Equine Herpesvirus Type 4 (EHV-4) Outbreak in Germany: Virological, Serological, and Molecular Investigations
Abstract
1. Introduction
2. Results
2.1. The Outbreak
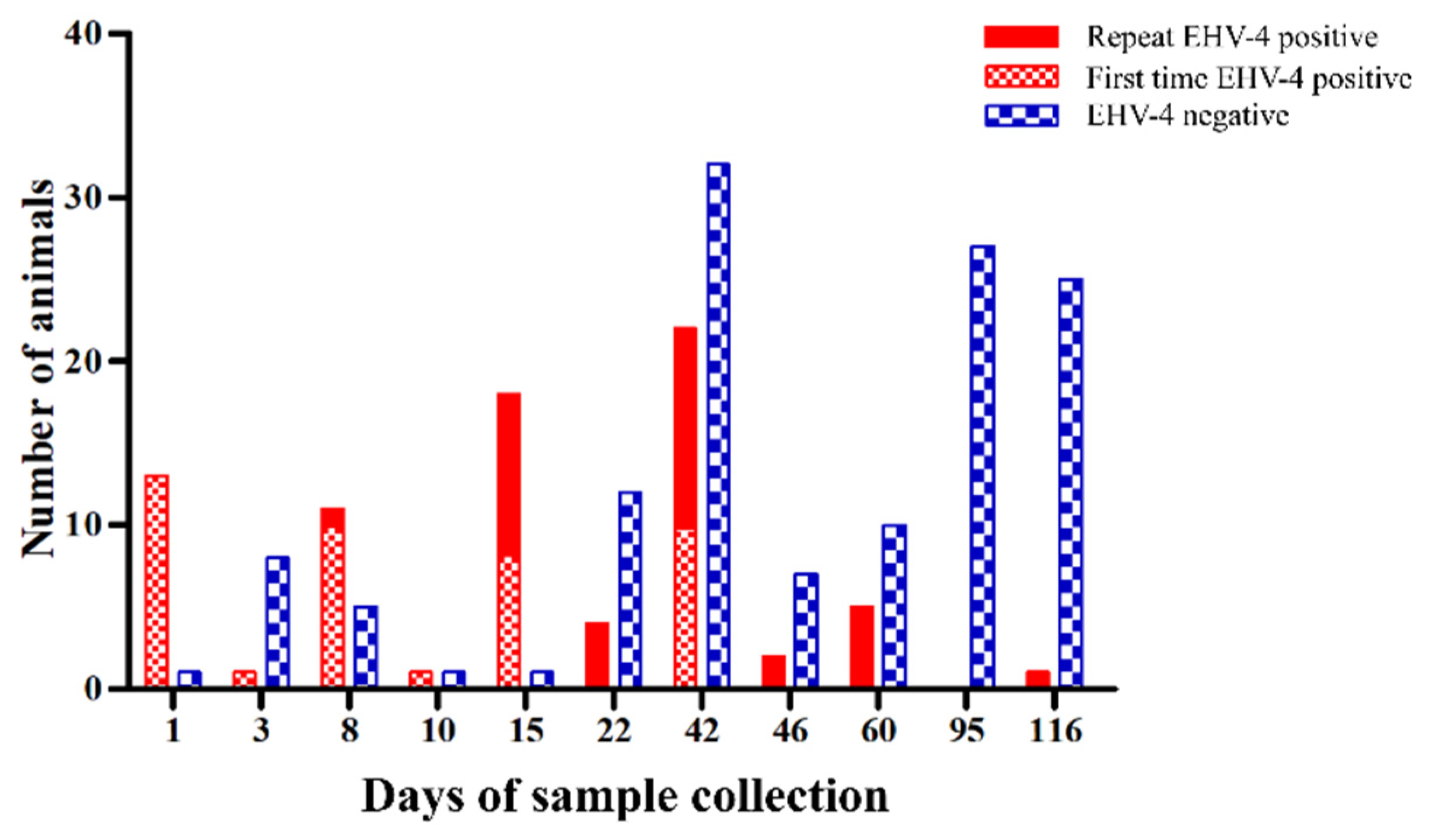
2.2. Virus Shedding
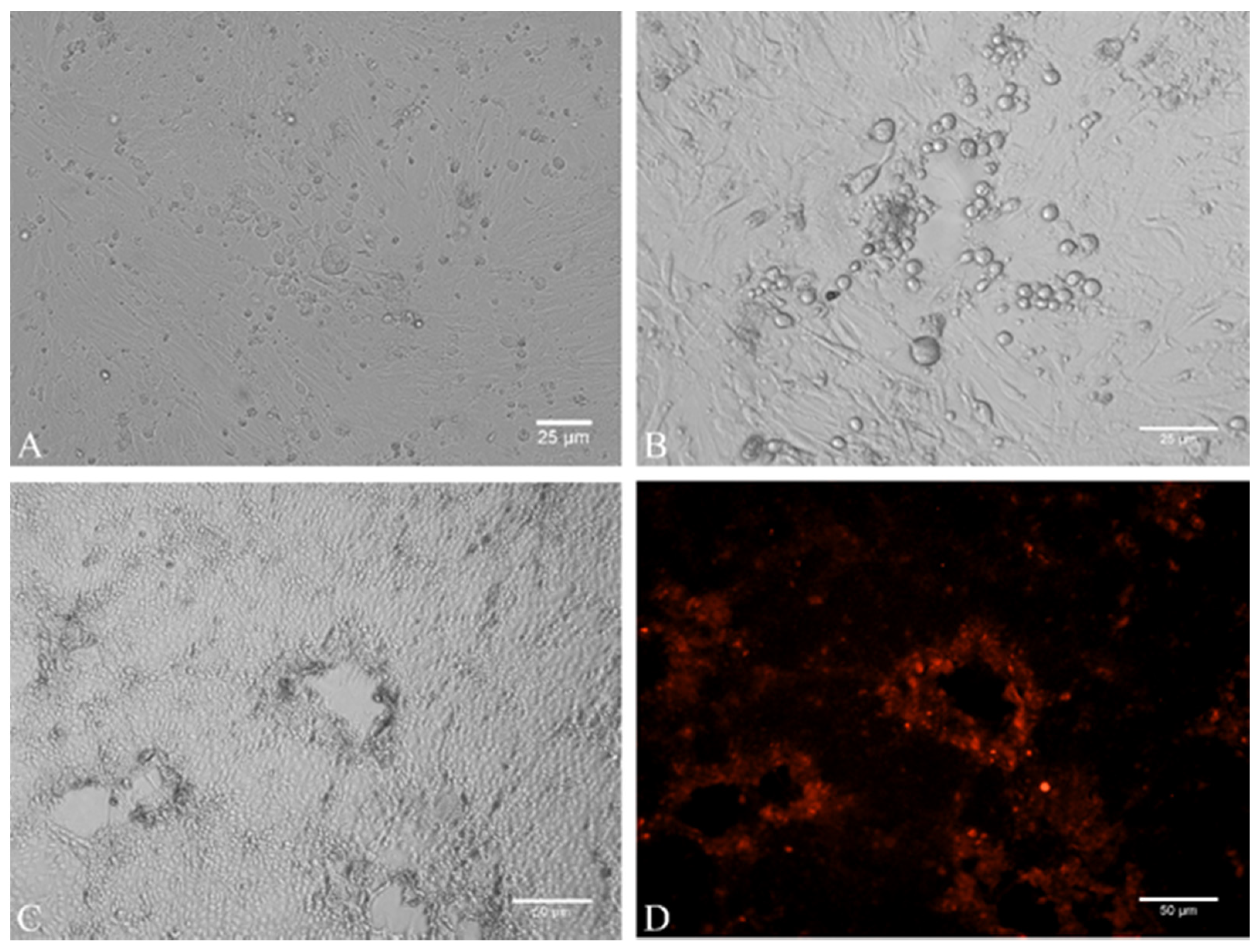
2.3. Virus Isolation
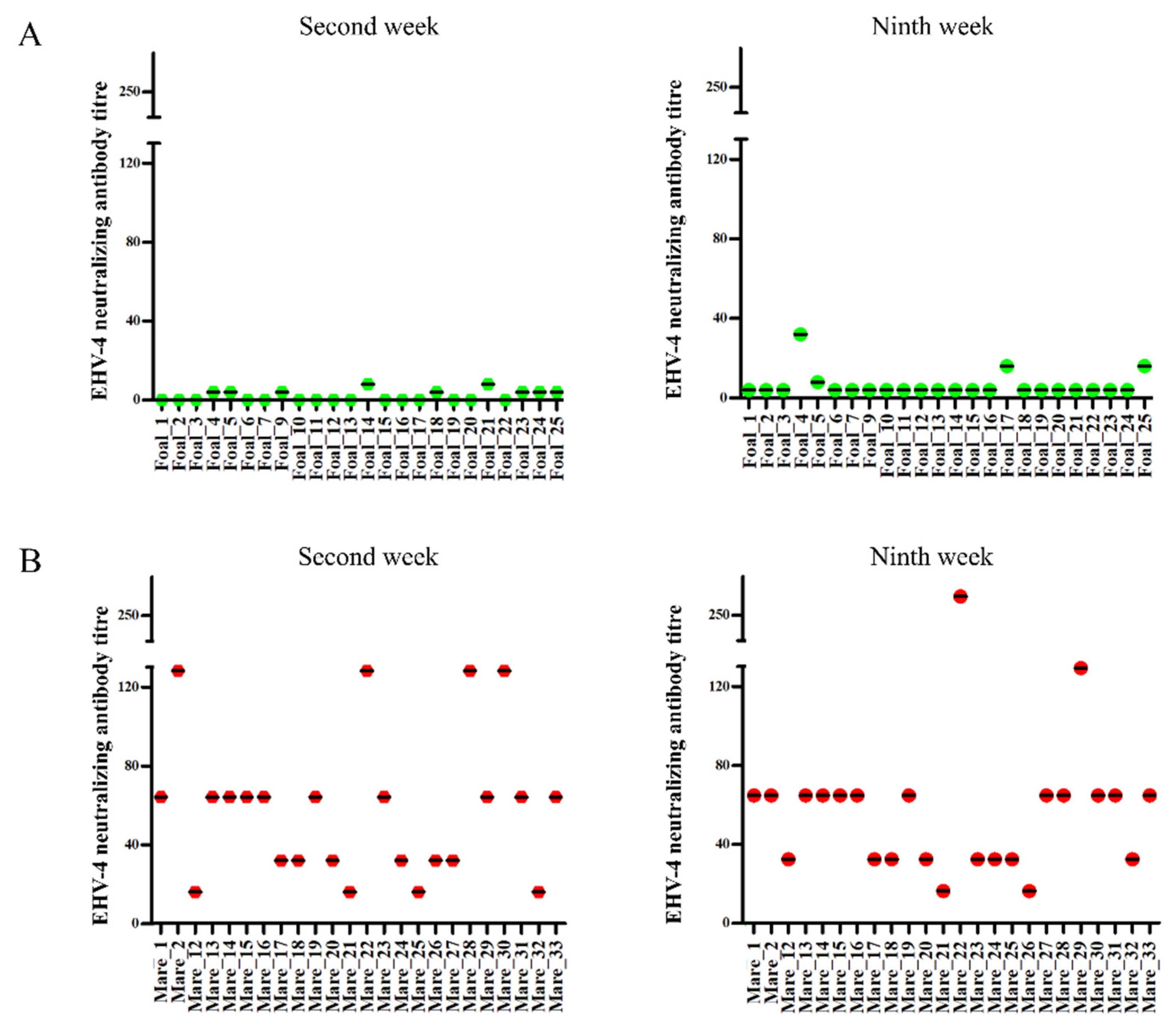
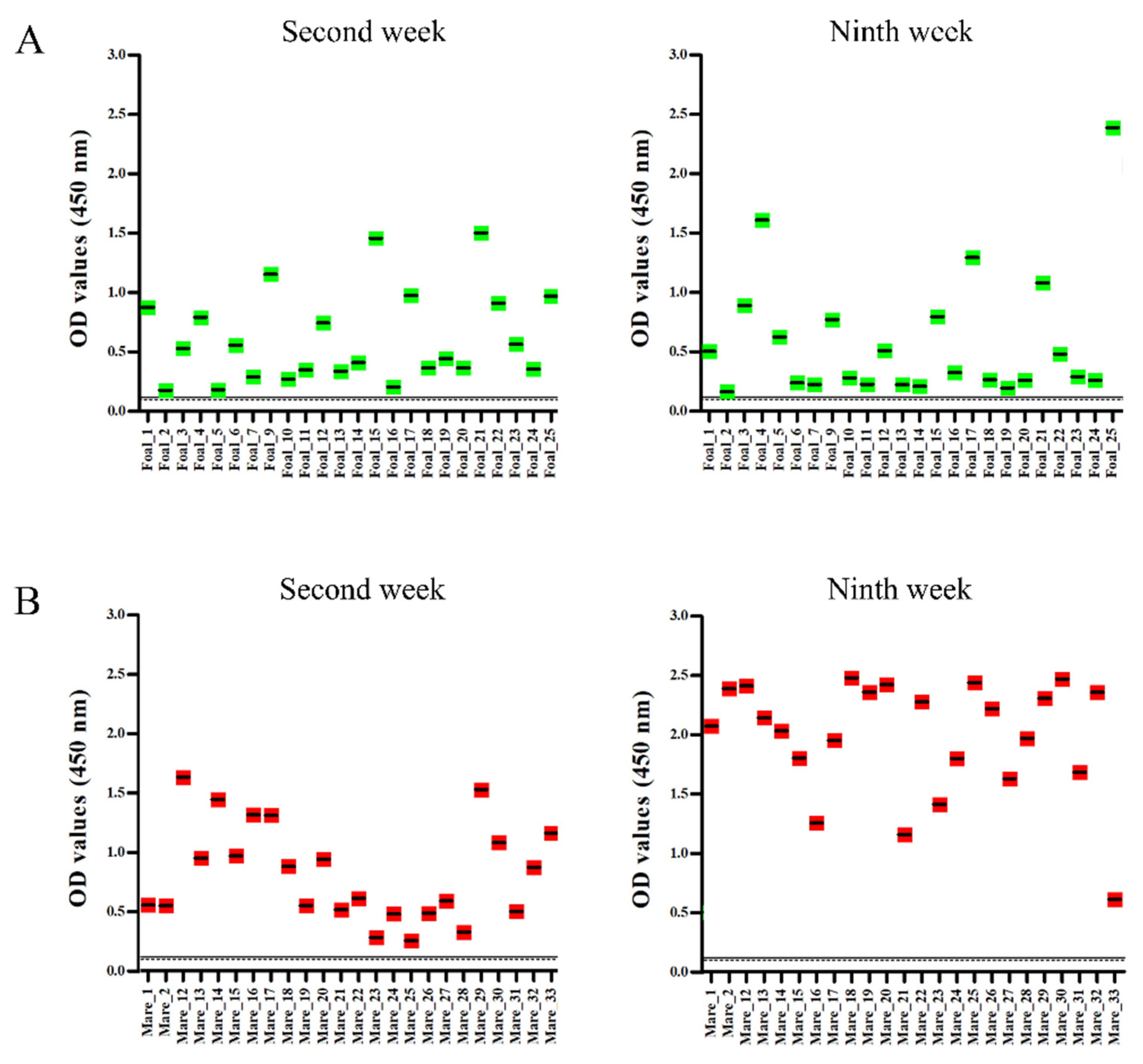
2.4. Serology
2.5. RFLP Analysis
2.6. Whole Genome Sequencing
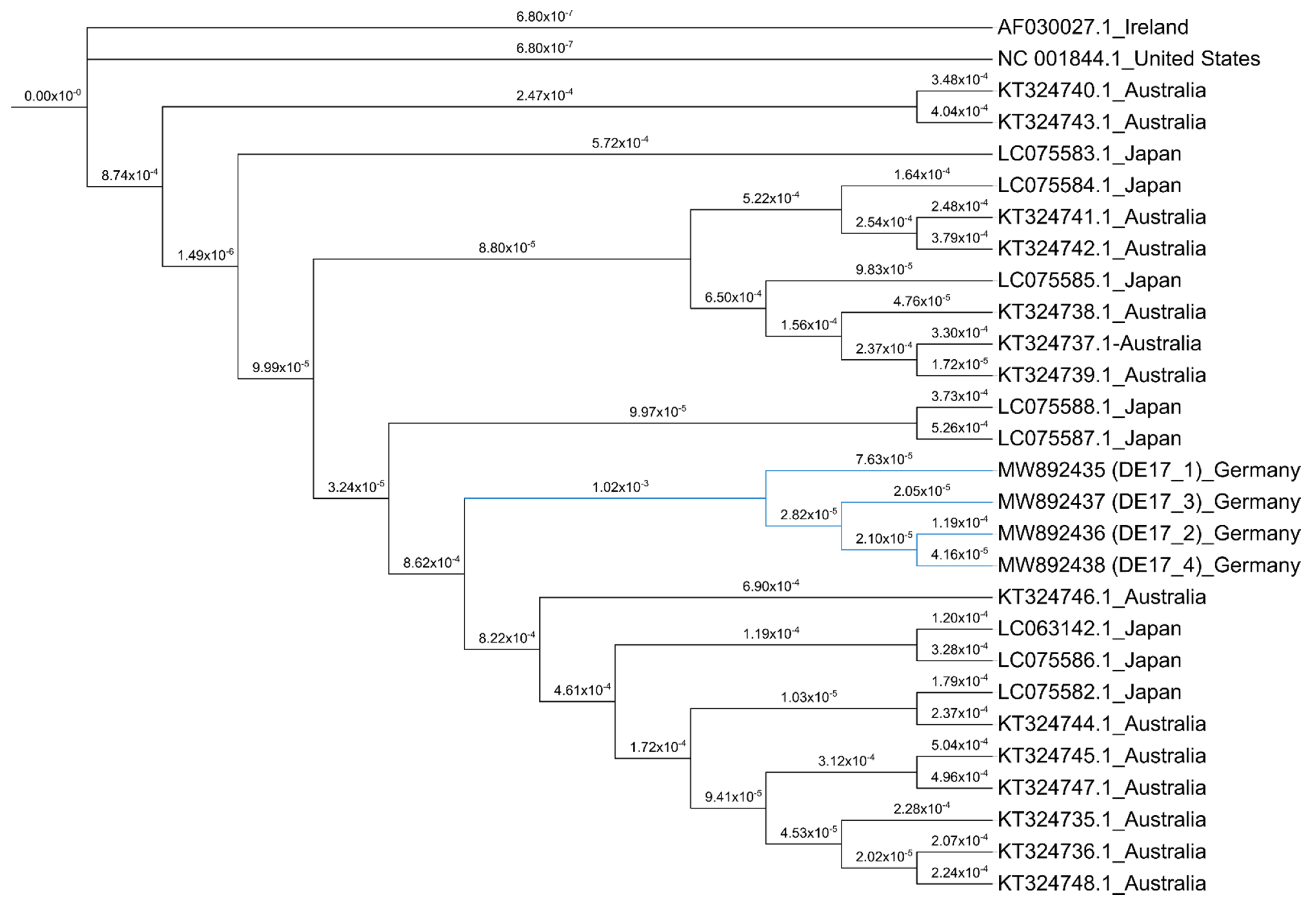
2.7. Phylogenetic Analysis
3. Discussion
4. Materials and Methods
4.1. Premise and Horses
4.2. Sample Collection
4.3. Quantitative (q)-PCR
4.4. Cell Culture and Virus Isolation
4.5. Indirect Immunofluorescence Assay (Indirect-IF)
4.6. Virus Neutralization Test (VNT)
4.7. Peptide ELISA
4.8. Restriction Fragment Length Polymorphism (RFLP)
4.9. Lllumina Library Preparation and Sequencing
4.10. Sequencing Data Analysis
4.11. Genome Sequencing and Phylogenetic Analysis
4.12. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Patel, J.R.; Heldens, J. Equine herpesviruses 1 (EHV-1) and 4 (EHV-4)--epidemiology, disease and immunoprophylaxis: A brief review. Vet. J. 2005, 170, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Crabb, B.S.; Studdert, M.J. Epitopes of glycoprotein G of equine herpesviruses 4 and 1 located near the C termini elicit type-specific antibody responses in the natural host. J. Virol. 1993, 67, 6332–6338. [Google Scholar] [CrossRef]
- Sabine, M.; Robertson, G.R.; Whalley, J.M. Differentiation of sub-types of equine herpesvirus I by restriction endonuclease analysis. Aust. Vet. J. 1981, 57, 148–149. [Google Scholar] [CrossRef]
- Allen, G.P.; Bryans, J.T. Molecular epizootiology, pathogenesis, and prophylaxis of equine herpesvirus-1 infections. Prog. Vet. Microbiol. Immunol. 1986, 2, 78–144. [Google Scholar]
- Zarski, L.M.; Giessler, K.S.; Jacob, S.I.; Weber, P.S.D.; McCauley, A.G.; Lee, Y.; Soboll Hussey, G. Identification of Host Factors Associated with the Development of Equine Herpesvirus Myeloencephalopathy by Transcriptomic Analysis of Peripheral Blood Mononuclear Cells from Horses. Viruses 2021, 13, 356. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.R.; Foldi, J.; Bateman, H.; Williams, J.; Didlick, S.; Stark, R. Equid herpesvirus (EHV-1) live vaccine strain C147: Efficacy against respiratory diseases following EHV types 1 and 4 challenges. Vet. Microbiol. 2003, 92, 1–17. [Google Scholar] [CrossRef]
- Mekonnen, A.; Eshetu, A.; Gizaw, D. Equine herpesvirus 1 and/or 4 in working equids: Seroprevalence and risk factors in North Shewa Zone, Ethiopia. Ethiop. Vet. J. 2017, 21, 28–39. [Google Scholar] [CrossRef][Green Version]
- Ataseven, V.S.; Bilge-Dagalp, S.; Oguzoglu, T.C.; Karapinar, Z.; Guzel, M.; Tan, M.T. Detection and sequence analysis of equine gammaherpesviruses from horses with respiratory tract disease in Turkey. Transbound. Emerg. Dis. 2010, 57, 271–276. [Google Scholar] [CrossRef]
- Crabb, B.S.; Studdert, M.J. Equine herpesviruses 4 (equine rhinopneumonitis virus) and 1 (equine abortion virus). Adv. Virus Res. 1995, 45, 153–190. [Google Scholar] [PubMed]
- Damiani, A.M.; de Vries, M.; Reimers, G.; Winkler, S.; Osterrieder, N. A severe equine herpesvirus type 1 (EHV-1) abortion outbreak caused by a neuropathogenic strain at a breeding farm in northern Germany. Vet. Microbiol. 2014, 172, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Gilkerson, J.R.; Whalley, J.M.; Drummer, H.E.; Studdert, M.J.; Love, D.N. Epidemiology of EHV-1 and EHV-4 in the mare and foal populations on a Hunter Valley stud farm: Are mares the source of EHV-1 for unweaned foals. Vet. Microbiol. 1999, 68, 27–34. [Google Scholar] [CrossRef]
- Yildirim, Y.; Yilmaz, V.; Kirmizigul, A.H. Equine herpes virus type 1 (EHV-1) and 4 (EHV-4) infections in horses and donkeys in northeastern Turkey. Iran. J. Vet. Res. 2015, 16, 341–344. [Google Scholar] [PubMed]
- Pusterla, N.; Bain, F.; James, K.; Mapes, S.; Kenelty, K.; Barnett, D.C.; Gaughan, E.; Craig, B.; Chappell, D.E.; Vaala, W. Frequency of molecular detection of equine herpesvirus-4 in nasal secretions of 3028 horses with upper airway infection. Vet. Rec. 2017, 180, 593. [Google Scholar] [CrossRef]
- Borchers, K.; Wolfinger, U.; Ludwig, H. Latency-associated transcripts of equine herpesvirus type 4 in trigeminal ganglia of naturally infected horses. J. Gen. Virol. 1999, 80, 2165–2171. [Google Scholar] [CrossRef][Green Version]
- Browning, G.F.; Ficorilli, N.; Studdert, M.J. Asinine herpesvirus genomes: Comparison with those of the equine herpesviruses. Arch. Virol. 1988, 101, 183–190. [Google Scholar] [CrossRef]
- Blunden, A.S.; Smith, K.C.; Binns, M.M.; Zhang, L.; Gower, S.M.; Mumford, J.A. Replication of equid herpesvirus 4 in endothelial cells and synovia of a field case of viral pneumonia and synovitis in a foal. J. Comp. Pathol. 1995, 112, 133–140. [Google Scholar] [CrossRef]
- Spiesschaert, B.; Goldenbogen, B.; Taferner, S.; Schade, M.; Mahmoud, M.; Klipp, E.; Osterrieder, N.; Azab, W. Role of gB and pUS3 in Equine Herpesvirus 1 Transfer between Peripheral Blood Mononuclear Cells and Endothelial Cells: A Dynamic In Vitro Model. J. Virol. 2015, 89, 11899–11908. [Google Scholar] [CrossRef]
- Pavulraj, S.; Kamel, M.; Stephanowitz, H.; Liu, F.; Plendl, J.; Osterrieder, N.; Azab, W. Equine Herpesvirus Type 1 Modulates Cytokine and Chemokine Profiles of Mononuclear Cells for Efficient Dissemination to Target Organs. Viruses 2020, 12, 999. [Google Scholar] [CrossRef]
- Bannai, H.; Nemoto, M.; Tsujimura, K.; Yamanaka, T.; Kondo, T.; Matsumura, T. Development of a Focus-Reduction Neutralizing test for Detecting Equine Herpesvirus Type-1-Neutralizing Antibodies. J. Vet. Med. Sci. 2013. [Google Scholar] [CrossRef] [PubMed]
- Lang, A.; de Vries, M.; Feineis, S.; Muller, E.; Osterrieder, N.; Damiani, A.M. Development of a peptide ELISA for discrimination between serological responses to equine herpesvirus type 1 and 4. J. Virol. Methods 2013, 193, 667–673. [Google Scholar] [CrossRef] [PubMed]
- OIE. OIE Manual for Terrestial Animals. 2018. Equine Rhinopneumonitis. (Chapter 2.5.9). Available online: http://www.oie.int/standard-setting/terrestrial-manual/access-online/2.05.09_EQUINE_RHINO.pdf (accessed on 12 December 2018).
- Studdert, M.J.; Simpson, T.; Roizman, B. Differentiation of respiratory and abortigenic isolates of equine herpesvirus 1 by restriction endonucleases. Science 1981, 214, 562–564. [Google Scholar] [CrossRef] [PubMed]
- Heldens, J.G.; Pouwels, H.G.; van Loon, A.A. Efficacy and duration of immunity of a combined equine influenza and equine herpesvirus vaccine against challenge with an American-like equine influenza virus (A/equi-2/Kentucky/95). Vet. J. 2004, 167, 150–157. [Google Scholar] [CrossRef]
- Burki, F.; Rossmanith, W.; Nowotny, N.; Pallan, C.; Mostl, K.; Lussy, H. Viraemia and abortions are not prevented by two commercial equine herpesvirus-1 vaccines after experimental challenge of horses. Vet. Q. 1990, 12, 80–86. [Google Scholar] [CrossRef]
- Pusterla, N.; Kass, P.H.; Mapes, S.; Johnson, C.; Barnett, D.C.; Vaala, W.; Gutierrez, C.; McDaniel, R.; Whitehead, B.; Manning, J. Surveillance programme for important equine infectious respiratory pathogens in the USA. Vet. Rec. 2011, 169, 12. [Google Scholar] [CrossRef] [PubMed]
- Studdert, M.J. Restriction endonuclease DNA fingerprinting of respiratory, foetal and perinatal foal isolates of equine herpesvirus type 1. Arch. Virol. 1983, 77, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, T.; Sugiura, T.; Imagawa, H.; Fukunaga, Y.; Kamada, M. Epizootiological aspects of type 1 and type 4 equine herpesvirus infections among horse populations. J. Vet. Med. Sci. 1992, 54, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Smith, F.L.; Watson, J.L.; Spier, S.J.; Kilcoyne, I.; Mapes, S.; Sonder, C.; Pusterla, N. Frequency of shedding of respiratory pathogens in horses recently imported to the United States. J. Vet. Intern. Med. 2018, 32, 1436–1441. [Google Scholar] [CrossRef]
- Afify, A.F.; Ahmed, B.M.; Salem, S.A.; El-Sanousi, A.A.; Shalaby, M.A. First Isolation and identification of EHV-4 during abortion outbreak among Arabian horses in Egypt reflects an alteration in pathogenic potentiality of EHV-4. J. Virol. Sci. 2017, 2, 92–101. [Google Scholar]
- Ostlund, E.N. The equine herpesviruses. Vet. Clin. N. Am. Equine Pract. 1993, 9, 283–294. [Google Scholar] [CrossRef]
- Whitwell, K.E.; Smith, K.; Sinclair, R.; Mumford, J. Fetal lesions in spontaneous 472 EHV-4 abortions in mares. In Proceedings of the Seventh International Conference on Equine Infectious Diseases, Newmarket, UK, 11 June 1998; p. 354. [Google Scholar]
- Kendrick, J.W.; Stevenson, W. Immunity to equine herpesvirus 1 infection in foals during the first year of life. J. Reprod. Fertil. Suppl. 1979, 27, 615–618. [Google Scholar]
- Mumford, J.A.; Rossdale, P.D.; Jessett, D.M.; Gann, S.J.; Ousey, J.; Cook, R.F. Serological and virological investigations of an equid herpesvirus 1 (EHV-1) abortion storm on a stud farm in 1985. J. Reprod. Fertil. Suppl. 1987, 35, 509–518. [Google Scholar] [PubMed]
- Pusterla, N.; Leutenegger, C.M.; Wilson, W.D.; Watson, J.L.; Ferraro, G.L.; Madigan, J.E. Equine herpesvirus-4 kinetics in peripheral blood leukocytes and nasopharyngeal secretions in foals using quantitative real-time TaqMan PCR. J. Vet. Diagn. Investig. 2005, 17, 578–581. [Google Scholar] [CrossRef] [PubMed]
- Azab, W.; Osterrieder, N. Glycoproteins D of equine herpesvirus type 1 (EHV-1) and EHV-4 determine cellular tropism independently of integrins. J. Virol. 2012, 86, 2031–2044. [Google Scholar] [CrossRef]
- Thein, P.; Huselstein, P. Untersuchungen zur Stammvariabilität von Pferdeherpesviren des Typs EHV-4. Pferdeheilkunde 2000, 16, 479–486. [Google Scholar] [CrossRef]
- Sinzger, C.; Knapp, J.; Schmidt, K.; Kahl, M.; Jahn, G. A simple and rapid method for preparation of viral DNA from cell associated cytomegalovirus. J. Virol. Methods 1999, 81, 115–122. [Google Scholar] [CrossRef]
- Pavulraj, S.; Eschke, K.; Prahl, A.; Flugger, M.; Trimpert, J.; van den Doel, P.B.; Andreotti, S.; Kaessmeyer, S.; Osterrieder, N.; Azab, W. Fatal Elephant Endotheliotropic Herpesvirus Infection of Two Young Asian Elephants. Microorganisms 2019, 7, 396. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Chevreux, B.; Wetter, T.; Suhai, S. Genome Sequence Assembly Using Trace Signals and Additional Sequence Information. In Computer Science and Biology, Proceedings of the German Conference on Bioinformatics (GCB), Braunschweig, Germany, 10 October 2001; German Research Center for Biotechnology: Braunschweig, Germany, 2001; pp. 45–56. [Google Scholar]
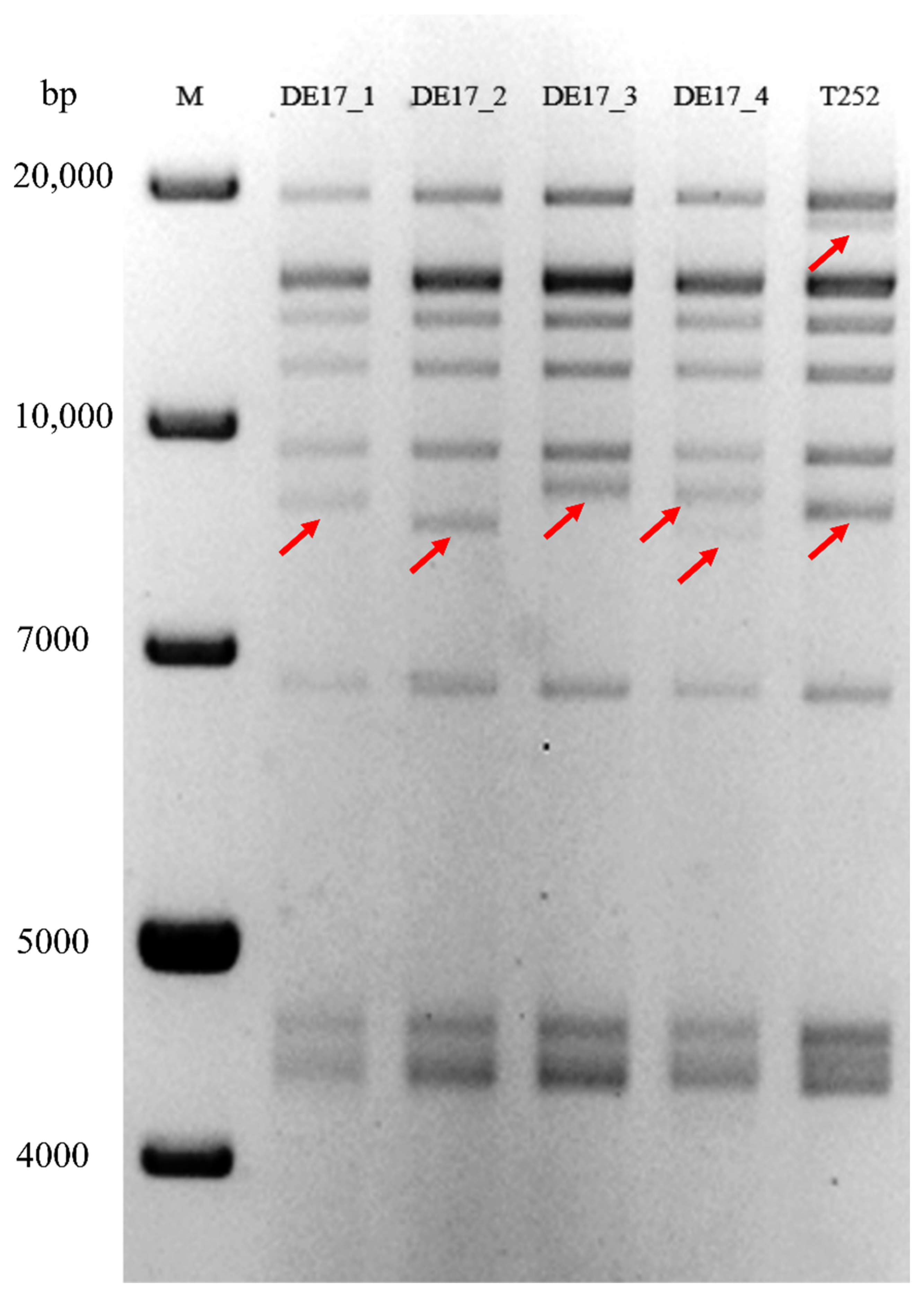
| S.No | Animal | Sex | Age | Day 1 | Day 3 | Day 8 | Day 10 | Day 15 | Day 22 | Day 42 | Day 46 | Day 60 | Day 95 | Day 116 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. | Mare_1 | Mare | 12 y 4 m | + | n/a | n/a | n/a | n/a | n/a | + | n/a | n/a | (−) | (−) |
| 2. | Foal_1 | Stallion | 164 d | ++ | n/a | n/a | n/a | ++ | n/a | (−) | n/a | +++ | n/a | (−) |
| 3. | Foal_2 | Stallion | 95 d | +++ | n/a | n/a | n/a | + | n/a | + | n/a | n/a | n/a | (−) |
| 4. | Foal_3 | Mare | 85 d | +/− | n/a | n/a | n/a | +/− | n/a | (−) | n/a | n/a | n/a | n/a |
| 5. | Foal_4 | Stallion | 126 d | +++ a | n/a | n/a | n/a | + | n/a | +/− | n/a | n/a | n/a | (−) |
| 6. | Mare_2 | Mare | 12 y 7 m | + | n/a | n/a | n/a | n/a | n/a | + | n/a | n/a | (−) | n/a |
| 7. | Foal_5 | Mare | 154 d | +++ | n/a | n/a | n/a | ++ | n/a | ++ | n/a | n/a | n/a | (−) |
| 8. | Foal_6 | Stallion | 121 d | ++ | n/a | n/a | n/a | + | n/a | + | n/a | n/a | n/a | (−) |
| 9. | Foal_7 | Mare | 87 d | + | n/a | n/a | n/a | + | n/a | (−) | n/a | n/a | n/a | n/a |
| 10. | Foal_8 | Mare | 95 d | + | n/a | n/a | n/a | n/a | n/a | + | n/a | n/a | n/a | (−) |
| 11. | Foal_9 | Stallion | 87 d | +/− | n/a | n/a | n/a | +/− | n/a | (−) | n/a | n/a | n/a | n/a |
| 12. | Foal_10 | Mare | 99 d | (−) | n/a | n/a | n/a | n/a | n/a | (−) | (−) | n/a | n/a | n/a |
| 13. | Foal_11 | Mare | 106 d | ++++ a | n/a | n/a | n/a | ++ | n/a | (−) | n/a | n/a | n/a | n/a |
| 14. | Foal_12 | Mare | 120 d | +++ | n/a | n/a | n/a | + | n/a | (−) | n/a | n/a | n/a | n/a |
| 15. | Mare_3 | Mare | 1 y 3 m | n/a | + | + | n/a | ++ | + | n/a | + | + | n/a | (−) |
| 16. | Stallion_1 | Stallion | 4 y 4 m | n/a | (−) | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| 17. | Gelding_1 | Gelding | 5 y 4 m | n/a | (−) | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| 18. | Gelding_2 | Gelding | 5 y 2 m | n/a | (−) | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| 19. | Mare_4 | Mare | 2 y 2 m | n/a | (−) | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| 20. | Mare_5 | Mare | 3 y 2 m | n/a | (−) | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| 21. | Stallion_2 | Stallion | 3 y 3 m | n/a | (−) | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| 22. | Mare_6 | Mare | 3 y 3 m | n/a | (−) | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| 23. | Stallion_3 | Stallion | 3 y 3 m | n/a | (−) | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| 24. | Stallion_4 | Stallion | 1 y 4 m | n/a | n/a | +/− | n/a | n/a | (−) | n/a | (−) | (−) | n/a | n/a |
| 25. | Stallion_5 | Stallion | 1 y 4 m | n/a | n/a | + | n/a | n/a | (−) | n/a | (−) | (−) | n/a | n/a |
| 26. | Stallion_6 | Stallion | 1 y 3 m | n/a | n/a | ++++ a | n/a | n/a | + | n/a | +/− | + | (−) | n/a |
| 27. | Stallion_7 | Stallion | 1 y 3 m | n/a | n/a | + | n/a | n/a | + | n/a | (−) | (−) | n/a | n/a |
| 28. | Stallion_8 | Stallion | 1 y 3 m | n/a | n/a | + | n/a | n/a | (−) | n/a | n/a | n/a | n/a | n/a |
| 29. | Stallion_9 | Stallion | 1 y 3 m | n/a | n/a | ++++ | n/a | n/a | (−) | + | (−) | n/a | n/a | n/a |
| 30. | Stallion_10 | Stallion | 1 y 2 m | n/a | n/a | ++++ | n/a | n/a | + | (−) | n/a | n/a | n/a | n/a |
| 31. | Stallion_11 | Stallion | 1 y 2 m | n/a | n/a | + | n/a | n/a | (−) | (−) | (−) | (−) | n/a | n/a |
| 32. | Stallion_12 | Stallion | 1 y 2 m | n/a | n/a | +++ | n/a | n/a | (−) | n/a | n/a | n/a | n/a | n/a |
| 33. | Stallion_13 | Stallion | 1 y 2 m | n/a | n/a | ++++ | n/a | n/a | (−) | n/a | (−) | (−) | n/a | n/a |
| 34. | Mare_7 | Mare | 1 y 3 m | n/a | n/a | (−) | n/a | n/a | (−) | n/a | n/a | n/a | n/a | n/a |
| 35. | Mare_8 | Mare | 1 y 3 m | n/a | n/a | (−) | n/a | n/a | (−) | n/a | n/a | n/a | n/a | n/a |
| 36. | Mare_9 | Mare | 1 y 3 m | n/a | n/a | (−) | n/a | n/a | (−) | n/a | n/a | n/a | n/a | n/a |
| 37. | Mare_10 | Mare | 1 y 4 m | n/a | n/a | (−) | n/a | n/a | (−) | n/a | n/a | n/a | n/a | n/a |
| 38. | Mare_11 | Mare | 1 y 3 m | n/a | n/a | (−) | n/a | n/a | (−) | n/a | n/a | n/a | n/a | n/a |
| 39. | Foal_13 | Stallion | 114 d | n/a | n/a | n/a | ++++ a | n/a | n/a | (−) | n/a | n/a | (−) | (−) |
| 40. | Foal_14 | Mare | 74 d | n/a | n/a | n/a | (−) | (−) | n/a | (−) | n/a | (−) | n/a | n/a |
| 41. | Foal_15 | Stallion | 147 d | n/a | n/a | n/a | n/a | + | n/a | + | n/a | ++ | n/a | (−) |
| 42. | Foal_16 | Stallion | 161 d | n/a | n/a | n/a | n/a | + | n/a | +/− | n/a | (−) | n/a | n/a |
| 43. | Foal_17 | Stallion | 209 d | n/a | n/a | n/a | n/a | +++ | n/a | + | n/a | +/− | n/a | (−) |
| 44. | Foal_18 | Stallion | 87 d | n/a | n/a | n/a | n/a | ++ | n/a | (−) | n/a | n/a | n/a | n/a |
| 45. | Foal_19 | Mare | 95 d | n/a | n/a | n/a | n/a | + | n/a | n/a | n/a | n/a | n/a | (−) |
| 46. | Foal_20 | Stallion | 99 d | n/a | n/a | n/a | n/a | ++ | n/a | +/− | n/a | n/a | n/a | (−) |
| 47. | Foal_21 | Stallion | 100 d | n/a | n/a | n/a | n/a | + | n/a | (−) | n/a | n/a | n/a | n/a |
| 48. | Mare_12 | Mare | 12 y 5 m | n/a | n/a | n/a | n/a | n/a | n/a | + | n/a | n/a | (−) | (−) |
| 49. | Mare_13 | Mare | 11 y 4 m | n/a | n/a | n/a | n/a | n/a | n/a | (−) | n/a | n/a | (−) | (−) |
| 50. | Mare_14 | Mare | 18 y 4 m | n/a | n/a | n/a | n/a | n/a | n/a | + | n/a | n/a | (−) | (−) |
| 51. | Mare_15 | Mare | 13 y 3 m | n/a | n/a | n/a | n/a | n/a | n/a | + | n/a | n/a | (−) | + |
| 52. | Mare_16 | Mare | 11 y 6 m | n/a | n/a | n/a | n/a | n/a | n/a | (−) | n/a | n/a | (−) | (−) |
| 53. | Mare_17 | Mare | 7 y 2 m | n/a | n/a | n/a | n/a | n/a | n/a | (−) | n/a | n/a | (−) | (−) |
| 54. | Mare_18 | Mare | 6 y 3 m | n/a | n/a | n/a | n/a | n/a | n/a | (−) | n/a | n/a | (−) | (−) |
| 55. | Mare_19 | Mare | 15 y 3 m | n/a | n/a | n/a | n/a | n/a | n/a | (−) | n/a | n/a | (−) | (−) |
| 56. | Mare_20 | Mare | 10 y 3 m | n/a | n/a | n/a | n/a | n/a | n/a | (−) | n/a | n/a | (−) | (−) |
| 57. | Mare_21 | Mare | 5 y 4 m | n/a | n/a | n/a | n/a | n/a | n/a | (−) | n/a | n/a | (−) | (−) |
| 58. | Mare_22 | Mare | 7 y 3 m | n/a | n/a | n/a | n/a | n/a | n/a | + | n/a | n/a | (−) | (−) |
| 59. | Mare_23 | Mare | 8 y 4 m | n/a | n/a | n/a | n/a | n/a | n/a | +/− | n/a | n/a | (−) | n/a |
| 60. | Mare_24 | Mare | 13 y 3 m | n/a | n/a | n/a | n/a | n/a | n/a | + | n/a | n/a | (−) | (−) |
| 61. | Mare_25 | Mare | 6 y 2 m | n/a | n/a | n/a | n/a | n/a | n/a | + | n/a | n/a | (−) | n/a |
| 62. | Mare_26 | Mare | 5 y 5 m | n/a | n/a | n/a | n/a | n/a | n/a | (−) | n/a | n/a | (−) | n/a |
| 63. | Mare_27 | Mare | 13 y 5 m | n/a | n/a | n/a | n/a | n/a | n/a | (−) | n/a | n/a | n/a | n/a |
| 64. | Mare_28 | Mare | 10 y 3 m | n/a | n/a | n/a | n/a | n/a | n/a | +/− | n/a | n/a | (−) | n/a |
| 65. | Mare_29 | Mare | 10 y 3 m | n/a | n/a | n/a | n/a | n/a | n/a | (−) | n/a | n/a | (−) | n/a |
| 66. | Mare_30 | Mare | 9 y 2 m | n/a | n/a | n/a | n/a | n/a | n/a | (−) | n/a | n/a | (−) | n/a |
| 67. | Mare_31 | Mare | 11 y 2 m | n/a | n/a | n/a | n/a | n/a | n/a | (−) | n/a | (−) | (−) | n/a |
| 68. | Mare_32 | Mare | 7 y 5 m | n/a | n/a | n/a | n/a | n/a | n/a | (−) | n/a | n/a | (−) | n/a |
| 69. | Mare_33 | Mare | 16 y 4 m | n/a | n/a | n/a | n/a | n/a | n/a | +/− | n/a | (−) | (−) | n/a |
| 70. | Foal_22 | Mare | 154 d | n/a | n/a | n/a | n/a | n/a | n/a | (−) | n/a | n/a | n/a | n/a |
| 71. | Foal_23 | Mare | 103 d | n/a | n/a | n/a | n/a | n/a | n/a | (−) | n/a | n/a | n/a | n/a |
| 72. | Foal_24 | Mare | 100 d | n/a | n/a | n/a | n/a | n/a | n/a | +/− | n/a | n/a | (−) | n/a |
| 73. | Foal_25 | Mare | 86 d | n/a | n/a | n/a | n/a | n/a | n/a | (−) | n/a | (−) | n/a | n/a |
| 74. | Stallion_14 | Stallion | 105 d | n/a | n/a | n/a | n/a | n/a | n/a | (−) | n/a | n/a | n/a | n/a |
| 75. | Stallion_15 | Stallion | 19 y 5 m | n/a | n/a | n/a | n/a | n/a | n/a | (−) | n/a | n/a | n/a | (−) |
| 76. | Mare_34 | Mare | 4 y 3 m | n/a | n/a | n/a | n/a | n/a | n/a | (−) | n/a | n/a | (−) | n/a |
| Animals | Number of Samples | VNT | ELISA | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 2nd Week | 9th Week | 2nd Week | 9th Week | ||||||
| Positive | Negative | Positive | Negative | Positive | Negative | Positive | Negative | ||
| Foals | 24 | 0 | 24 | 3 | 21 | 24 | 0 | 24 | 0 |
| Mares | 24 | 24 | 0 | 24 | 0 | 24 | 0 | 24 | 0 |
| Total | 48 | 24 | 24 | 27 | 21 | 48 | 0 | 48 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pavulraj, S.; Eschke, K.; Theisen, J.; Westhoff, S.; Reimers, G.; Andreotti, S.; Osterrieder, N.; Azab, W. Equine Herpesvirus Type 4 (EHV-4) Outbreak in Germany: Virological, Serological, and Molecular Investigations. Pathogens 2021, 10, 810. https://doi.org/10.3390/pathogens10070810
Pavulraj S, Eschke K, Theisen J, Westhoff S, Reimers G, Andreotti S, Osterrieder N, Azab W. Equine Herpesvirus Type 4 (EHV-4) Outbreak in Germany: Virological, Serological, and Molecular Investigations. Pathogens. 2021; 10(7):810. https://doi.org/10.3390/pathogens10070810
Chicago/Turabian StylePavulraj, Selvaraj, Kathrin Eschke, Jana Theisen, Stephanie Westhoff, Gitta Reimers, Sandro Andreotti, Nikolaus Osterrieder, and Walid Azab. 2021. "Equine Herpesvirus Type 4 (EHV-4) Outbreak in Germany: Virological, Serological, and Molecular Investigations" Pathogens 10, no. 7: 810. https://doi.org/10.3390/pathogens10070810
APA StylePavulraj, S., Eschke, K., Theisen, J., Westhoff, S., Reimers, G., Andreotti, S., Osterrieder, N., & Azab, W. (2021). Equine Herpesvirus Type 4 (EHV-4) Outbreak in Germany: Virological, Serological, and Molecular Investigations. Pathogens, 10(7), 810. https://doi.org/10.3390/pathogens10070810






