Abstract
Multi-host pathogens are challenging to control and are responsible for some of the most important diseases of humans, livestock, and wildlife. Leptospira spp. are some of the most common multi-host pathogens and represent an important cause of zoonotic infections and livestock productivity loss in the developing world, where contact with wildlife species is common. Although there is increasing evidence that cattle in Africa harbour a broad diversity of Leptospira genotypes and serovars, little is known about the epidemiology of these pathogens in wild bovids, such as African buffaloes (Syncerus caffer). Using microscopic agglutination testing (MAT) on serum samples collected from free-ranging buffaloes (n = 98) captured in the Hluhluwe-iMfolozi Park (HiP), South Africa, we demonstrated an overall seroprevalence of 21% with seropositivity almost exclusively limited to serovar Tarassovi (serogroup Tarassovi). Moreover, we found no evidence of seropositivity in unweaned calves and showed temporal- or herd-specific variation in exposure risk, and increased probability of seropositivity (OR = 5.44, 95% CI = 1.4–27) in female buffaloes. Together, these findings demonstrate that free-ranging African buffaloes are exposed to Leptospira spp. infections, providing insights into the epidemiology of an emerging Leptospira serovar in herds with an absence of any disease control and minimal management.
1. Introduction
Wildlife can play an important role in the epidemiology of multi-host pathogens of humans and livestock by maintaining pathogens in reservoirs of epidemiologically connected reservoir host populations or environments [1] where traditional control measures, such as vaccination, may be challenging. Moreover, the transmission of pathogens from domestic animals [2] or livestock [3] to wildlife has also been shown to have important implications for the conservation of iconic wildlife species. Therefore, understanding the epidemiology of these pathogens in their wildlife reservoir hosts is critical to mitigating their effects in humans, livestock, and wildlife.
Globally, Leptospira spp. are one of the most common and diverse multi-host pathogens and are an important cause of zoonotic infections and livestock productivity loss. Currently, there are 64 Leptospira species described, 38 of which belong to pathogenic or potentially pathogenic subclades [4]. However, two species, L. interrogans and L. borgpetersenii, are responsible for most pathogenic infections [5]. While rodents are frequently implicated as sources of human infection, there is increasing evidence that, in some contexts, livestock may play a key role as reservoir hosts in Africa [6] where molecular [7] and serological [8] studies have demonstrated a broad diversity of Leptospira genotypes and serovars. In livestock, Leptospira spp. may be transmitted through venereal transmission [9] or contact with environments contaminated with urine from infected hosts [10]. However, laboratory and genomic studies have demonstrated that the relative importance of environmental and direct host-to-host (e.g., venereal) transmission may differ between L. interrogans and L. borgpetersenii, respectively [5].
While there is increasing evidence that leptospirosis poses a significant public health threat [11] in South Africa and that rodents [12], horses [13], and livestock [8] may play a role in the epidemiology of this neglected zoonosis, there are relatively few studies examining Leptospira spp. exposure in wild bovids, such as African buffaloes (Syncerus caffer), which may act as wildlife reservoir hosts [14]. Using microscopic agglutination typing (MAT) on serum samples collected from free-ranging buffaloes captured in 2016, 2018, and 2019 in the Hluhluwe-iMfolozi Park (HiP) in KwaZulu-Natal (KZN), South Africa, we aimed to (i) characterise the seroprevalence and serological diversity of Leptospira spp. infections, (ii) explore the dynamics in MAT titres in paired samples collected three days apart, and (iii) identify temporal, age, and sex patterns in exposure.
2. Results
2.1. Seroprevalence and Serological Diversity
While most buffaloes were only sampled once, in 2016, 25 buffaloes were sampled twice at a three-day interval. When including animals that tested positive for Leptospira on either sample in 2016, the MAT results revealed a combined seroprevalence, for all years and age groups, of 21% (21 in 98, 95% CI 14–31%). Evidence for exposure was limited to adult and subadult buffaloes, with 95% (20 in 21) of positive samples reacting to serovar Tarassovi (serogroup Tarassovi) and one adult animal, sampled in 2018, reacting to serovar Szwajizak (serogroup Mini) (Table 1).

Table 1.
Number of animals in each age category positive for the eight Leptospira serovars representing eight serogroups (Australis, Canicola, Icterohaemorrhagiae, Pomona, Tarassovi, Mini, Grippotyphosa, Sejroe) included in the MAT panel across the three sampled years.
2.2. MAT Titre Dynamics
In the 25 paired samples (17 adult, seven subadult, and one calf) collected in 2016, titres varied between samples, despite a relatively short interval (three days) between samples (Figure 1). Although discrepancies between the identification of seropositive animals were noted in 64% (seven out of 11; four adult and three subadult) of paired samples, 71% (five out of seven; three adult and two subadult) of animals that switched between positive and negative results exhibited only a single fold dilution change between titres of 1/100 and negative results (Figure 1), suggesting that MAT lacks sensitivity at low titres. However, two samples did show three- and four-fold titre changes between positive and negative samples.
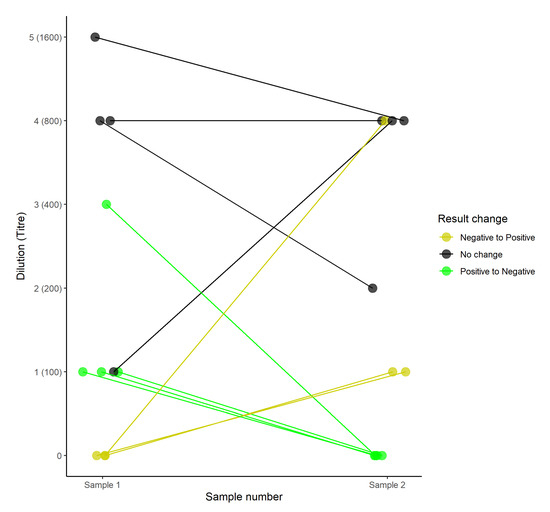
Figure 1.
The titre dynamics in paired samples taken three days apart from individual buffaloes in which at least one sample tested seropositive (agglutination at a titre of 1/100). Colours indicate whether there were discrepancies in classification of the animal as seropositive or seronegative.
2.3. Temporal, Age, and Sex Patterns in Exposure Risk
Using only the results from the first sample taken in 2016 to facilitate comparisons with other years, where only a single sample was obtained, bivariate analyses demonstrated a significantly lower seroprevalence in calves when compared to subadults or adults (χ2 = 6.69, p = 0.03, p value simulated based on 1 × 106 replicates). Overall seroprevalence appeared to decline from 2016 to 2019 (χ2 = 5.11, df = 2, p = 0.08), largely due to a reduction in the seroprevalence in female buffaloes (Figure 2). Overall seroprevalence tended to be higher in female compared to male buffaloes (χ2 = 2.99, df = 1, p = 0.08).
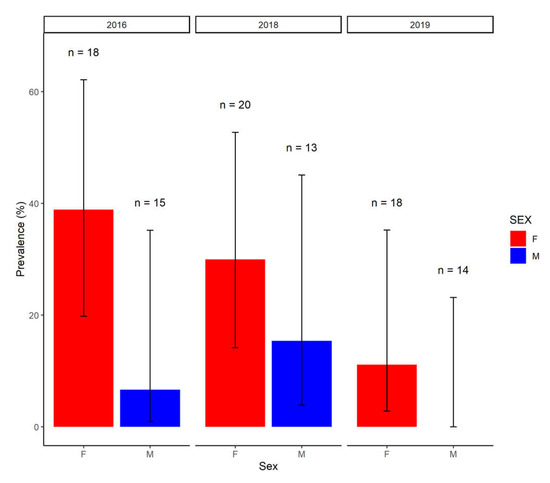
Figure 2.
The seroprevalence in male (M) and female (F) buffaloes in each year of sampling. Error bars represent 95% confidence intervals (logit) and the number (n) of male and female buffaloes tested in each year are shown. In 2016, only results from the first sample were considered.
Using a Bayesian multivariate generalised linear model approach, findings confirmed that calves were over 100 times less likely to be infected than adult or subadult animals (OR = 0.01, 95% CI ≤ 0.001–0.37) (Figure 3). By accounting for sex, year, and age, female buffaloes were shown to be over five times more likely to be seropositive than male buffaloes (OR = 5.44, 95% CI = 1.4–27) and animals sampled in 2019 were almost seven times less likely to be infected than animals captured in 2016 and 2018 (OR = 0.15, 95% CI = 0.02–0.71) (Figure 3).
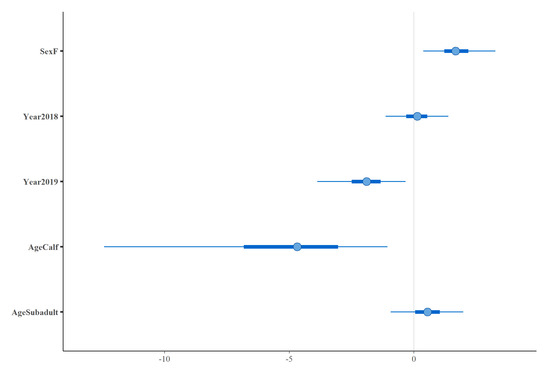
Figure 3.
The posterior distributions for temporal (Year), sex (Sex), and age (Age) covariates included in a Bayesian multivariate generalised linear model. Points indicate the posterior mean, thick bars represent 50% confidence intervals and thin bars represent 95% confidence intervals. The reference level for sex was male, 2016 for year of sampling and adult for age.
3. Discussion
In free-ranging African buffaloes in South Africa, we identified evidence of exposure to L. borgpeterseni serovar Tarassovi with temporal differences in exposure risk, increased exposure risk in female buffaloes, and no evidence of exposure in unweaned calves, providing unique insights into the epidemiology of an important zoonotic and livestock pathogen in the absence of control measures. The seroprevalence noted in this study (21%) is higher than that previously identified in free-ranging African buffaloes (1.7%, seven in 406) in the Kruger National Park (KNP) in South Africa [15] and lower than that (42%, 39 in 92) identified in Uganda [14]. However, the seroprevalence is similar to that (19.4%, 392 in 2021) previously identified in cattle in the same province (KZN) in 2001–2003 [8]. The methodological differences between studies, such as the use of ELISA in Uganda [14] versus MAT in South Africa and heat inactivation of samples from the KNP [15], may account for some of the differences in seroprevalence estimates in buffaloes. Although MAT is considered the gold standard serological assay, it has been shown to lack sensitivity in African contexts [16]. Therefore, the seroprevalence estimates presented in this study represent the apparent seroprevalence, since they were not adjusted for test performance. Furthermore, the discrepancies between results from paired samples suggest that seroprevalence within these herds may be higher than estimated here.
It is also possible that the difficulties of processing samples under challenging field conditions, the prolonged storage of samples, or the stress of capture may explain the larger (three- and four-fold) variation in MAT titres between paired samples. While MAT may lack sensitivity, particularly at low titres, it is highly specific [16] and allows the presumptive identification of the infecting serovar. Apart from a single animal that reacted to L. interrogans serovar Szwajizak (serogroup Mini), all positive samples in this study reacted to L. borgpetersenii serovar Tarassovi (serogroup Tarassovi). Serovar Tarassovi is increasingly recognised in a variety of hosts in Africa. For example, it has been detected in cattle in South Africa [8,17] and Uganda [16], horses in South Africa [13], and has historically been detected, at low seroprevalence, in buffaloes in other regions of South Africa [15]. In New Zealand, evidence of exposure to serovar Tarassovi in a broad range of livestock species, and its implication in human cases of leptospirosis [18], suggest that this serovar may be an underrecognized cause of livestock productivity loss and zoonotic disease [19].
Seroprevalence decreased between 2016 and 2019, largely driven by a reduction in seroprevalence in female buffaloes from a high of 39% (seven in 18) in 2016 to 11% (two in 18) in 2019. However, the sampling of different herds in each year makes it difficult to disentangle temporal trends from herd-specific differences in exposure risk. In arid or semi-arid areas, the aggregation of wildlife and livestock around water sources may drive the transmission of environmentally transmitted pathogens [20] and Leptospira spp. can survive for prolonged periods in surface water, which may act as a source of infection for livestock [10]. Therefore, it is possible that the severe drought conditions experienced in South Africa over the earlier sampling period [21], which had significant impacts on HiP [22], may have resulted in increased environmental contamination due to the increased aggregation of wildlife around scarce water sources and increased utilisation of the rivers upstream of HiP by livestock. An environmental, rather than venereal, route of transmission is supported by the broadly similar infection probability in sexually immature subadults and sexually mature adults.
After accounting for age and temporal trends in seroprevalence, female buffaloes were nearly five times more likely to be infected than male buffaloes. In large scale studies of domesticated water buffaloes in Thailand (n = 1376) [23] and slaughter cattle in Uganda (n = 500) [16], no effects of sex on seroprevalence were noted. Therefore, our findings may reflect differences in the social structure of free-ranging buffaloes and domesticated bovids. Female buffaloes in HiP are found in large (n = 30–250), mixed-sex herds whereas males are predominantly found in smaller (n = 1–12), all-male sub-herds [24]. Even when found within mixed-sex herds, the majority of males do not spend the entire year in the herd, with the proportion of males in these herds ranging between 5% and 25% in the breeding and non-breeding season, respectively [24]. Therefore, it is possible that the aggregation of female buffaloes in larger herds increases their risk of contact with environments contaminated by infected animals. Alternatively, it is possible that the capture process did not allow for the representative sampling of adult male buffalo. For example, older males that have left the breeding herds may be underrepresented in the captured animals.
While these findings demonstrate clear trends in exposure in free-ranging buffaloes, the relatively small sample size in this study, the lack of contemporaneous seroprevalence data in livestock, and the inability to obtain clinical data, such as abortion and stillbirth incidence, from buffaloes raise further questions regarding cross-species infection, the clinical relevance of these findings for buffaloes, and the epidemiology of this emerging serovar. Moreover, the reliance on serological data and the lack of suitable tissues, such as kidney samples, for the direct detection and molecular typing of Leptospira spp. infections in buffaloes suggest that conclusions on Leptospira spp. diversity in wild buffaloes should be treated with some caution. To address these limitations, further studies are needed to generate molecular typing data from wildlife and livestock, to identify potential routes and drivers of environmental transmission between livestock and wildlife and to clarify the clinical and zoonotic importance of serovar Tarassovi. However, it is clear that, in the absence of any disease control and minimal management of wild bovids, exposure to Leptospira spp. can vary widely either temporally or between herds. This finding has implications for the evaluation of control measures in livestock herds and highlights the need to consider these natural variations in exposure when evaluating the efficacy of control measures.
4. Materials and Methods
Serum samples were obtained from 98 buffaloes from three different herds captured at three different locations within the Hluhluwe-iMfolozi Park (HiP) in 2016, 2018 and 2019 (Figure 4). The ~100,000 ha Hluhluwe-iMfolozi Park is situated in KZN province and is the third largest game reserve in South Africa. It has an African buffalo population of 4544 [25], split into herds that change in number seasonally and in response to resource availability, with mixed sex herds that range between 30 and 250 individuals and all male groups that range between 1 and 12 individuals [24]. Buffaloes are free to range within the confines of the HiP but game fencing largely prevents contact with livestock in surrounding communal farms. Management of the buffalo population is restricted to yearly culls and disease surveillance for bovine tuberculosis (bTB). Capture operations for bTB surveillance [26] frequently capture entire herds, resulting in representative sampling of mixed sex herds present at the time of capture [27]. However, as male animals spend the majority of their time segregated from mixed sex herds [24], sera collected during annual bTB surveillance were selected to provide a broadly similar age and sex profile between years. In 2016, animals were captured and held in temporary holding pens to allow interpretation of single intradermal comparative tuberculin tests (SICTT) [26], allowing for the collection of twenty-five paired serum samples collected at three-day intervals. In 2018 and 2019, single serum samples were obtained from captured buffaloes. Ten millilitres of whole blood were drawn from each animal into serum vacutainer tubes (Fisher Scientific, Suwanee, GA, USA) by venipuncture of the jugular vein. Serum samples were left to clot for 2 h at ambient temperature. Thereafter, sera were centrifuged at 1000× g for 15 min, and the supernatant was harvested and stored at −80 ℃ until further analysis.
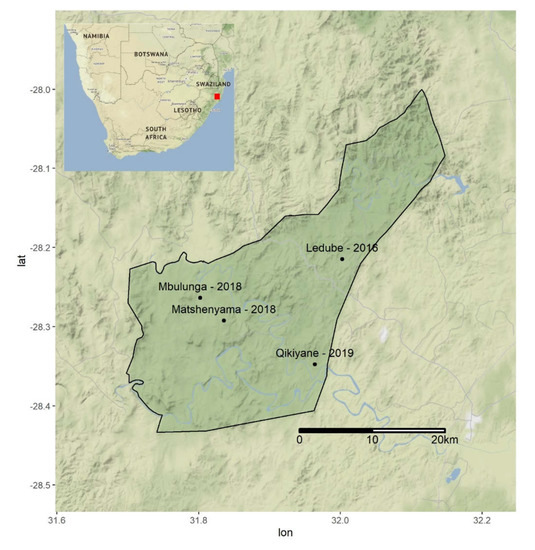
Figure 4.
The locations of buffalo sampling efforts in 2016, 2018, and 2019 within the Hluhluwe-iMfolozi Park (HiP). The location of the HiP within South Africa is indicated by the red square in Table 1. All sera were initially screened at a dilution of 1:80 for antibodies against 8 Leptospira spp. serovars (8 serogroups) using live antigens (Table 2).
Live cultures grown in Ellinghausen–McCullough–Johnson–Harris (EMJH) medium with densities of approximately 2 × 108 leptospires per ml were used as the antigens. An agglutination of 50–100% was taken as a reaction. Titration tests on reacting sera were performed by doubling dilutions of sera using phosphate buffered saline (Sorensen’s buffer), resulting in final dilutions of 1/100 through to 1/3200. The endpoint was defined as the dilution of serum that shows 50% agglutination, leaving 50% free leptospires compared with a control culture diluted 1:2 in Sorensen’s buffer. Results are available in Supplementary data (File S1).
As all positive samples collected in 2016 only reacted to serovar Tarassovi, animals that tested positive in either of the paired samples were considered positive for the calculation of overall population level seroprevalence. However, to enable comparison between 2016 and years where paired samples were not available, only the result from the first sample from 2016 was used for subsequent statistical analyses. Initial exploratory analyses utilised bivariate Chi square tests to examine associations between seroprevalence and age, sex, and year of sampling. Where low expected values violated assumptions, Chi square statistics were simulated using 1 × 106 replicates.
To quantify exposure risk while accounting for variance associated with other covariates, a generalised linear model (glm) was fitted with an individual’s age, sex, and year of sampling as covariates and exposure status as a Bernoulli response (seropositive/seronegative). The glm was implemented in a Bayesian framework using the rstanarm R package using default (weakly informative) priors. To facilitate comparisons between years, the response variable only included the results from the first sample collected in 2016.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/pathogens10091072/s1, File S1: MAT results and metadata.csv.
Author Contributions
Conceptualization, M.H.M., W.G., and M.M.; methodology, M.H.M., W.G., A.P., and M.M.; formal analysis, M.H.M. and W.G.; writing—original draft preparation, M.H.M.; writing—review and editing, W.G., A.P., T.J.K., and M.M.; visualization, M.H.M.; supervision, M.M.; project administration, M.H.M. and M.M.; funding acquisition, M.H.M. and M.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Wellcome Trust, grant number 216634/Z/19/Z to M.H.M. and grant number 222941/Z/21/Z to W.G. Sample collection and W.G., T.K., and M.M. were funded by the South African government through the South African Medical Research Council and the National Research Foundation South African Research Chair Initiative [grant #86949]. The APC was funded by the Wellcome Trust.
Institutional Review Board Statement
Ethical approval for this study was granted by the Stellenbosch University Animal Care and Use committee (ACU-2019-9081 and ACU-2021-23032). Permission to perform animal research in terms of section 20 of the Animal Diseases Act was granted by the South African Department of Agriculture, Land Reform and Rural Development (DALRRD), South Africa (12/11/1/7/2). All buffaloes were handled by the Enzemvelo KZN Wildlife veterinarians and game capture teams according to their standard operating procedures.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available in supplementary material.
Acknowledgments
We thank David Cooper, Alicia and Warren McCall, Rowan Leeming, Dumisani Zwane, JP van Heerden, and the Game Capture staff from KwaZulu-Natal Ezemvelo Wildlife for their assistance with this study. Shapefiles for the HiP boundary were kindly provided by KwaZulu-Natal Ezemvelo Wildlife.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Viana, M.; Mancy, R.; Biek, R.; Cleaveland, S.; Cross, P.; Lloyd-Smith, J.; Haydon, D. Assembling evidence for identifying reservoirs of infection. Trends Ecol. Evol. 2014, 29, 270–279. [Google Scholar] [CrossRef] [Green Version]
- Viana, M.; Cleaveland, S.; Matthiopoulos, J.; Halliday, J.; Packer, C.; Craft, M.E.; Hampson, K.; Czupryna, A.; Dobson, A.P.; Dubovi, E.J.; et al. Dynamics of a morbillivirus at the domestic-wildlife interface: Canine distemper virus in domestic dogs and lions. Proc. Natl. Acad. Sci. USA 2015, 112, 1464–1469. [Google Scholar] [CrossRef] [Green Version]
- Roeder, P.; Mariner, J.; Kock, R. Rinderpest: The veterinary perspective on eradication. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20120139. [Google Scholar] [CrossRef]
- Vincent, A.T.; Schiettekatte, O.; Goarant, C.; Neela, V.K.; Bernet, E.; Thibeaux, R.; Ismail, N.; Khalid, M.K.N.M.; Amran, F.; Masuzawa, T.; et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl. Trop. Dis. 2019, 13, e0007270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bulach, D.; Zuerner, R.L.; Wilson, P.; Seemann, T.; McGrath, A.; Cullen, P.A.; Davis, J.; Johnson, M.; Kuczek, E.; Alt, D.; et al. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc. Natl. Acad. Sci. USA 2006, 103, 14560–14565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allan, K.J.; Halliday, J.; Moseley, M.; Carter, R.W.; Ahmed, A.; Goris, M.G.A.; Hartskeerl, R.A.; Keyyu, J.; Kibona, T.; Maro, V.P.; et al. Assessment of animal hosts of pathogenic Leptospira in northern Tanzania. PLoS Negl. Trop. Dis. 2018, 12, e0006444. [Google Scholar] [CrossRef] [PubMed]
- Rahelinirina, S.; Moseley, M.H.; Allan, K.J.; Ramanohizakandrainy, E.; Ravaoarinoro, S.; Rajerison, M.; Rakotoharinome, V.; Telfer, S. Leptospira in livestock in Madagascar: Uncultured strains, mixed infections and small mammal-livestock transmission highlight challenges in controlling and diagnosing leptospirosis in the developing world. Parasitology 2019, 146, 1707–1713. [Google Scholar] [CrossRef]
- Hesterberg, U.W.; Bagnall, R.; Bosch, B.; Perrett, K.; Horner, R.; Gummow, B. A serological prevalence survey of leptospirosis in cattle of rural communities in the province of KwaZulu-Natal, South Africa. J. S. Afr. Vet. Assoc. 2008, 79, 15–18. [Google Scholar] [CrossRef] [Green Version]
- Hamond, C.; Martins, G.; Medeiros, M.A.; Lilenbaum, W. Presence of Leptospiral DNA in Semen Suggests Venereal Transmission in Horses. J. Equine Vet. Sci. 2013, 33, 1157–1159. [Google Scholar] [CrossRef] [Green Version]
- Martins, G.; Penna, B.; Lilenbaum, W. Maintenance of Leptospira infection in cattle under tropical conditions. Vet. Rec. 2010, 167, 629–630. [Google Scholar] [PubMed]
- Naidoo, K.; Moseley, M.; McCarthy, K.; Chingonzoh, R.; Lawrence, C.; Setshedi, G.M.; Frean, J.; Rossouw, J. Fatal Rodentborne Leptospirosis in Prison Inmates, South Africa, 2015. Emerg. Infect. Dis. 2020, 26, 1031–1033. [Google Scholar] [CrossRef]
- Moseley, M.; Naidoo, K.; Bastos, A.; Retief, L.; Frean, J.; Telfer, S.; Rossouw, J. Multi-locus sequence analyses reveal a clonal L. borgpetersenii genotype in a heterogeneous invasive Rattus spp. community across the City of Johannesburg, South Africa. Parasites Vectors 2020, 13, 570. [Google Scholar] [CrossRef]
- Simbizi, V.; Saulez, M.N.; Potts, A.; Lötter, C.; Gummow, B. A study of leptospirosis in South African horses and associated risk factors. Prev. Vet. Med. 2016, 134, 6–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atherstone, C.; Picozzi, K.; Kalema-Zikusoka, G. Seroprevalence of Leptospira hardjo in cattle and African buffalos in southwestern Uganda. Am. J. Trop. Med. Hyg. 2014, 90, 288–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myburgh, J.G.; Bengis, R.G.; Bester, C.J.; Chaparro, F. Serological reactions to Leptospira species in buffalo (Syncerus caffer) from the Kruger National Park. Onderstepoort. J. Vet. Res. 1990, 57, 281–282. [Google Scholar]
- Alinaitwe, L.; Kankya, C.; Namanya, D.; Pithua, P.; Dreyfus, A. Leptospira Seroprevalence Among Ugandan Slaughter Cattle: Comparison of Sero-Status With Renal Leptospira Infection. Front. Veter. Sci. 2020, 7, 106. [Google Scholar] [CrossRef]
- Dogonyaro, B.B.; van Heerden, H.; Potts, A.D.; Kolo, B.F.; Lotter, C.; Katsande, C.; Fasina, F.O.; Ko, A.I.; Wunder, E.A.; Adesiyun, A.A. Seroepidemiology of Leptospira infection in slaughtered cattle in Gauteng province, South Africa. Trop. Anim. Health Prod. 2020, 52, 3789–3798. [Google Scholar] [CrossRef]
- Wilson, P.; Mannewald, A.; Collins-Emerson, J.; Dreyfus, A.; Sanhueza, J.; Benschop, J.; Verdugo, C.; Emanuelson, U.; Boqvist, S.; Heuer, C. Serological study of Leptospira interrogans serovar Copenhageni and L. borgpetersenii serovars Tarassovi and Ballum in beef cattle, sheep and deer in New Zealand. N. Z. Veter. J. 2021, 69, 83–92. [Google Scholar] [CrossRef]
- Yupiana, Y.; Vallee, E.; Wilson, P.; Collins-Emerson, J.; Weston, J.; Benschop, J.; Heuer, C. Emerging Leptospira strain poses public health risk for dairy farmers in New Zealand. Prev. Vet. Med. 2019, 170, 104727. [Google Scholar] [CrossRef] [PubMed]
- Barasona, J.A.; Vicente, J.; Díez-Delgado, I.; Aznar, J.; Gortázar, C.; Torres, M.J. Environmental Presence of Mycobacterium tuberculosis Complex in Aggregation Points at the Wildlife/Livestock Interface. Transbound. Emerg. Dis. 2017, 64, 1148–1158. [Google Scholar] [CrossRef]
- Archer, E.R.M.; Landman, W.A.; Tadross, M.A.; Malherbe, J.; Weepener, H.; Maluleke, P.; Marumbwa, F.M. Understanding the evolution of the 2014–2016 summer rainfall seasons in southern Africa: Key lessons. Clim. Risk Manag. 2017, 16, 22–28. [Google Scholar] [CrossRef]
- Mbatha, N.; Xulu, S. Time series analysis of MODIS-Derived NDVI for the Hluhluwe-Imfolozi Park, South Africa: Impact of recent intense drought. Climate 2018, 6, 95. [Google Scholar] [CrossRef] [Green Version]
- Suwancharoen, D.; Chaisakdanugull, Y.; Thanapongtharm, W.; Yoshida, S. Serological survey of leptospirosis in livestock in Thailand. Epidemiol. Infect. 2013, 141, 2269–2277. [Google Scholar] [CrossRef] [Green Version]
- Turner, W.C.; Jolles, A.E.; Owen-Smith, N. Alternating sexual segregation during the mating season by male African buffalo (Syncerus caffer). J. Zool. 2005, 267, 291–299. [Google Scholar] [CrossRef] [Green Version]
- EKZNW. Annual Report 2018/19. 2019. Available online: http://www.kznwildlife.com/Documents/AnnualReport/EzemveloAR2019.pdf (accessed on 11 August 2021).
- Goosen, W.; Miller, M.; Chegou, N.N.; Cooper, D.; Warren, R.; van Helden, P.D.; Parsons, S.D. Agreement between assays of cell-mediated immunity utilizing Mycobacterium bovis-specific antigens for the diagnosis of tuberculosis in African buffaloes (Syncerus caffer). Vet. Immunol. Immunopathol. 2014, 160, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Jolles, A.E. Population biology of African buffalo (Syncerus caffer) at Hluhluwe-iMfolozi Park, South Africa. Afr. J. Ecol. 2007, 45, 398–406. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).