Proteomic Analysis of Fasciola hepatica Excretory and Secretory Products Co-Immunoprecipitated Using Time Course Infection Sera
Abstract
1. Introduction
2. Materials and Methods
2.1. FhESPs Preparation
2.2. Preparation of Serum
2.3. Co-IP of FhESPs-Antibody Binding Proteins
2.4. In-Gel Trypsin Digestion
2.5. LC–MS/MS Analysis
2.6. Data Analysis
3. Results and Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
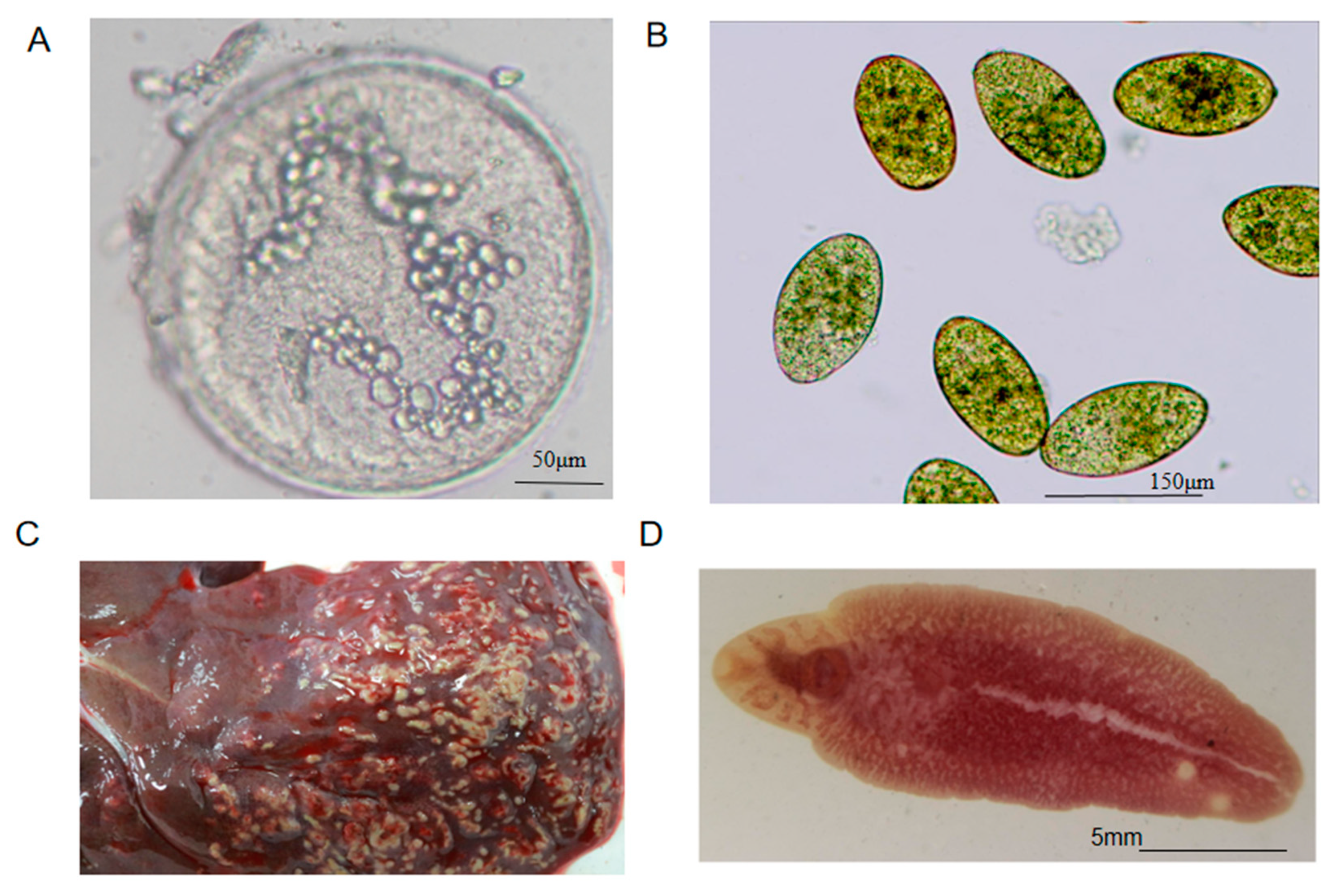
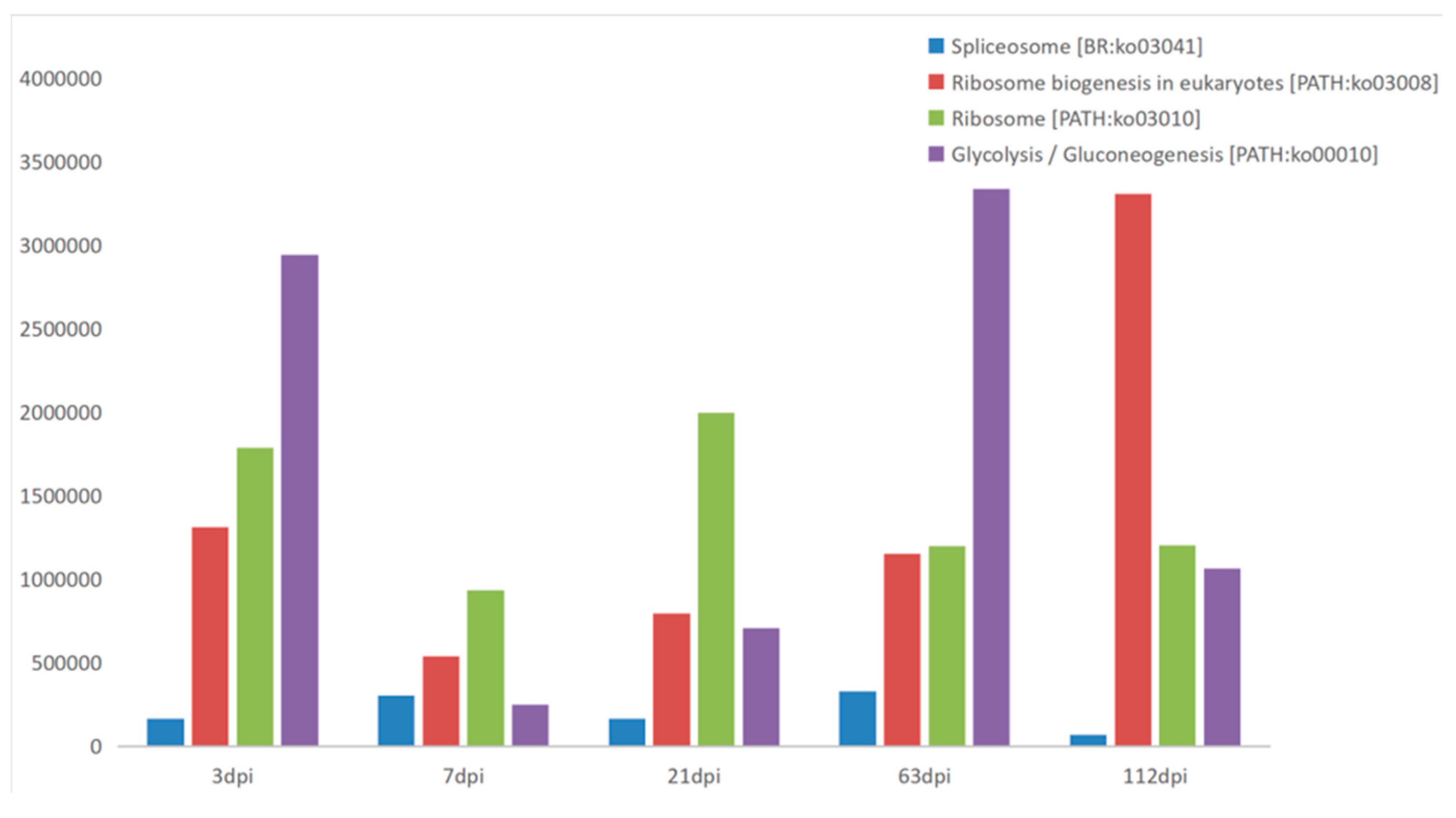
Appendix B
| GenBank | Protein Description | Abundance | ||||
|---|---|---|---|---|---|---|
| 3 dpi | 7 dpi | 21 dpi | 63 dpi | 112 dpi | ||
| THD21074.1 | Gelsolin repeat protein | 24,022.21289 | — | — | — | — |
| THD22536.1 | Fructose-bisphosphate aldolase | 86,937.625 | — | 172,559.1563 | — | — |
| THD27725.1 | Uncharacterized protein | 553,797.5 | — | 793,536.625 | — | 504,179.2188 |
| THD23308.1 | Dynein light chain | 11,747.43164 | 14,691.50586 | — | 7394.90625 | 21,221.78906 |
| THD24816.1 | Serpin protein | 1,646,911.188 | — | 134,091.1719 | 333,078.8438 | 906,792.8125 |
| THD27667.1 | Phosphoglucomutase | 341,188.4844 | — | — | 417,641.75 | — |
| THD20135.1 | Legumain | 2,782,378 | — | — | — | 17,424,414.5 |
| THD20526.1 | Uncharacterized protein | 77,763.89844 | 53,412.98438 | 79,610.59375 | 44,011.75781 | 116,365.3672 |
| THD22651.1 | Glucose-6-phosphate 1-dehydrogenase | 161,910.9531 | — | 170,645.4531 | 536,310.6875 | — |
| THD26293.1 | Annexin | 488,116.9375 | — | — | — | — |
| THD21694.1 | Fructose-bisphosphatase | 2,940,358.5 | 250,116.7344 | 706,553.3125 | 3,337,365.5 | 1,063,860 |
| THD23211.1 | Tubulin alpha chain | 330,362.1563 | — | 130,189.7266 | 768,367.25 | 243,900.8594 |
| THD28435.1 | HIV Tat-specific factor 1 protein | 163,905.5781 | 302,068.5313 | 164,833.7969 | 328,743.75 | 67,329.42188 |
| THD24727.1 | Heparosan-N-sulfate-glucuronate 5-epimerase | 409,968.5 | 85,834.78125 | 160,129.6875 | 155,940 | 156,170.7813 |
| THD22918.1 | Synaptophysin | 119,084.1016 | 68,156.35938 | 103,948.9063 | 222,138.8828 | 87,811.85156 |
| THD28852.1 | Ubiquitin-conjugating enzyme | 668,924.625 | 118,031.6016 | NA | 505,793.9375 | 343,043.6172 |
| THD27516.1 | Uncharacterized protein | 4,501,626 | 926,071.1875 | 2,201,982.75 | 1,899,086.625 | 1,882,076.625 |
| THD24445.1 | Alpha subunit of casein kinase II | 1,313,028.719 | 537,602.3906 | 793,033.75 | 1,153,075.125 | 3,306,170.313 |
| THD19712.1 | ATP-dependent Clp protease ATP-binding subunit clpX mitochondrial | 3,510,484.25 | — | — | — | — |
| THD22278.1 | Tubulin--tyrosine ligase-like protein 9 | 473,730.9688 | — | — | — | — |
| THD28860.1 | Uncharacterized protein | 49,954,940 | 14,922,535 | 18,942,136 | 40,814,652 | 38,499,836 |
| THD26528.1 | rRNA-processing protein FCF1 isogeny | 263,285.7188 | 110,388.4141 | — | 325,662.4688 | — |
| THD25229.1 | Hexosyltransferase | 100,760.8984 | 29,143.81445 | — | 26,410.00195 | — |
| THD22369.1 | Uncharacterized protein | 188,394.8125 | 62,918.48828 | — | 93,628.02344 | 101,414.2734 |
| THD24171.1 | Uncharacterized protein | 499,705.5625 | 93,921.17969 | 293,650.7813 | 1,253,055.875 | 694,243.1875 |
| THD21345.1 | Uncharacterized protein | 389,435 | 503,797.4063 | 2,321,797.25 | 602,599.5625 | — |
| THD21879.1 | Inhibitor of apoptosis protein | 1,977,777.75 | 1,209,314.875 | 1,346,054 | 1,589,138.125 | — |
| THD28280.1 | 40S ribosomal protein S3a | 1,786,953.672 | 934,144.1875 | 1,994,597.219 | 1,198,627.375 | 1,202,573 |
| THD21305.1 | Acyl-coenzyme A thioesterase 8 | 1,959,986.75 | 162,016.5469 | 185,430.125 | 1,942,750.313 | 1051,204.156 |
| THD28550.1 | Uncharacterized protein | 335,545.5625 | 39,628.06641 | — | 181,182.0156 | 118,531.6094 |
| THD28078.1 | Guanine nucleotide exchange factor | — | 44,768.47266 | 466,165.8438 | — | 236,903.5234 |
| THD19712.1 | ATP-dependent Clp protease ATP-binding subunit clpX mitochondrial | — | 221,215.0469 | 985,376.8125 | 2,183,246.25 | 2,075,125.5 |
| THD20617.1 | ATP-dependent DNA helicase | — | 417,392.3438 | 442,980.4375 | 286,577.8125 | 481,421.625 |
| THD19133.1 | VIT domain-containing protein | — | — | 93,653.66406 | 51,209.55859 | — |
| THD19784.1 | ML domain-containing protein | — | — | 65,019.27344 | — | — |
| THD28903.1 | Isocitrate dehydrogenase [NADP] | — | — | — | 770,771.625 | — |
References
- Mas-Coma, S.; Bargues, M.D.; Valero, M.A. Human fascioliasis infection sources, their diversity, incidence factors, analytical methods and prevention measures. Parasitology 2020, 145, 1665–1699. [Google Scholar] [CrossRef]
- Mehmood, K.; Zhang, H.; Sabir, A.J.; Abbas, R.Z.; Ijaz, M.; Durrani, A.Z.; Saleem, M.H.; Ur-Rehman, M.; Iqbal, M.K.; Wang, Y.; et al. A review on epidemiology, global prevalence and economical losses of fasciolosis in ruminants. Microb. Pathog. 2017, 109, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Pinilla, J.C.; Florez-Muñoz, A.A.; Uribe-Delgado, N. Prevalence and risk factors associated with liver fluke Fasciola hepatica in cattle and sheep in three municipalities in the Colombian Northeastern Mountains. Vet. Parasitol. Reg. Stud. Rep. 2020, 19, 100364. [Google Scholar] [CrossRef]
- Machicado, C.; Machicado, J.D.; Maco, V.; Terashima, A.; Marcos, L.A. Association of Fasciola hepatica Infection with Liver Fibrosis, Cirrhosis, and Cancer: A Systematic Review. PLoS. Negl. Trop. Dis. 2016, 10, e0004962. [Google Scholar] [CrossRef]
- Abuzeid, A.M.I.; Zhou, X.; Huang, Y.; Li, G. Twenty-five-year research progress in hookworm excretory/secretory products. Parasit. Vectors 2020, 13, 136. [Google Scholar] [CrossRef] [PubMed]
- MartÍnez-Pérez, J.M.; Robles-Pérez, D.; Rojo-VÁzquez, F.A.; Martínez-Valladares, M. Comparison of three different techniques to diagnose Fasciola hepatica infection in experimentally and naturally infected sheep. Vet. Parasitol. 2012, 190, 80–86. [Google Scholar] [CrossRef]
- Ma, Z.; Mutashar-Alhameed, A.M.; Kaminga, A.C.; Lu, B.; Li, X.; Zhang, J.; Wu, X. Bioinformatics of excretory/secretory proteins of Toxoplasma gondii strain ME49. Microb. Pathog. 2020, 140, 103951. [Google Scholar] [CrossRef]
- Gupta, S.C.; Yadav, S.C. Emergence of Fasciola gigantica cercariae from naturally infected Lymnaea auricularia. J. Parasit. Dis. 1994, 18, 53–56. [Google Scholar]
- Rotmans, J.P.; Van, M.J.; Looze, M.; Mooij, G.W.; Deelder, A.M. Schistosoma mansoni: Use of antigens from excretions and secretions in immunodiagnosis. Exp. Parasitol. 1981, 52, 319–330. [Google Scholar] [CrossRef]
- Chehayeb, J.F.; Robertson, A.P.; Martin, R.J.; Geary, T.G. Proteomic analysis of adult Ascaris suum fluid compartments and secretory products. PLoS. Negl. Trop. Dis. 2014, 8, e2939. [Google Scholar] [CrossRef]
- Gadahi, J.; Yongqian, B.; Ehsan, M.; Zhang, Z.; Wang, S.; Yan, R.F.; Song, X.K.; Xu, L.X.; Li, X.R. Haemonchus contortus excretory and secretory proteins (HcESPs) suppress functions of goat PBMCs in vitro. Oncotarget 2016, 7, 35670–35679. [Google Scholar] [CrossRef]
- Sotillo, J.; Sanchez-Flores, A.; Cantacessi, C.; Harcus, Y.; Pickering, D.; Bouchery, T.; Camberis, M.; Tang, S.C.; Giacomin, P.; Mulvenna, J.; et al. Secreted proteomes of different developmental stages of the gastrointestinal nematode Nippostrongylus brasiliensis. Mol. Cell. Proteom. 2014, 13, 2736–2751. [Google Scholar] [CrossRef]
- Li, M.C.; Pan, J.D.; Fan, Z.H.; Chen, W.L.; Chen, C.L.; Wu, H.L.; Li, T.G.; Sun, Y.J.; Du, L.Y. Changes of protein components in small cell lung cancer H446 cells treated with ESPs of Trichinella spiralis. Chin. J. Zoonoses 2020, 36, 821–826. [Google Scholar]
- Gourbal, B.E.; Guillou, F.; Mitta, G.; Sibille, P.; Thèron, A.; Pointier, J.P.; Coustau, C. Excretory-secretory products of larval Fasciola hepatica investigated using a two-dimensional proteomic approach. Mol. Biochem. Parasitol. 2008, 161, 63–66. [Google Scholar] [CrossRef] [PubMed]
- Jefferies, J.R.; Campbell, A.M.; van-Rossum, A.; Barrett, J.; Brophy, P.M. Proteomic analysis of Fasciola hepatica excretory secretory products. Proteomics 2001, 1, 1128–1132. [Google Scholar] [CrossRef]
- Huang, S.Y.; Yue, D.M.; Hou, J.L.; Zhang, X.X.; Zhang, F.K.; Wang, C.R.; Zhu, X.Q. Proteomic analysis of Fasciola gigantica excretory and secretory products (FgESPs) interacting with buffalo serum of different infection periods by shotgun LC-MS/MS. Parasitol. Res. 2019, 118, 453–460. [Google Scholar] [CrossRef]
- Cwiklinski, K.; Dalton, J.P.; Dufresne, P.J.; La-Course, J.; Williams, D.J.; Hodgkinson, J.; Paterson, S. The Fasciola hepatica genome: Gene duplication and polymorphism reveals adaptation to the host environment and the capacity for rapid evolution. Genome Biol. 2015, 16, 71. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Fontenla, S.; Fischer, P.U.; Le, T.H.; Costábile, A.; Blair, D.; Brindley, P.J.; Tort, J.F.; Cabada, M.M.; Mitreva, M. Adaptive Radiation of the Flukes of the Family Fasciolidae Inferred from Genome-Wide Comparisons of Key Species. Mol. Biol. Evol. 2020, 37, 84–99. [Google Scholar] [CrossRef]
- Novobilsky, A.; Kasny, M.; Mikes, L.; Kovarcik, K.; Koudela, B. Humoral immune responses during experim ental infection with Fascioloides magna and Fasciola hepatica in goats and comparison. Parasitol. Res. 2007, 101, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Rim, H.J.; Faraq, H.F.; Sornmani, S.; Cross, J.H. Food-borne trematodes: Ignored or emerging. Parasitol. Today 1994, 10, 207–209. [Google Scholar] [CrossRef]
- Salizzato, V.; Zanin, S.; Borgo, C.; Lidron, E.; Salvi, M.; Rizzuto, R.; Pallafacchina, G.; Donella-Deana, A. Protein kinase CK2 subunits exert specific and coordinated functions in skeletal muscle differentiation and fusogenic activity. FASEB J. 2019, 33, 10648–10667. [Google Scholar] [CrossRef]
- Miller, H.B.; Saunders, K.O.; Tomaras, G.D.; Garcia-Blanco, M.A. Tat-SF1 is not required for Tat transactivation but does regulate the relative levels of unspliced and spliced HIV-1 RNAs. PLoS ONE 2009, 27, e5710. [Google Scholar] [CrossRef] [PubMed]
- Kochanowski, M.; Dąbrowska, J.; Różycki, M.; Karamon, J.; Sroka, J.; Cencek, T. Proteomic Profiling Reveals New Insights into the Allergomes of Anisakis simplex, Pseudoterranova decipiens, and Contracaecum osculatum. J. Parasitol. 2020, 106, 572–588. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein. Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Cervi, L.; Masih, D.T. Inhibition of spleen cell proliferative response to mitogens by excretory-secretory antigens of Fasciola hepatica. Int. J. Parasitol. 1997, 27, 573–579. [Google Scholar] [CrossRef]
- Guasconi, L.; Chiapello, L.S.; Masih, D.T. Fasciola hepatica excretorysecretory products induce CD4+T cell anergy via selective upregulation of PD-L2 expression on macrophages in a Dectin-1 dependent way. Immunobiology 2015, 220, 934–939. [Google Scholar] [CrossRef] [PubMed]
- Mezo, M.; Gonzalez-Warleta, M.; Castro-Hermida, J.A.; Muiño, L.; Ubeira, F.M. Field evaluation of the MM3-SERO ELISA for detection of anti-Fasciola IgG antibodies in milk samples from individual cows and bulk milk tanks. Parasitol. Int. 2010, 59, 610–615. [Google Scholar] [CrossRef]



| Protein Description | Species | Genome Mapping Gene ID a | Peptides | Unique Peptides | Cover Percent | GeneBank Bank ID | MW b(KDa) | calc. pI c |
|---|---|---|---|---|---|---|---|---|
| Uncharacterized protein 1 | Fasciola hepatica | D915_008782 | 2 | 2 | 12 | THD20526.1 | 25.9 | 6.24 |
| Heparosan-N-sulfate-glucuronate 5-epimerase | Fasciola hepatica | D915_004542 | 1 | 1 | 1 | THD24727.1 | 80 | 9.03 |
| Synaptophysin | Fasciola hepatica | D915_006334 | 1 | 1 | 3 | THD22918.1 | 26.1 | 5.34 |
| Uncharacterized protein 2 | Fasciola hepatica | D915_001733 | 1 | 1 | 4 | THD27516.1 | 15.5 | 7.03 |
| Acyl-coenzyme A thioesterase 8 | Fasciola hepatica | D915_007699 | 1 | 1 | 2 | THD21305.1 | 37.1 | 6.87 |
| ATP-dependent Clp protease ATP-binding subunit clpX mitochondrial | Fasciola hepatica | D915_009354 | 1 | 1 | 1 | THD19712.1 | 60.5 | 6.35 |
| Uncharacterized protein 3 | Fasciola hepatica | D915_000294 | 1 | 1 | 2 | THD28860.1 | 30.9 | 6.99 |
| Uncharacterized protein 4 | Fasciola hepatica | D915_005102 | 1 | 1 | 1 | THD24171.1 | 55.6 | 8.76 |
| 40S ribosomal protein S3a | Fasciola hepatica | D915_000892 | 1 | 1 | 3 | THD28280.1 | 25.3 | 9.85 |
| HIV Tat-specific factor 1 protein | Fasciola hepatica | D915_000726 | 1 | 1 | 1 | THD28435.1 | 67.5 | 5 |
| Fructose-bisphosphatase | Fasciola hepatica | D915_007520 | 1 | 1 | 5 | THD21694.1 | 29.1 | 6.16 |
| Alpha subunit of casein kinase II | Fasciola hepatica | D915_004801 | 1 | 1 | 2 | THD24445.1 | 44.2 | 8.48 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lan, Z.; Liu, X.-L.; Lv, Q.-B.; Zeng, M.-H.; Gao, J.-F.; Chang, Q.-C.; Chen, Y.-Y.; Wang, C.-R. Proteomic Analysis of Fasciola hepatica Excretory and Secretory Products Co-Immunoprecipitated Using Time Course Infection Sera. Pathogens 2021, 10, 749. https://doi.org/10.3390/pathogens10060749
Lan Z, Liu X-L, Lv Q-B, Zeng M-H, Gao J-F, Chang Q-C, Chen Y-Y, Wang C-R. Proteomic Analysis of Fasciola hepatica Excretory and Secretory Products Co-Immunoprecipitated Using Time Course Infection Sera. Pathogens. 2021; 10(6):749. https://doi.org/10.3390/pathogens10060749
Chicago/Turabian StyleLan, Zhuo, Xiao-Lei Liu, Qing-Bo Lv, Min-Hao Zeng, Jun-Feng Gao, Qiao-Cheng Chang, Yuan-Yuan Chen, and Chun-Ren Wang. 2021. "Proteomic Analysis of Fasciola hepatica Excretory and Secretory Products Co-Immunoprecipitated Using Time Course Infection Sera" Pathogens 10, no. 6: 749. https://doi.org/10.3390/pathogens10060749
APA StyleLan, Z., Liu, X.-L., Lv, Q.-B., Zeng, M.-H., Gao, J.-F., Chang, Q.-C., Chen, Y.-Y., & Wang, C.-R. (2021). Proteomic Analysis of Fasciola hepatica Excretory and Secretory Products Co-Immunoprecipitated Using Time Course Infection Sera. Pathogens, 10(6), 749. https://doi.org/10.3390/pathogens10060749






