.; Redwine, E. Prostate-specific antigen as a serum marker for adenocarcinoma of the prostate. N. Engl. J. Med. 1987, 317, 909–916. [Google Scholar] [CrossRef]
- Mavroudis, D. Circulating cancer cells. Ann. Oncol. 2010, 21 (Suppl. 7), vii95–vii100. [Google Scholar] [CrossRef] [PubMed]
- Save the Tasmanian Devil Program. Risk Categorisation Guidelines for the Keeping and Movement of Captive Tasmanian Devils; Department of Primary Industries, Park, Water & Environment: Hobart, Australia, 2017.
- Karu, N.; Wilson, R.; Hamede, R.; Jones, M.; Woods, G.M.; Hilder, E.F.; Shellie, R.A. Discovery of biomarkers for Tasmanian Devil Cancer (DFTD) by metabolic profiling of serum. J. Proteome Res. 2016, 15, 3827–3840. [Google Scholar] [CrossRef] [PubMed]
- Hayes, D.A.; Kunde, D.A.; Taylor, R.L.; Pyecroft, S.B.; Sohal, S.S.; Snow, E.T. ERBB3: A potential serum biomarker for early detection and therapeutic target for devil facial tumour 1 (DFT1). PLoS ONE 2017, 12, e0177919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campos, C.D.; Jackson, J.M.; Witek, M.A.; Soper, S.A. Molecular profiling of liquid biopsy samples for precision medicine. Cancer J. 2018, 24, 93. [Google Scholar] [CrossRef] [PubMed]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.; Jutzy, J.M.; Valenzuela, M.M.A.; Turay, D.; Aspe, J.R.; Ashok, A.; Mirshahidi, S.; Mercola, D.; Lilly, M.B.; Wall, N.R. Plasma-derived exosomal survivin, a plausible biomarker for early detection of prostate cancer. PLoS ONE 2012, 7, e46737. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.; Bennit, H.F.; Turay, D.; Perez, M.; Mirshahidi, S.; Yuan, Y.; Wall, N.R. Early diagnostic value of survivin and its alternative splice variants in breast cancer. BMC Cancer 2014, 14, 176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, P.-G.; Lee, J.-E.; Cho, Y.-E.; Lee, S.J.; Chae, Y.S.; Jung, J.H.; Kim, I.-S.; Park, H.Y.; Baek, M.-C. Fibronectin on circulating extracellular vesicles as a liquid biopsy to detect breast cancer. Oncotarget 2016, 7, 40189. [Google Scholar] [CrossRef] [Green Version]
- Save the Tasmanian Devil Program. Save the Tasmanian Devil Program. Available online: https://dpipwe.tas.gov.au/wildlife-management/save-the-tasmanian-devil-program (accessed on 16 June 2021).
- Espejo, C.; Wilson, R.; Pye, R.J.; Ractcliffe, J.C.; Ruiz-Aravena, M.; Willms, E.; Wolfe, B.W.; Hamede, R.; Hill, A.F.; Jones, M.E.; et al. Cathelicidin-3 associated with serum extracellular vesicles enables early diagnosis of a transmissible cancer. bioRxiv 2021. [Google Scholar] [CrossRef]
- Flies, A.S.; Flies, E.J.; Fox, S.; Gilbert, A.; Johnson, S.R.; Liu, G.-S.; Lyons, A.B.; Patchett, A.L.; Pemberton, D.; Pye, R.J. An oral bait vaccination approach for the Tasmanian devil facial tumor diseases. Expert Rev. Vaccines 2020, 19, 1–10. [Google Scholar] [CrossRef]
- Wang, D.; Eraslan, B.; Wieland, T.; Hallström, B.; Hopf, T.; Zolg, D.P.; Zecha, J.; Asplund, A.; Li, L.H.; Meng, C.; et al. A deep proteome and transcriptome abundance atlas of 29 healthy human tissues. Mol. Syst. Biol. 2019, 15, e8503. [Google Scholar] [CrossRef]
- Patchett, A.L.; Wilson, R.; Charlesworth, J.C.; Corcoran, L.M.; Papenfuss, A.T.; Lyons, B.A.; Woods, G.M.; Tovar, C. Transcriptome and proteome profiling reveals stress-induced expression signatures of imiquimod-treated Tasmanian devil facial tumor disease (DFTD) cells. Oncotarget 2018, 9, 15895–15914. [Google Scholar] [CrossRef] [Green Version]
- Mann, M.; Kumar, C.; Zeng, W.F.; Strauss, M.T. Artificial intelligence for proteomics and biomarker discovery. Cell Syst. 2021, 12, 759–770. [Google Scholar] [CrossRef] [PubMed]
- Picard, M.; Scott-Boyer, M.P.; Bodein, A.; Périn, O.; Droit, A. Integration strategies of multi-omics data for machine learning analysis. Comput. Struct. Biotechnol. J. 2021, 19, 3735–3746. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Espejo, C.; Patchett, A.L.; Wilson, R.; Lyons, A.B.; Woods, G.M. Challenges of an Emerging Disease: The Evolving Approach to Diagnosing Devil Facial Tumour Disease. Pathogens 2022, 11, 27. https://doi.org/10.3390/pathogens11010027
Espejo C, Patchett AL, Wilson R, Lyons AB, Woods GM. Challenges of an Emerging Disease: The Evolving Approach to Diagnosing Devil Facial Tumour Disease. Pathogens. 2022; 11(1):27. https://doi.org/10.3390/pathogens11010027
Chicago/Turabian StyleEspejo, Camila, Amanda L. Patchett, Richard Wilson, A. Bruce Lyons, and Gregory M. Woods. 2022. "Challenges of an Emerging Disease: The Evolving Approach to Diagnosing Devil Facial Tumour Disease" Pathogens 11, no. 1: 27. https://doi.org/10.3390/pathogens11010027
APA StyleEspejo, C., Patchett, A. L., Wilson, R., Lyons, A. B., & Woods, G. M. (2022). Challenges of an Emerging Disease: The Evolving Approach to Diagnosing Devil Facial Tumour Disease. Pathogens, 11(1), 27. https://doi.org/10.3390/pathogens11010027





