Variations in the Abortive HIV-1 RNA Hairpin Do Not Impede Viral Sensing and Innate Immune Responses
Abstract
:1. Introduction
2. Results
2.1. Variation in TAR Sequences Affects RNA Structure and Thermodynamic Stability
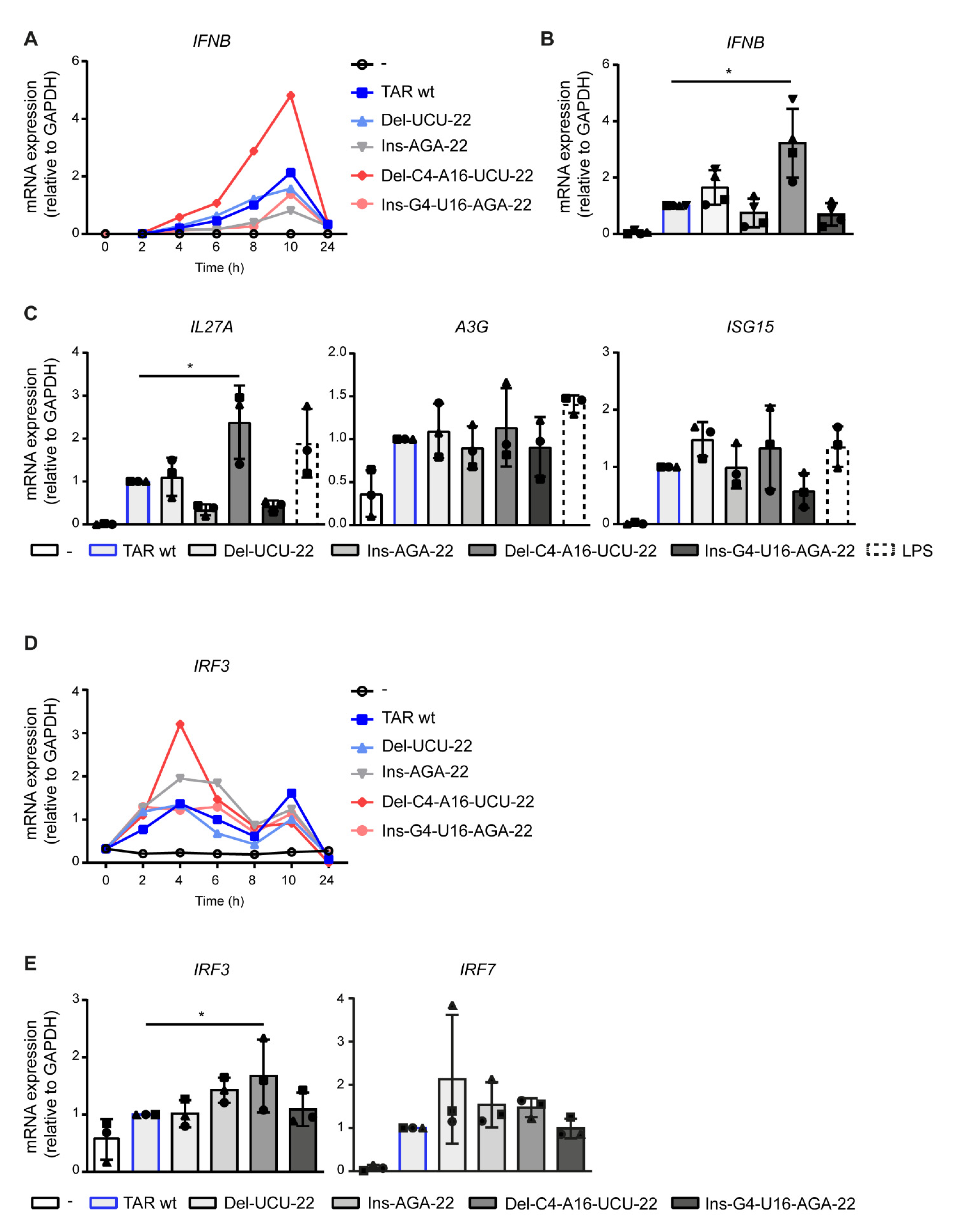
2.2. Deletions in the TAR Hairpin That Enhance Its Thermodynamic Stability Increase Antiviral Immunity
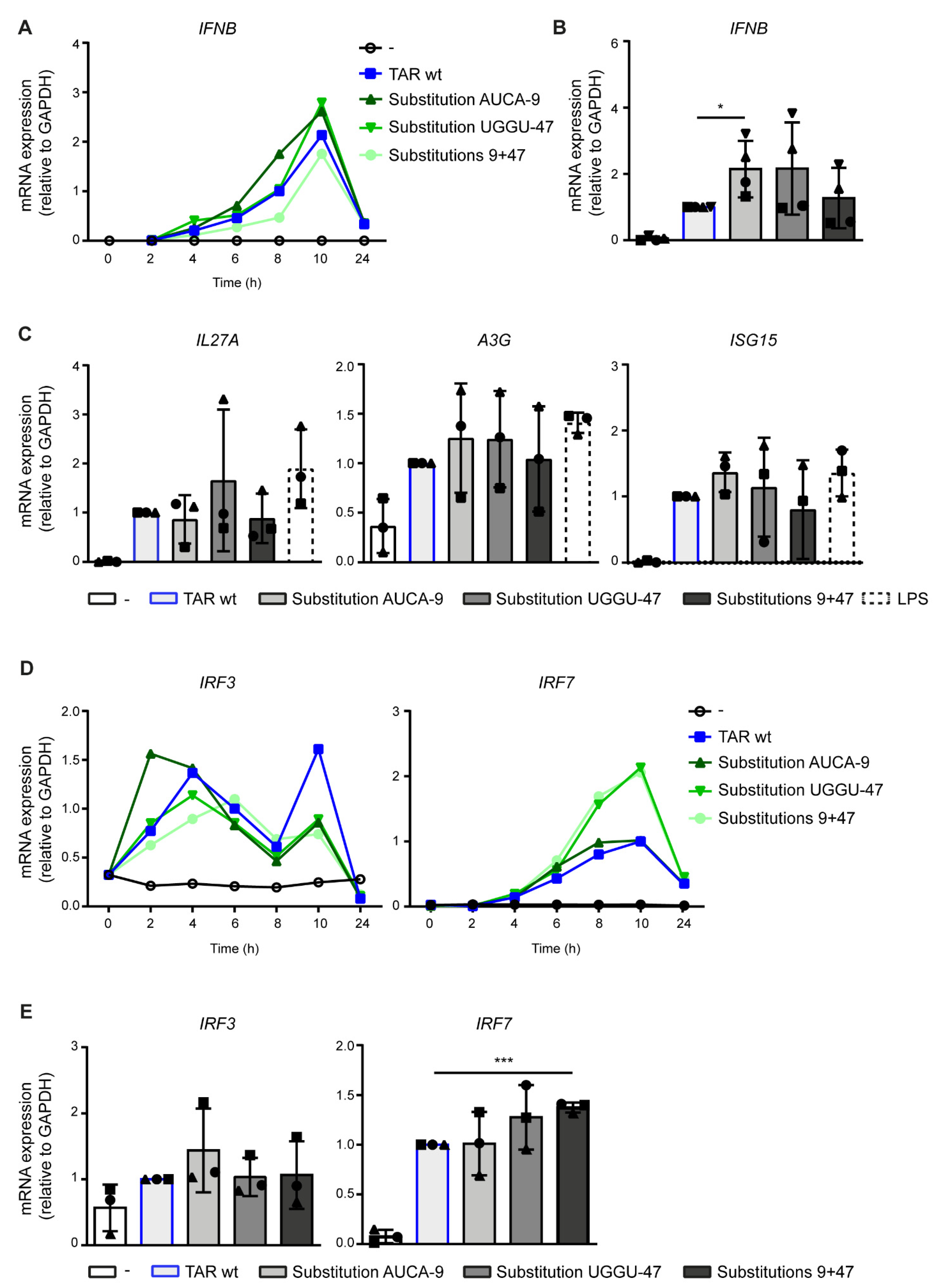
2.3. Destabilization of the Lower TAR Stem Enhances Antiviral Immunity
2.4. PKR Senses TAR Independent of Variation in Stability and Hairpin Integrity
3. Discussion
4. Materials and Methods
4.1. TAR RNA Sequences of People Living with HIV-1
4.2. TAR RNA Mutants
4.3. Cells and Reagents
4.4. Quantitative Real-Time PCR
4.5. ELISA
4.6. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Berkhout, B. Structural features in TAR RNA of human and simian immunodeficiency viruses: A phylogenetic analysis. Nucleic Acids Res. 1992, 20, 27–31. [Google Scholar] [CrossRef] [Green Version]
- Kao, S.; Calman, A.F.; Luciw, P.A.; Peterlin, B.M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 1987, 330, 489–493. [Google Scholar] [CrossRef] [PubMed]
- Harrich, D.; Hooker, C.W.; Parry, E. The Human Immunodeficiency Virus Type 1 TAR RNA Upper Stem-Loop Plays Distinct Roles in Reverse Transcription and RNA Packaging. J. Virol. 2000, 74, 5639–5646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berkhout, B. Multiple biological roles associated with the repeat (R) Region of the HIV-I RNA genome. Adv. Pharmacol. 2000, 48, 29–73. [Google Scholar] [CrossRef] [PubMed]
- Bannwarth, S.; Gatignol, A. HIV-1 TAR RNA: The Target of Molecular Interactions Between the Virus and its Host. Curr. HIV Res. 2005, 3, 61–71. [Google Scholar] [CrossRef]
- Tan, R.; Brodsky, A.; Williamson, J.R.; Frankel, A.D. RNA Recognition by HIV-1 Tat and Rev. In Seminars in VIROLOGY; Academic Press: Cambridge, MA, USA, 1997; Volume 8, pp. 186–193. [Google Scholar]
- Fischer, W.H.; Fang, S.-M.; Jones, K.A.; Wei, P.; Garber, M.E. A Novel CDK9-Associated C-Type Cyclin Interacts Directly with HIV-1 Tat and Mediates Its High-Affinity, Loop-Specific Binding to TAR RNA. Cell 1998, 92, 451–462. [Google Scholar]
- Richter, S.; Ping, Y.H.; Rana, T.M. TAR RNA loop: A scaffold for the assembly of a regulatory switch in HIV replication. Proc. Natl. Acad. Sci. USA 2002, 99, 7928–7933. [Google Scholar] [CrossRef] [Green Version]
- Bieniasz, P.D.; Grdina, T.A.; Bogerd, H.P.; Cullen, B.R. Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription. Proc. Natl. Acad. Sci. USA 1999, 96, 7791–7796. [Google Scholar] [CrossRef] [Green Version]
- Das, A.T.; Klaver, B.; Berkhout, B. The 5′ and 3′ TAR Elements of Human Immunodeficiency Virus Exert Effects at Several Points in the Virus Life Cycle. J. Virol. 1998, 72, 9217–9223. [Google Scholar] [CrossRef] [Green Version]
- Chavali, S.S.; Bonn-Breach, R.; Wedekind, J.E. Face-time with TAR: Portraits of an HIV-1 RNA with diverse modes of effector recognition relevant for drug discovery. J. Biol. Chem. 2019, 294, 9326–9341. [Google Scholar] [CrossRef] [Green Version]
- Gringhuis, S.I.; Hertoghs, N.; Kaptein, T.M.; Zijlstra-Willems, E.M.; Sarrami-Fooroshani, R.; Sprokholt, J.K.; van Teijlingen, N.H.; Kootstra, N.A.; Booiman, T.; van Dort, K.A.; et al. HIV-1 blocks the signaling adaptor MAVS to evade antiviral host defense after sensing of abortive HIV-1 RNA by the host helicase DDX3. Nat. Immunol. 2017, 18, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Stunnenberg, M.; Sprokholt, J.K.; van Hamme, J.L.; Kaptein, T.M.; Zijlstra-Willems, E.M.; Gringhuis, S.I.; Geijtenbeek, T.B.H. Synthetic Abortive HIV-1 RNAs Induce Potent Antiviral Immunity. Front. Immunol. 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Maitra, R.; McMillan, N.A.; Desai, S.; McSwiggen, J.; Hivanessian, A.G.; Sen, G.; Williams, B.R.; Silverman, R.H. HIV-1 TAR RNA has an intrinsic ability to activate interferon-inducible enzmes. Virology 1994, 204, 823–827. [Google Scholar] [CrossRef] [PubMed]
- Clerzius, G.; Gélinas, J.-F.; Gatignol, A. Multiple levels of PKR inhibition during HIV-1 replication. Rev. Med. Virol. 2010, 21, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Stunnenberg, M.; Geijtenbeek, T.B.H.; Gringhuis, S.I. DDX3 in HIV-1 infection and sensing: A paradox. Cytokine Growth Factor Rev. 2018, 40, 32–39. [Google Scholar] [CrossRef]
- Soto-Rifo, R.; Rubilar, P.S.; Limousin, T.; De Breyne, S.; Décimo, D.; Ohlmann, T. DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J. 2012, 31, 3745–3756. [Google Scholar] [CrossRef] [Green Version]
- Heinicke, L.A.; Wong, C.J.; Lary, J.; Nallagatla, S.R.; Diegelman-Parente, A.; Zheng, X.; Cole, J.L.; Bevilacqua, P.C. RNA Dimerization Promotes PKR Dimerization and Activation. J. Mol. Biol. 2009, 390, 319–338. [Google Scholar] [CrossRef] [Green Version]
- Kim, I.; Liu, C.W.; Puglisi, J.D. Specific Recognition of HIV TAR RNA by the dsRNA Binding Domains (dsRBD1-dsRBD2) of PKR. J. Mol. Biol. 2006, 358, 430–442. [Google Scholar] [CrossRef]
- Carpick, B.W.; Graziano, V.; Schneider, D.; Maitra, R.K.; Lee, X.; Williams, B.R.G. Characterization of the solution complex between the interferon-induced, double-stranded RNA-activated protein kinase and HIV-I trans-activating region RNA. J. Biol. Chem. 1997, 272, 9510–9516. [Google Scholar] [CrossRef] [Green Version]
- Clemens, M.J.; Elia, A. The double-stranded RNA-dependent protein kinase PKR: Structure and function. J. Interf. Cytokine Res. 1997, 17, 503–524. [Google Scholar] [CrossRef]
- Roy, S.; Agy, M.; Hovanessian, A.G.; Sonenberg, N.; Katze, M.G. The integrity of the stem structure of human immunodeficiency virus type 1 Tat-responsive sequence of RNA is required for interaction with the interferon-induced 68,000-Mr protein kinase. J. Virol. 1991, 65, 632–640. [Google Scholar] [CrossRef] [Green Version]
- Stunnenberg, M.; van Hamme, J.L.; Trimp, M.; Gringhuis, S.I.; Geijtenbeek, T.B.H. Abortive HIV-1 RNA induces pro-IL-1β maturation via protein kinase PKR and inflammasome activation in humans. Eur. J. Cell Biol. 2021. [Google Scholar] [CrossRef]
- Berkhout, B.; Silverman, R.H.; Jeang, K.T. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 1989, 59, 273–282. [Google Scholar] [CrossRef]
- Berkhout, B.; Jeang, K. tech Detailed mutational analysis of TAR RNA: Critical spacing between the bulge and loop recognition domains. Nucleic Acids Res. 1991, 19, 6169–6176. [Google Scholar] [CrossRef] [Green Version]
- Ho, Y.C.; Shan, L.; Hosmane, N.N.; Wang, J.; Laskey, S.B.; Rosenbloom, D.I.S.; Lai, J.; Blankson, J.N.; Siliciano, J.D.; Siliciano, R.F. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 2013, 155, 540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruner, K.M.; Murray, A.J.; Pollack, R.A.; Soliman, M.G.; Sarah, B.; Capoferri, A.A.; Lai, J.; Strain, M.C.; Lada, S.M.; Ho, Y.; et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat. Med. 2016, 22, 1043–1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, J.D.; Kharytonchyk, S.; Chaudry, I.; Iyer, A.S.; Carter, H.; Becker, G.; Desai, Y.; Glang, L.; Choi, S.H.; Singh, K.; et al. Structural basis for transcriptional start site control of HIV-1 RNA fate. Science 2020, 368, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Levy, D.E.; Marié, I.; Prakash, A. Ringing the interferon alarm: Differential regulation of gene expression at the interface between innate and adaptive immunity. Curr. Opin. Immunol. 2003, 15, 52–58. [Google Scholar] [CrossRef]
- Edery, I.; Petryshyn, R.; Sonenberg, N. Activation of double-stranded RNA-dependent kinase (dsl) by the TAR region of HIV-1 mRNA: A novel translational control mechanism. Cell 1989, 56, 303–312. [Google Scholar] [CrossRef]
- Klaver, B.; Berkhout, B. Evolution of a disrupted TAR RNA hairpin structure in the HIV-1 virus. EMBO J. 1994, 13, 2650–2659. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Mesman, A.W.; Zijlstra-Willems, E.M.; Kaptein, T.M.; De Swart, R.L.; Davis, M.E.; Ludlow, M.; Duprex, W.P.; Gack, M.U.; Gringhuis, S.I.; Geijtenbeek, T.B.H. Measles virus suppresses RIG-I-like receptor activation in dendritic cells via DC-SIGN-mediated inhibition of PP1 phosphatases. Cell Host Microbe 2014, 16, 31–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stunnenberg, M.; van Hamme, J.L.; Das, A.T.; Berkhout, B.; Geijtenbeek, T.B.H. Variations in the Abortive HIV-1 RNA Hairpin Do Not Impede Viral Sensing and Innate Immune Responses. Pathogens 2021, 10, 897. https://doi.org/10.3390/pathogens10070897
Stunnenberg M, van Hamme JL, Das AT, Berkhout B, Geijtenbeek TBH. Variations in the Abortive HIV-1 RNA Hairpin Do Not Impede Viral Sensing and Innate Immune Responses. Pathogens. 2021; 10(7):897. https://doi.org/10.3390/pathogens10070897
Chicago/Turabian StyleStunnenberg, Melissa, John L. van Hamme, Atze T. Das, Ben Berkhout, and Teunis B. H. Geijtenbeek. 2021. "Variations in the Abortive HIV-1 RNA Hairpin Do Not Impede Viral Sensing and Innate Immune Responses" Pathogens 10, no. 7: 897. https://doi.org/10.3390/pathogens10070897
APA StyleStunnenberg, M., van Hamme, J. L., Das, A. T., Berkhout, B., & Geijtenbeek, T. B. H. (2021). Variations in the Abortive HIV-1 RNA Hairpin Do Not Impede Viral Sensing and Innate Immune Responses. Pathogens, 10(7), 897. https://doi.org/10.3390/pathogens10070897





