Regulation of the Human Papillomavirus Lifecyle through Post-Translational Modifications of the Viral E2 Protein
Abstract
1. Introduction
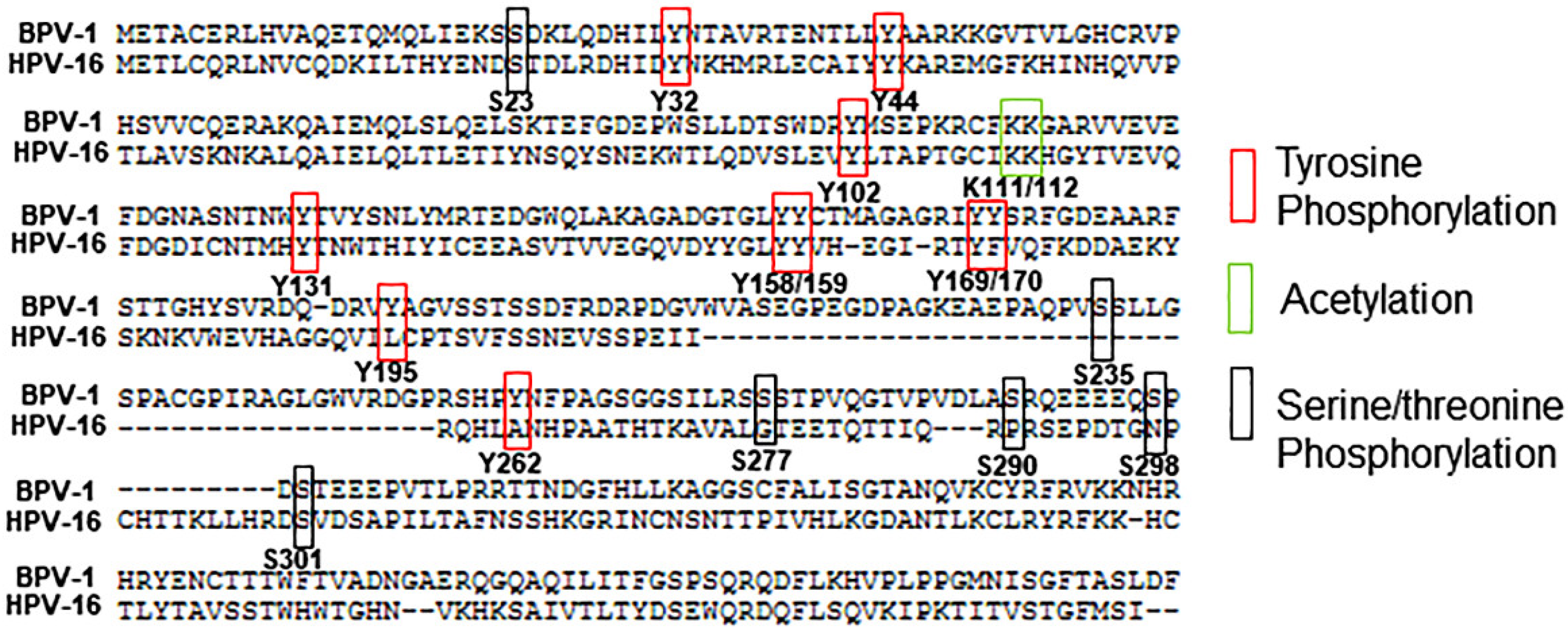
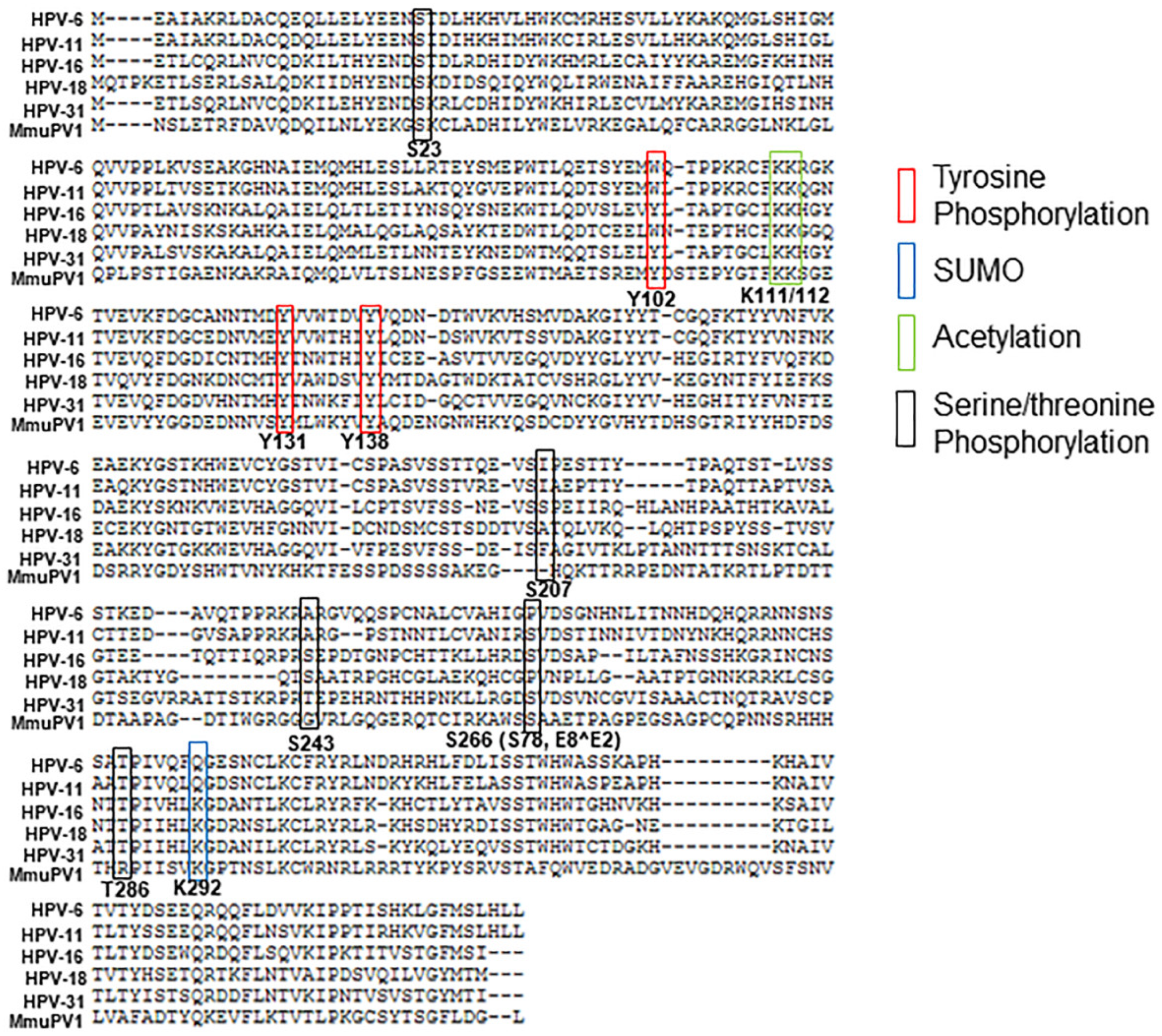
2. Serine/Threonine Phosphorylation
3. Tyrosine Phosphorylation
4. Acetylation
5. Sumoylation
6. Ubiquitination
7. Future Perspectives
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Frattini, M.G.; Laimins, L.A. The role of the E1 and E2 proteins in the replication of human papillomavirus type 31b. Virology 1994, 204, 799–804. [Google Scholar] [CrossRef]
- Reinson, T.; Henno, L.; Toots, M.; Ustav, M., Jr.; Ustav, M. The Cell Cycle Timing of Human Papillomavirus DNA Replication. PLoS ONE 2015, 10, e0131675. [Google Scholar] [CrossRef]
- Hoffmann, R.; Hirt, B.; Bechtold, V.; Beard, P.; Raj, K. Different modes of human papillomavirus DNA replication during maintenance. J. Virol. 2006, 80, 4431–4439. [Google Scholar] [CrossRef]
- McKinney, C.; Hussmann, K.; McBride, A. The Role of the DNA Damage Response throughout the Papillomavirus Life Cycle. Viruses 2015, 7, 2450–2469. [Google Scholar] [CrossRef]
- Hong, S.; Laimins, L.A. Regulation of the life cycle of HPVs by differentiation and the DNA damage response. Future Microbiol. 2013, 8, 1547–1557. [Google Scholar] [CrossRef]
- McBride, A.A. The Papillomavirus E2 proteins. Virology 2013, 445, 57–79. [Google Scholar] [CrossRef]
- Graham, S.V. Human Papillomavirus E2 Protein: Linking Replication, Transcription, and RNA Processing. J. Virol. 2016, 90, 8384–8388. [Google Scholar] [CrossRef]
- Sanders, C.M.; Stern, P.L.; Maitland, N.J. Characterization of human papillomavirus type 16 E2 protein and subdomains expressed in insect cells. Virology 1995, 211, 418–433. [Google Scholar] [CrossRef][Green Version]
- Chang, S.W.; Liu, W.C.; Liao, K.Y.; Tsao, Y.P.; Hsu, P.H.; Chen, S.L. Phosphorylation of HPV-16 E2 at serine 243 enables binding to Brd4 and mitotic chromosomes. PLoS ONE 2014, 9, e110882. [Google Scholar] [CrossRef] [PubMed]
- Johansson, C.; Graham, S.V.; Dornan, E.S.; Morgan, I.M. The human papillomavirus 16 E2 protein is stabilised in S phase. Virology 2009, 394, 194–199. [Google Scholar] [CrossRef]
- DeSmet, M.; Kanginakudru, S.; Jose, L.; Xie, F.; Gilson, T.; Androphy, E.J. Papillomavirus E2 protein is regulated by specific fibroblast growth factor receptors. Virology 2018, 521, 62–68. [Google Scholar] [CrossRef]
- Xie, F.; DeSmet, M.; Kanginakudru, S.; Jose, L.; Culleton, S.P.; Gilson, T.; Li, C.; Androphy, E.J. Kinase Activity of Fibroblast Growth Factor Receptor 3 Regulates Activity of the Papillomavirus E2 Protein. J. Virol. 2017, 91, e01066-17. [Google Scholar] [CrossRef]
- Ma, T.; Zou, N.; Lin, B.Y.; Chow, L.T.; Harper, J.W. Interaction between cyclin-dependent kinases and human papillomavirus replication-initiation protein E1 is required for efficient viral replication. Proc. Natl. Acad. Sci. USA 1999, 96, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, M.S.; Wettstein, F.O. E2 of cottontail rabbit papillomavirus is a nuclear phosphoprotein translated from an mRNA encoding multiple open reading frames. J. Virol. 1988, 62, 3242–3249. [Google Scholar] [CrossRef] [PubMed]
- Sekhar, V.; McBride, A.A. Phosphorylation regulates binding of the human papillomavirus type 8 E2 protein to host chromosomes. J. Virol. 2012, 86, 10047–10058. [Google Scholar] [CrossRef] [PubMed]
- McBride, A.A.; Bolen, J.B.; Howley, P.M. Phosphorylation sites of the E2 transcriptional regulatory proteins of bovine papillomavirus type 1. J. Virol. 1989, 63, 5076–5085. [Google Scholar] [CrossRef]
- Penrose, K.J.; Garcia-Alai, M.; de Prat-Gay, G.; McBride, A.A. Casein Kinase II phosphorylation-induced conformational switch triggers degradation of the papillomavirus E2 protein. J. Biol. Chem. 2004, 279, 22430–22439. [Google Scholar] [CrossRef] [PubMed]
- Schuck, S.; Ruse, C.; Stenlund, A. CK2 phosphorylation inactivates DNA binding by the papillomavirus E1 and E2 proteins. J. Virol. 2013, 87, 7668–7679. [Google Scholar] [CrossRef] [PubMed]
- Penrose, K.J.; McBride, A.A. Proteasome-Mediated Degradation of the Papillomavirus E2-TA Protein Is Regulated by Phosphorylation and Can Modulate Viral Genome Copy Number. J. Virol. 2000, 74, 6031–6038. [Google Scholar] [CrossRef]
- McBride, A.A.; Howley, P.M. Bovine papillomavirus with a mutation in the E2 serine 301 phosphorylation site replicates at a high copy number. J. Virol. 1991, 65, 6528–6534. [Google Scholar] [CrossRef]
- Lehman, C.W.; King, D.S.; Botchan, M.R. A papillomavirus E2 phosphorylation mutant exhibits normal transient replication and transcription but is defective in transformation and plasmid retention. J. Virol. 1997, 71, 3652–3665. [Google Scholar] [CrossRef]
- Sekhar, V.; Reed, S.C.; McBride, A.A. Interaction of the betapapillomavirus E2 tethering protein with mitotic chromosomes. J. Virol. 2010, 84, 543–557. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.K.; Anderson, D.E.; van Doorslaer, K.; McBride, A.A. A proteomic approach to discover and compare interacting partners of papillomavirus E2 proteins from diverse phylogenetic groups. Proteomics 2015, 15, 2038–2050. [Google Scholar] [CrossRef] [PubMed]
- Prabhakar, A.T.; James, C.D.; Das, D.; Otoa, R.; Day, M.; Burgner, J.; Fontan, C.T.; Wang, X.; Wieland, A.; Donaldson, M.M.; et al. CK2 phosphorylation of human papillomavirus 16 E2 on serine 23 promotes interaction with TopBP1 and is critical for E2 plasmid retention function. bioRxiv 2021, 431757. [Google Scholar] [CrossRef]
- Lai, M.C.; Lin, R.I.; Huang, S.Y.; Tsai, C.W.; Tarn, W.Y. A human importin-beta family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J. Biol. Chem. 2000, 275, 7950–7957. [Google Scholar] [CrossRef]
- Chang, S.W.; Tsao, Y.P.; Lin, C.Y.; Chen, S.L. NRIP, a novel calmodulin binding protein, activates calcineurin to dephosphorylate human papillomavirus E2 protein. J. Virol. 2011, 85, 6750–6763. [Google Scholar] [CrossRef]
- Prescott, E.L.; Brimacombe, C.L.; Hartley, M.; Bell, I.; Graham, S.; Roberts, S. Human papillomavirus type 1 E1^E4 protein is a potent inhibitor of the serine-arginine (SR) protein kinase SRPK1 and inhibits phosphorylation of host SR proteins and of the viral transcription and replication regulator E2. J. Virol. 2014, 88, 12599–12611. [Google Scholar] [CrossRef]
- Stubenrauch, F.; Hummel, M.; Iftner, T.; Laimins, L.A. The E8E2C protein, a negative regulator of viral transcription and replication, is required for extrachromosomal maintenance of human papillomavirus type 31 in keratinocytes. J. Virol. 2000, 74, 1178–1186. [Google Scholar] [CrossRef]
- Van de Poel, S.; Dreer, M.; Velic, A.; Macek, B.; Baskaran, P.; Iftner, T.; Stubenrauch, F. Identification and Functional Characterization of Phosphorylation Sites of the Human Papillomavirus 31 E8^E2 Protein. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Hunter, T. Tyrosine phosphorylation: Thirty years and counting. Curr. Opin. Cell Biol. 2009, 21, 140–146. [Google Scholar] [CrossRef]
- Hunter, T. The genesis of tyrosine phosphorylation. Cold Spring Harb. Perspect. Biol. 2014, 6, a020644. [Google Scholar] [CrossRef]
- Culleton, S.P.; Kanginakudru, S.; DeSmet, M.; Gilson, T.; Xie, F.; Wu, S.Y.; Chiang, C.M.; Qi, G.; Wang, M.; Androphy, E.J. Phosphorylation of the Bovine Papillomavirus E2 Protein on Tyrosine Regulates Its Transcription and Replication Functions. J. Virol. 2017, 91, e01854-16. [Google Scholar] [CrossRef]
- Abbate, E.A.; Berger, J.M.; Botchan, M.R. The X-ray structure of the papillomavirus helicase in complex with its molecular matchmaker E2. Genes Dev. 2004, 18, 1981–1996. [Google Scholar] [CrossRef]
- Abbate, E.A.; Voitenleitner, C.; Botchan, M.R. Structure of the papillomavirus DNA-tethering complex E2:Brd4 and a peptide that ablates HPV chromosomal association. Mol. Cell 2006, 24, 877–889. [Google Scholar] [CrossRef]
- McBride, A.A.; Jang, M.K. Current understanding of the role of the Brd4 protein in the papillomavirus lifecycle. Viruses 2013, 5, 1374–1394. [Google Scholar] [CrossRef]
- Jang, M.K.; Mochizuki, K.; Zhou, M.; Jeong, H.S.; Brady, J.N.; Ozato, K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 2005, 19, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Yik, J.H.; Chen, R.; He, N.; Jang, M.K.; Ozato, K.; Zhou, Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 2005, 19, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Gilson, T.; Culleton, S.; Xie, F.; DeSmet, M.; Androphy, E.J. HPV-31 tyrosine 102 regulates interaction with E2 binding partners and episomal maintenance. J. Virol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Touat, M.; Ileana, E.; Postel-Vinay, S.; André, F.; Soria, J.C. Targeting FGFR Signaling in Cancer. Clin. Cancer Res. 2015, 21, 2684–2694. [Google Scholar] [CrossRef]
- Gong, S.G. Isoforms of receptors of fibroblast growth factors. J. Cell. Physiol. 2014, 229, 1887–1895. [Google Scholar] [CrossRef]
- DeSmet, M.; Jose, L.; Isaq, N.; Androphy, E.J. Phosphorylation of a Conserved Tyrosine in the Papillomavirus E2 Protein Regulates Brd4 Binding and Viral Replication. J. Virol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Van Doorslaer, K.; Li, Z.; Xirasagar, S.; Maes, P.; Kaminsky, D.; Liou, D.; Sun, Q.; Kaur, R.; Huyen, Y.; McBride, A.A. The Papillomavirus Episteme: A major update to the papillomavirus sequence database. Nucleic Acids Res. 2017, 45, D499–D506. [Google Scholar] [CrossRef] [PubMed]
- Muller, M.; Jacob, Y.; Jones, L.; Weiss, A.; Brino, L.; Chantier, T.; Lotteau, V.; Favre, M.; Demeret, C. Large scale genotype comparison of human papillomavirus E2-host interaction networks provides new insights for e2 molecular functions. PLoS Pathog. 2012, 8, e1002761. [Google Scholar] [CrossRef] [PubMed]
- Jose, L.; DeSmet, M.; Androphy, E.J. Pyk2 Regulates Human Papillomavirus Replication by Tyrosine Phosphorylation of the E2 Protein. J. Virol. 2020. [Google Scholar] [CrossRef]
- Jose, L.; Androphy, E.J.; DeSmet, M. Phosphorylation of the Human Papillomavirus E2 Protein at Tyrosine 138 Regulates Episomal Replication. J. Virol. 2020. [Google Scholar] [CrossRef]
- Wu, S.Y.; Nin, D.S.; Lee, A.Y.; Simanski, S.; Kodadek, T.; Chiang, C.M. BRD4 Phosphorylation Regulates HPV E2-Mediated Viral Transcription, Origin Replication, and Cellular MMP-9 Expression. Cell Rep. 2016, 16, 1733–1748. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Xia, C.; Tao, Y.; Li, M.; Che, T.; Qu, J. Protein acetylation and deacetylation: An important regulatory modification in gene transcription (Review). Exp. Ther. Med. 2020, 20, 2923–2940. [Google Scholar] [CrossRef]
- Quinlan, E.J.; Culleton, S.P.; Wu, S.Y.; Chiang, C.M.; Androphy, E.J. Acetylation of conserved lysines in bovine papillomavirus E2 by p300. J. Virol. 2013, 87, 1497–1507. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.; Yigitliler, A.; Stubenrauch, F.; Iftner, T. Cottontail Rabbit Papillomavirus E1 and E2 Proteins Mutually Influence Their Subcellular Localizations. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed]
- Thomas, Y.; Androphy, E.J. Human Papillomavirus Replication Regulation by Acetylation of a Conserved Lysine in the E2 Protein. J. Virol. 2018, 92, 01912–01917. [Google Scholar] [CrossRef]
- Thomas, Y.; Androphy, E.J. Acetylation of E2 by P300 Mediates Topoisomerase Entry at the Papillomavirus Replicon. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Ritzkowsky, A.; Steger, G. Cooperative Activation of Human Papillomavirus Type 8 Gene Expression by the E2 Protein and the Cellular Coactivator p300. J. Virol. 2002, 76, 11042–11053. [Google Scholar] [CrossRef]
- Wong, P.-P.; Pickard, A.; McCance, D.J. p300 alters keratinocyte cell growth and differentiation through regulation of p21(Waf1/CIP1). PLoS ONE 2010, 5, e8369. [Google Scholar] [CrossRef]
- Langsfeld, E.S.; Bodily, J.M.; Laimins, L.A. The Deacetylase Sirtuin 1 Regulates Human Papillomavirus Replication by Modulating Histone Acetylation and Recruitment of DNA Damage Factors NBS1 and Rad51 to Viral Genomes. PLoS Pathog. 2015, 11, e1005181. [Google Scholar] [CrossRef] [PubMed]
- Allison, S.J.; Jiang, M.; Milner, J. Oncogenic viral protein HPV E7 up-regulates the SIRT1 longevity protein in human cervical cancer cells. Aging Albany N. Y. 2009, 1, 316–327. [Google Scholar] [CrossRef]
- Das, D.; Smith, N.; Wang, X.; Morgan, I.M. The Deacetylase SIRT1 Regulates the Replication Properties of Human Papillomavirus 16 E1 and E2. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Ammermann, I.; Bruckner, M.; Matthes, F.; Iftner, T.; Stubenrauch, F. Inhibition of transcription and DNA replication by the papillomavirus E8-E2C protein is mediated by interaction with corepressor molecules. J. Virol. 2008, 82, 5127–5136. [Google Scholar] [CrossRef]
- Geiss-Friedlander, R.; Melchior, F. Concepts in sumoylation: A decade on. Nat. Rev. Mol. Cell Biol. 2007, 8, 947–956. [Google Scholar] [CrossRef]
- Wu, Y.C.; Roark, A.A.; Bian, X.L.; Wilson, V.G. Modification of papillomavirus E2 proteins by the small ubiquitin-like modifier family members (SUMOs). Virology 2008, 378, 329–338. [Google Scholar] [CrossRef]
- Deyrieux, A.F.; Rosas-Acosta, G.; Ozbun, M.A.; Wilson, V.G. Sumoylation dynamics during keratinocyte differentiation. J. Cell Sci. 2007, 120, 125–136. [Google Scholar] [CrossRef]
- Wu, Y.C.; Bian, X.L.; Heaton, P.R.; Deyrieux, A.F.; Wilson, V.G. Host cell sumoylation level influences papillomavirus E2 protein stability. Virology 2009, 387, 176–183. [Google Scholar] [CrossRef]
- Zheng, N.; Shabek, N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu. Rev. Biochem. 2017, 86, 129–157. [Google Scholar] [CrossRef]
- Wilson, V.G. The role of ubiquitin and ubiquitin-like modification systems in papillomavirus biology. Viruses 2014, 6, 3584–3611. [Google Scholar] [CrossRef]
- Bellanger, S.; Demeret, C.; Goyat, S.; Thierry, F. Stability of the human papillomavirus type 18 E2 protein is regulated by a proteasome degradation pathway through its amino-terminal transactivation domain. J. Virol. 2001, 75, 7244–7251. [Google Scholar] [CrossRef]
- Hubbert, N.L.; Schiller, J.T.; Lowy, D.R.; Androphy, E.J. Bovine papilloma virus-transformed cells contain multiple E2 proteins. Proc. Natl. Acad. Sci. USA 1988, 85, 5864–5868. [Google Scholar] [CrossRef]
- Bellanger, S.; Tan, C.L.; Nei, W.; He, P.P.; Thierry, F. The human papillomavirus type 18 E2 protein is a cell cycle-dependent target of the SCFSkp2 ubiquitin ligase. J. Virol. 2010, 84, 437–444. [Google Scholar] [CrossRef]
- Spurgeon, M.E.; Lambert, P.F. Mus musculus Papillomavirus 1: A New Frontier in Animal Models of Papillomavirus Pathogenesis. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Uberoi, A.; Lambert, P.F. Rodent Papillomaviruses. Viruses 2017, 9, 362. [Google Scholar] [CrossRef]
- Stevenson, M.; Hudson, L.C.; Burns, J.E.; Stewart, R.L.; Wells, M.; Maitland, N.J. Inverse relationship between the expression of the human papillomavirus type 16 transcription factor E2 and virus DNA copy number during the progression of cervical intraepithelial neoplasia. J. Gen. Virol. 2000, 81, 1825–1832. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Sudol, M.; Hanafusa, H.; Krueger, J. Increased tyrosine kinase activity of c-Src during calcium-induced keratinocyte differentiation. Proc. Natl. Acad. Sci. USA 1992, 89, 8298–8302. [Google Scholar] [CrossRef]
- Padilla-Mendoza, J.R.; Contis-Montes de Oca, A.; Rodríguez, M.A.; López-Casamichana, M.; Bolaños, J.; Quintas-Granados, L.I.; Reyes-Hernández, O.D.; Fragozo-Sandoval, F.; Reséndiz-Albor, A.A.; Arellano-Gutiérrez, C.V.; et al. Protein Phosphorylation in Serine Residues Correlates with Progression from Precancerous Lesions to Cervical Cancer in Mexican Patients. BioMed Res. Int. 2020, 2020, 5058928. [Google Scholar] [CrossRef]
| HPV-31 | 12 High | 26 Low | BPV-1 | Mm/Mmu |
|---|---|---|---|---|
| Y19 | 92%Y 8%F | 100%Y | Y19 | Y/Y |
| Y32 | 75%Y | 100%H | Y32 | H/Y |
| Y44 | 75%Y 25%F | 38%Y 46%F | Y44 | Y/F |
| Y87 | 92%Y 8%F | 77%Y | F87 | Y/F |
| Y102 | 25%Y | 100%W | Y102 | W/Y |
| F121 | 33%Y 66%F | 39%Y 62%F | Y121 | F/Y |
| Y131 | 100%Y | 96%Y | Y131 | Y/Y |
| Y138 | 100%Y | 85%Y 4%F | Y138 | Y/Y |
| Y158 | 100%Y | 85%Y 8%F | Y158 | Y/V |
| Y159 | 100%Y | 92%Y 4%F | Y159 | Y/Y |
| Y167 | 92%Y 8%F | 65%Y 15%F | Y169 | Y/Y |
| F168 | 58%Y 42%F | 100%Y | Y170 | Y/Y |
| Y178 | 92%Y 8%F | 100%Y | F180 | Y/Y |
| Y310 | 100%Y | 96%Y | F343 | F/N |
| Y315 | 50%Y 8%F | 15%Y | H349 | H/T |
| Y319 | 75%Y 25%F | 30%Y 70%F | Y353 | Y/Y |
| Y344 | 83%Y 17%F | 92%Y | F380 | F/F |
| Y369 | 50%Y 17%F | 46%Y 19%F | T405 | S/T |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jose, L.; Gilson, T.; Androphy, E.J.; DeSmet, M. Regulation of the Human Papillomavirus Lifecyle through Post-Translational Modifications of the Viral E2 Protein. Pathogens 2021, 10, 793. https://doi.org/10.3390/pathogens10070793
Jose L, Gilson T, Androphy EJ, DeSmet M. Regulation of the Human Papillomavirus Lifecyle through Post-Translational Modifications of the Viral E2 Protein. Pathogens. 2021; 10(7):793. https://doi.org/10.3390/pathogens10070793
Chicago/Turabian StyleJose, Leny, Timra Gilson, Elliot J. Androphy, and Marsha DeSmet. 2021. "Regulation of the Human Papillomavirus Lifecyle through Post-Translational Modifications of the Viral E2 Protein" Pathogens 10, no. 7: 793. https://doi.org/10.3390/pathogens10070793
APA StyleJose, L., Gilson, T., Androphy, E. J., & DeSmet, M. (2021). Regulation of the Human Papillomavirus Lifecyle through Post-Translational Modifications of the Viral E2 Protein. Pathogens, 10(7), 793. https://doi.org/10.3390/pathogens10070793







