Dysregulation of Glycerophosphocholines in the Cutaneous Lesion Caused by Leishmania major in Experimental Murine Models
Abstract
1. Introduction
2. Results
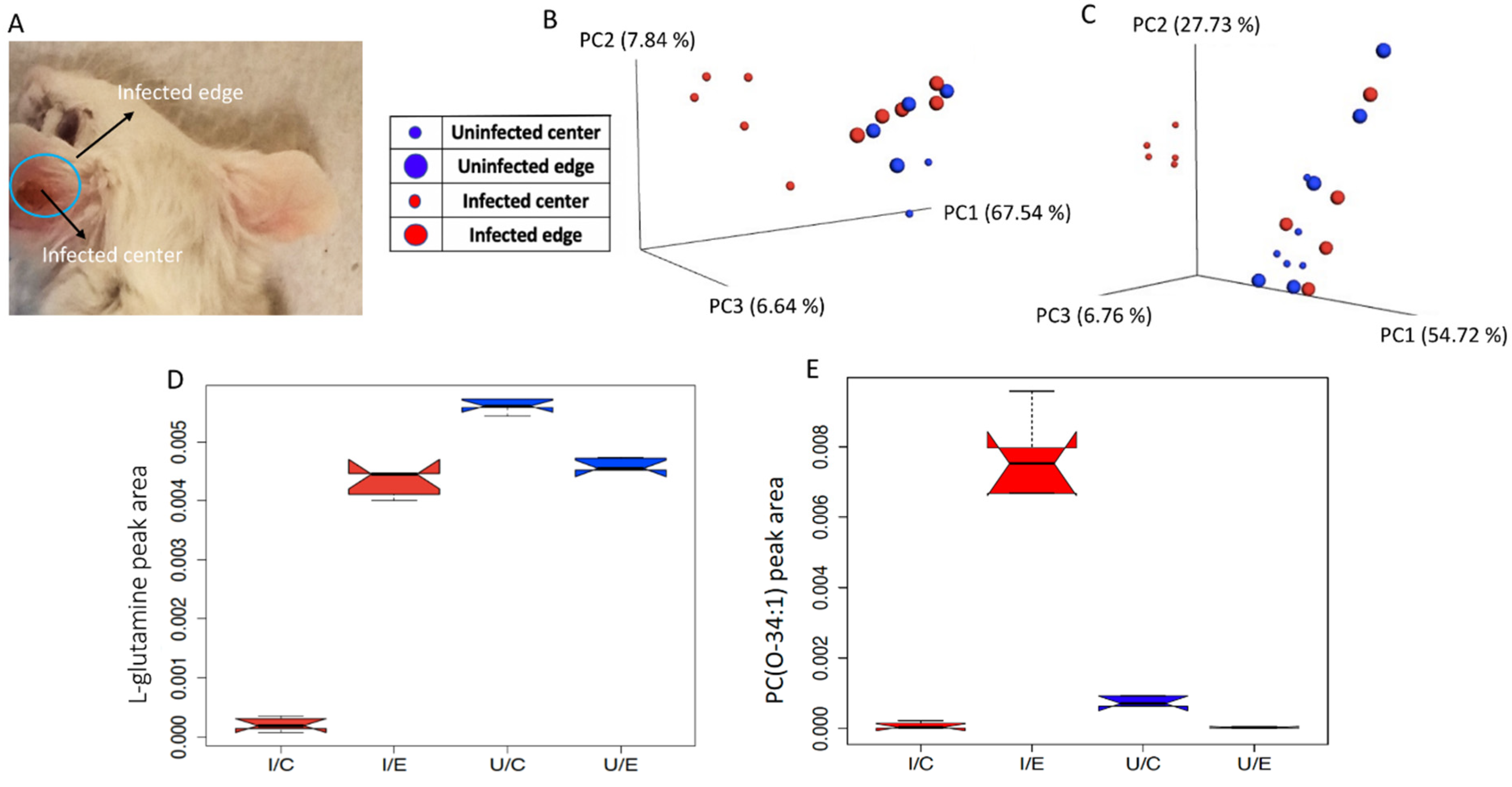
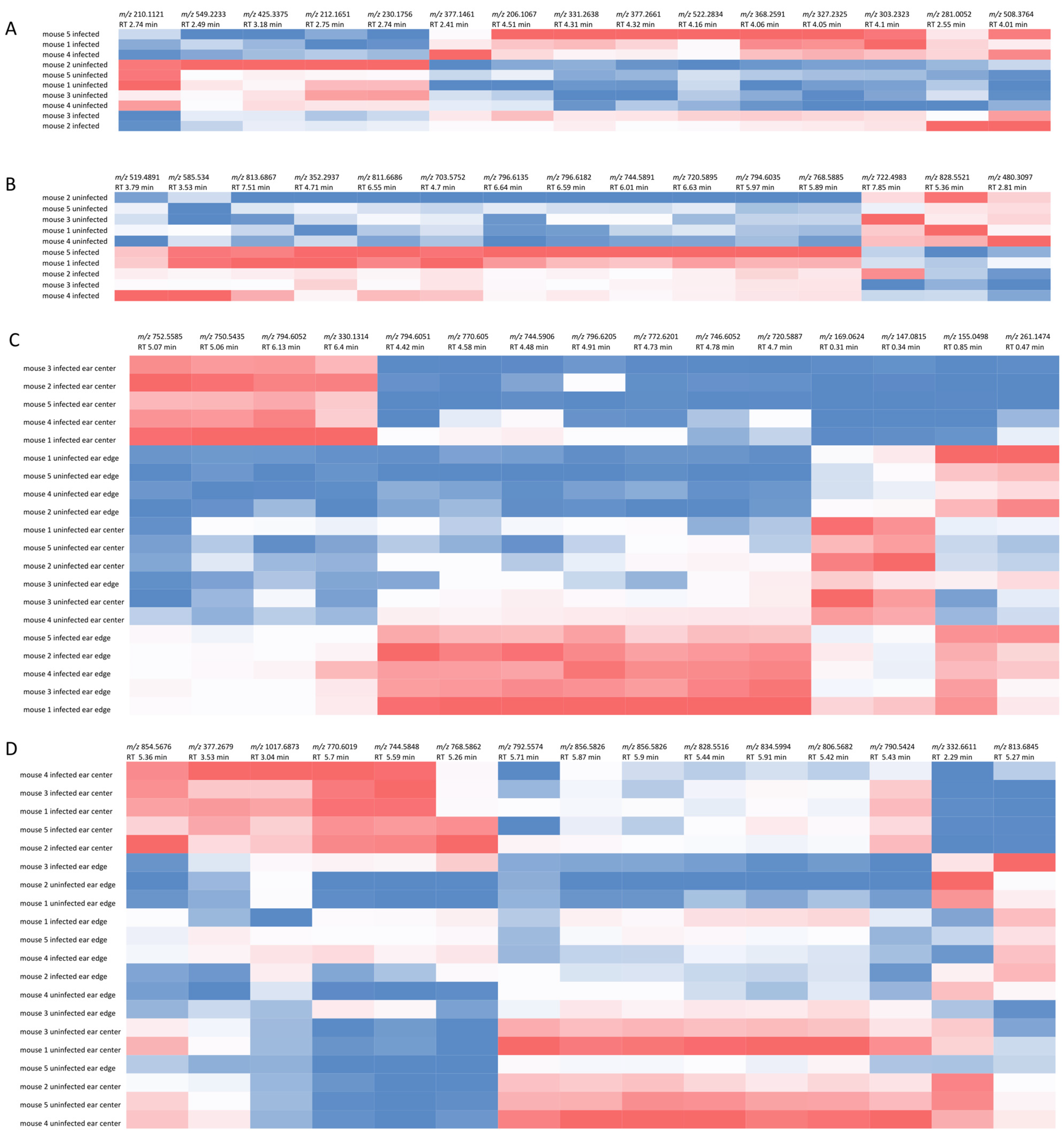
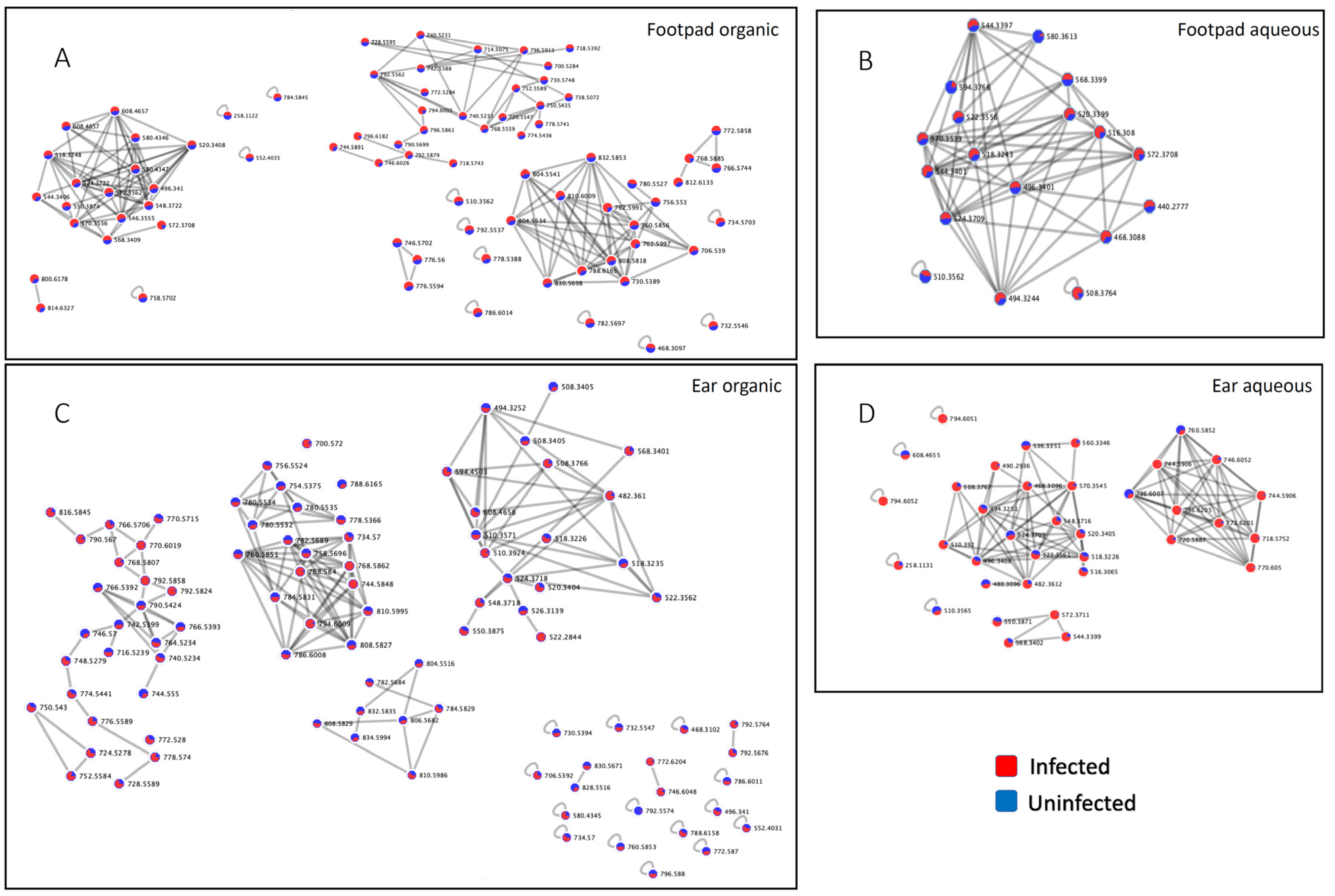
2.1. Overall Impact of L. major Infection on Ear and Footpad Metabolism
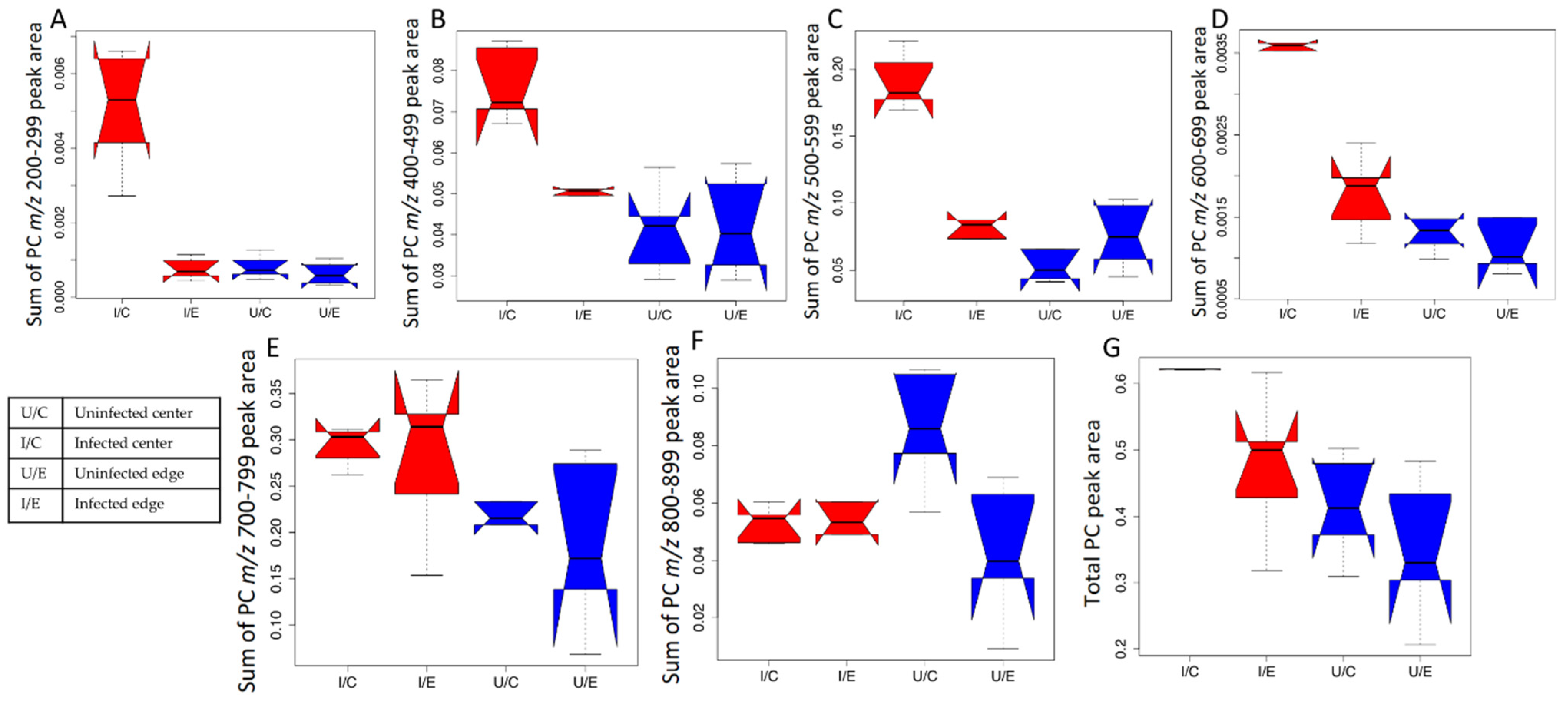
2.2. Impact of L. major Infection on Tissue PCs
3. Discussion
4. Materials and Methods
4.1. In Vivo Experimentation
4.2. LC-MS/MS
4.3. LC-MS/MS Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Georgiadou, S.P.; Makaritsis, K.P.; Dalekos, G.N. Leishmaniasis Revisited: Current Aspects on Epidemiology, Diagnosis and Treatment. J. Transl. Int. Med. 2015, 3, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Alvar, J.; Yactayo, S.; Bern, C. Leishmaniasis and Poverty. Trends Parasitol. 2006, 22, 552–557. [Google Scholar] [CrossRef]
- Bern, C.; Maguire, J.H.; Alvar, J. Complexities of Assessing the Disease Burden Attributable to Leishmaniasis. PLoS Negl. Trop. Dis. 2008, 2, e313. [Google Scholar] [CrossRef]
- McCall, L.-I.; Zhang, W.-W.; Matlashewski, G. Determinants for the Development of Visceral Leishmaniasis Disease. PLoS Pathog. 2013, 9, e1003053. [Google Scholar] [CrossRef]
- McCall, L.-I.; McKerrow, J.H. Determinants of Disease Phenotype in Trypanosomatid Parasites. Trends Parasitol. 2014, 30, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Burza, S.; Croft, S.L.; Boelaert, M. Leishmaniasis. Lancet 2018, 392, 951–970. [Google Scholar] [CrossRef]
- Ponte-Sucre, A.; Gamarro, F.; Dujardin, J.-C.; Barrett, M.P.; López-Vélez, R.; García-Hernández, R.; Pountain, A.W.; Mwenechanya, R.; Papadopoulou, B. Drug Resistance and Treatment Failure in Leishmaniasis: A 21st Century Challenge. PLoS Negl. Trop. Dis. 2017, 11, e0006052. [Google Scholar] [CrossRef]
- Bueno-Marí, R.; Jiménez-Peydró, R. Global Change and Human Vulnerability to Vector-Borne Diseases. Front. Physiol. 2013, 4, 158. [Google Scholar] [CrossRef]
- Deidda, M.; Piras, C.; Bassareo, P.P.; Dessalvi, C.C.; Mercuro, G. Metabolomics, a Promising Approach to Translational Research in Cardiology. IJC Metab. Endocr. 2015, 9, 31–38. [Google Scholar] [CrossRef]
- Vinayavekhin, N.; Saghatelian, A. Untargeted Metabolomics. Curr. Protoc. Mol. Biol. 2010. [Google Scholar] [CrossRef]
- Vincent, I.M.; Weidt, S.; Rivas, L.; Burgess, K.; Smith, T.K.; Ouellette, M. Untargeted Metabolomic Analysis of Miltefosine Action in Leishmania Infantum Reveals Changes to the Internal Lipid Metabolism. Int. J. Parasitol. Drugs Drug Resist. 2014, 4, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-López, J.M.; Ríos-Marco, P.; Marco, C.; Segovia, J.L.; Carrasco, M.P. Alterations in the Homeostasis of Phospholipids and Cholesterol by Antitumor Alkylphospholipids. Lipids Health Dis. 2010, 9, 33. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, V.B.; Rossjohn, J.; Wakelam, M.J.O. Phospholipid Signaling in Innate Immune Cells. J. Clin. Investig. 2018, 128, 2670–2679. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Newsom, S.N.; McCall, L.-I. Metabolomics: Eavesdropping on Silent Conversations between Hosts and Their Unwelcome Guests. PLoS Pathog. 2018, 14, e1006926. [Google Scholar] [CrossRef]
- Hossain, E.; Khanam, S.; Wu, C.; Lostracco-Johnson, S.; Thomas, D.; Katemauswa, M.; Gosmanov, C.; Li, D.; Woelfel-Monsivais, C.; Sankaranarayanan, K.; et al. Mapping of Host-Parasite-Microbiome Interactions Reveals Metabolic Determinants of Tropism and Tolerance in Chagas Disease. Sci. Adv. 2020, 6, eaaz2015. [Google Scholar] [CrossRef]
- Varikuti, S.; Jha, B.K.; Volpedo, G.; Ryan, N.M.; Halsey, G.; Hamza, O.M.; McGwire, B.S.; Satoskar, A.R. Host-Directed Drug Therapies for Neglected Tropical Diseases Caused by Protozoan Parasites. Front. Microbiol. 2018, 9, 2655. [Google Scholar] [CrossRef]
- Vargas, D.A.; Prieto, M.D.; Martínez-Valencia, A.J.; Cossio, A.; Burgess, K.E.V.; Burchmore, R.J.S.; Gómez, M.A. Pharmacometabolomics of Meglumine Antimoniate in Patients with Cutaneous Leishmaniasis. Front. Pharm. 2019, 10. [Google Scholar] [CrossRef]
- Saunders, E.C.; Ng, W.W.; Kloehn, J.; Chambers, J.M.; Ng, M.; McConville, M.J. Induction of a Stringent Metabolic Response in Intracellular Stages of Leishmania mexicana Leads to Increased Dependence on Mitochondrial Metabolism. PLoS Pathog. 2014, 10, e1003888. [Google Scholar] [CrossRef]
- Ty, M.C.; Loke, P.; Alberola, J.; Rodriguez, A.; Rodriguez-Cortes, A. Immuno-Metabolic Profile of Human Macrophages after Leishmania and Trypanosoma cruzi Infection. PLoS ONE 2019, 14, e0225588. [Google Scholar] [CrossRef]
- Muxel, S.M.; Mamani-Huanca, M.; Aoki, J.I.; Zampieri, R.A.; Floeter-Winter, L.M.; López-Gonzálvez, Á.; Barbas, C. Metabolomic Profile of BALB/c Macrophages Infected with Leishmania amazonensis: Deciphering L-Arginine Metabolism. Int. J. Mol. Sci. 2019, 20, 6248. [Google Scholar] [CrossRef]
- Negrão, F.; Abánades, D.R.; Jaeeger, C.F.; Rocha, D.F.O.; Belaz, K.R.A.; Giorgio, S.; Eberlin, M.N.; Angolini, C.F.F. Lipidomic Alterations of in Vitro Macrophage Infection by L. infantum and L. amazonensis. Mol. Biosyst. 2017, 13, 2401–2406. [Google Scholar] [CrossRef]
- Kloehn, J.; Saunders, E.C.; O’Callaghan, S.; Dagley, M.J.; McConville, M.J. Characterization of Metabolically Quiescent Leishmania Parasites in Murine Lesions Using Heavy Water Labeling. PLoS Pathog. 2015, 11, e1004683. [Google Scholar] [CrossRef]
- Moitra, S.; Basu, S.; Pawlowic, M.; Hsu, F.-F.; Zhang, K. Synthesis of Phosphatidylcholine Is Essential for the Promastigote but Not Amastigote Stage in. Front. Cell. Infect. Microbiol. 2021, 11, 647870. [Google Scholar] [CrossRef]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The Human Metabolome Database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The Human Metabolome Database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- Henriques, C.; Atella, G.C.; Bonilha, V.L.; de Souza, W. Biochemical Analysis of Proteins and Lipids Found in Parasitophorous Vacuoles Containing Leishmania amazonensis. Parasitol. Res. 2003, 89, 123–133. [Google Scholar]
- Tounsi, N.; Meghari, S.; Moser, M.; Djerdjouri, B. Lysophosphatidylcholine Exacerbates Leishmania major-Dendritic Cell Infection through Interleukin-10 and a Burst in arginase1 and Indoleamine 2,3-Dioxygenase Activities. Int. Immunopharmacol. 2015, 25, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bohdanowicz, M.; Grinstein, S. Role of Phospholipids in Endocytosis, Phagocytosis, and Macropinocytosis. Physiol. Rev. 2013, 93, 69–106. [Google Scholar] [CrossRef]
- Tian, Y.; Pate, C.; Andreolotti, A.; Wang, L.; Tuomanen, E.; Boyd, K.; Claro, E.; Jackowski, S. Cytokine Secretion Requires Phosphatidylcholine Synthesis. J. Cell Biol. 2008, 181, 945–957. [Google Scholar] [CrossRef] [PubMed]
- Kropf, P.; Freudenberg, M.A.; Modolell, M.; Price, H.P.; Herath, S.; Antoniazi, S.; Galanos, C.; Smith, D.F.; Müller, I. Toll-like Receptor 4 Contributes to Efficient Control of Infection with the Protozoan Parasite Leishmania major. Infect. Immun. 2004, 72, 1920–1928. [Google Scholar] [CrossRef]
- Ecker, J.; Liebisch, G.; Englmaier, M.; Grandl, M.; Robenek, H.; Schmitz, G. Induction of Fatty Acid Synthesis Is a Key Requirement for Phagocytic Differentiation of Human Monocytes. Proc. Natl. Acad. Sci. USA 2010, 107, 7817–7822. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, K.; Liu, Z.; Hossain, E.; Bottazzi, M.E.; Hotez, P.J.; Jones, K.M.; McCall, L.I. Alterations to the Cardiac Metabolome Induced by Chronic T. cruzi Infection Relate to the Degree of Cardiac Pathology. ACS Infect. Dis. 2021. [Google Scholar] [CrossRef]
- Dean, D.A.; Gautham, J.L.; Siqueira-Neto, J.H.; McKerrow, J.H.; Dorrestein, P.C.; McCall, L.-I. Spatial Metabolomics Identifies Localized Chemical Changes in Heart Tissue during Chronic Cardiac Chagas Disease. bioRxiv 2020. [Google Scholar] [CrossRef]
- Rakotomanga, M.; Blanc, S.; Gaudin, K.; Chaminade, P.; Loiseau, P.M. Miltefosine Affects Lipid Metabolism in Leishmania donovani Promastigotes. Antimicrob. Agents Chemother. 2007, 51, 1425–1430. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, C.; Mesquita, I.; Barbosa, A.M.; Osório, N.S.; Torrado, E.; Beauparlant, C.-J.; Droit, A.; Cunha, C.; Carvalho, A.; Saha, B.; et al. Glutamine Supplementation Improves the Efficacy of Miltefosine Treatment for Visceral Leishmaniasis. PLoS Negl. Trop. Dis. 2020, 14, e0008125. [Google Scholar] [CrossRef] [PubMed]
- Scorza, B.M.; Carvalho, E.M.; Wilson, M.E. Cutaneous Manifestations of Human and Murine Leishmaniasis. Int. J. Mol. Sci. 2017, 18, 1296. [Google Scholar] [CrossRef] [PubMed]
- Gimblet, C.; Meisel, J.S.; Loesche, M.A.; Cole, S.D.; Horwinski, J.; Novais, F.O.; Misic, A.M.; Bradley, C.W.; Beiting, D.P.; Rankin, S.C.; et al. Cutaneous Leishmaniasis Induces a Transmissible Dysbiotic Skin Microbiota That Promotes Skin Inflammation. Cell Host Microbe 2017, 22, 13–24.e4. [Google Scholar] [CrossRef] [PubMed]
- Loeuillet, C.; Bañuls, A.-L.; Hide, M. Study of Leishmania Pathogenesis in Mice: Experimental Considerations. Parasit. Vectors 2016, 9, 144. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro-Gomes, F.L.; Roma, E.H.; Carneiro, M.B.H.; Doria, N.A.; Sacks, D.L.; Peters, N.C. Site-Dependent Recruitment of Inflammatory Cells Determines the Effective Dose of Leishmania major. Infect. Immun. 2014, 82, 2713–2727. [Google Scholar] [CrossRef] [PubMed]
- Tabbara, K.S.; Peters, N.C.; Afrin, F.; Mendez, S.; Bertholet, S.; Belkaid, Y.; Sacks, D.L. Conditions Influencing the Efficacy of Vaccination with Live Organisms against Leishmania major Infection. Infect. Immun. 2005, 73, 4714–4722. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed Minimum Reporting Standards for Chemical Analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Roy, G.; Dumas, C.; Sereno, D.; Wu, Y.; Singh, A.K.; Tremblay, M.J.; Ouellette, M.; Olivier, M.; Papadopoulou, B. Episomal and Stable Expression of the Luciferase Reporter Gene for Quantifying Leishmania Spp. Infections in Macrophages and in Animal Models. Mol. Biochem. Parasitol. 2000, 110, 195–206. [Google Scholar] [CrossRef]
- Côrtes, D.F.; Carneiro, M.B.H.; Santos, L.M.; de Souza, T.C.O.; Maioli, T.U.; Duz, A.L.C.; Ramos-Jorge, M.L.; Afonso, L.C.C.; Carneiro, C.; Vieira, L.Q. Low and High-Dose Intradermal Infection with Leishmania major and Leishmania amazonensis in C57BL/6 Mice. Mem. Inst. Oswaldo Cruz 2010, 105, 736–745. [Google Scholar] [CrossRef]
- Zhang, W.W.; Ramasamy, G.; McCall, L.-I.; Haydock, A.; Ranasinghe, S.; Abeygunasekara, P.; Sirimanna, G.; Wickremasinghe, R.; Myler, P.; Matlashewski, G. Genetic Analysis of Leishmania donovani Tropism Using a Naturally Attenuated Cutaneous Strain. PLoS Pathog. 2014, 10, e1004244. [Google Scholar] [CrossRef]
- McCall, L.-I.; El Aroussi, A.; Choi, J.Y.; Vieira, D.F.; De Muylder, G.; Johnston, J.B.; Chen, S.; Kellar, D.; Siqueira-Neto, J.L.; Roush, W.R.; et al. Targeting Ergosterol Biosynthesis in Leishmania donovani: Essentiality of Sterol 14alpha-Demethylase. PLOS Negl. Trop. Dis. 2015, 9, e0003588. [Google Scholar] [CrossRef]
- McCall, L.-I.; Morton, J.T.; Bernatchez, J.A.; de Siqueira-Neto, J.L.; Knight, R.; Dorrestein, P.C.; McKerrow, J.H. Mass Spectrometry-Based Chemical Cartography of a Cardiac Parasitic Infection. Anal. Chem. 2017, 89, 10414–10421. [Google Scholar] [CrossRef]
- Melnik, A.V.; Vázquez-Baeza, Y.; Aksenov, A.A.; Hyde, E.; McAvoy, A.C.; Wang, M.; da Silva, R.R.; Protsyuk, I.; Wu, J.V.; Bouslimani, A.; et al. Molecular and Microbial Microenvironments in Chronically Diseased Lungs Associated with Cystic Fibrosis. mSystems 2019, 4. [Google Scholar] [CrossRef]
- Quinn, R.A.; Melnik, A.V.; Vrbanac, A.; Fu, T.; Patras, K.A.; Christy, M.P.; Bodai, Z.; Belda-Ferre, P.; Tripathi, A.; Chung, L.K.; et al. Global Chemical Effects of the Microbiome Include New Bile-Acid Conjugations. Nature 2020, 579, 123–129. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular Framework for Processing, Visualizing, and Analyzing Mass Spectrometry-Based Molecular Profile Data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- Perez, F.; Granger, B.E. IPython: A System for Interactive Scientific Computing. Comput. Sci. Eng. 2007, 9, 21–29. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Bray, J.R.; Roger Bray, J.; Curtis, J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Faith, D.P.; Minchin, P.R.; Belbin, L. Compositional Dissimilarity as a Robust Measure of Ecological Distance. Theory Models Veg. Sci. 1987, 69, 57–68. [Google Scholar]
- Vázquez-Baeza, Y.; Pirrung, M.; Gonzalez, A.; Knight, R. EMPeror: A Tool for Visualizing High-Throughput Microbial Community Data. Gigascience 2013, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- Phelan, V.V. Feature-Based Molecular Networking for Metabolite Annotation. In Computational Methods and Data Analysis for Metabolomics; Humana: New York, NY, USA, 2020; pp. 227–243. [Google Scholar]
- Nothias, L.F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Fahy, E.; Sud, M.; Cotter, D.; Subramaniam, S. LIPID MAPS Online Tools for Lipid Research. Nucleic Acids Res. 2007, 35, W606–W612. [Google Scholar] [CrossRef]
- Chambers, J.M. Graphical Methods for Data Analysis; Chapman and Hall/CRC: Wadsworth International Group: Boca Raton, FL, USA, 1983; ISBN 9780534980528. [Google Scholar]
- Dührkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Böcker, S. SIRIUS 4: A Rapid Tool for Turning Tandem Mass Spectra into Metabolite Structure Information. Nat. Methods 2019, 16, 299–302. [Google Scholar] [CrossRef]
| m/z | RT (min) | Predicted Molecular Formula (SIRIUS) or Spectral Match Formula | Spectral Match on GNPS/LIPID MAPS/Molecular Networking | Mass Difference | PPM Error | Cosine Score | Number of Matched Peaks for GNPS Spectral Match | p Value (Infected Center vs Uninfected Center) 1 | p Value (Infected Edge vs Uninfected Edge) | p Values (Infected vs Uninfected) |
|---|---|---|---|---|---|---|---|---|---|---|
| Dysregulated in ear center | ||||||||||
| 147.0815 | 0.34 | C5H10N2O3 | Glutamine | 0.01 | 31 | 1 | 4 | 0.008 | 0.421 | 0.001 |
| 169.0624 | 0.31 | C4H12N2O3S | NA 2 | NA | NA | NA | NA | 0.011 | 0.548 | 0.011 |
| 330.1314 | 6.4 | C14H21N5O2 ([M+K]+) | NA | NA | NA | NA | NA | 0.008 | 0.008 | 1.08 × 10−5 |
| 750.5435 3 | 5.06 | C34H17N9O9 | NA | NA | NA | NA | NA | 0.008 | 0.008 | 2.17 × 10−5 |
| 752.5585 | 5.07 | C40H73N5O8 | NA | NA | NA | NA | NA | 0.008 | 0.008 | 1.08 × 10−5 |
| 794.6052 | 6.13 | C46H84NO7P | PC O-38:5 | 0 | 0 | 1 | 5 | 0.008 | 0.008 | 1.08 × 10−5 |
| Dysregulated in ear edge | ||||||||||
| 261.1474 | 0.47 | C9H24N3O2S ([M+Na]+) | NA | NA | NA | NA | NA | 0.095 | 0.31 | 0.315 |
| 720.5887 | 4.7 | C40H82NO7P | LPC 32:0 or LPC O-32:1;O or PC O-32:0 | NA | 2 | NA | NA | 0.095 | 0.008 | 0.28 |
| 744.5906 | 4.48 | C42H82NO7P | LPC 34:2 or LPC O-34:3;O or PC O-34:2 | NA | 0.54 | NA | NA | 0.008 | 0.008 | 0.052 |
| 746.6052 | 4.78 | C42H84NO7P | PC O-16:0/18:1 | 0 | 4 | 1 | 14 | 0.032 | 0.008 | 0.28 |
| 770.605 | 4.58 | C44H84NO7P | PC O-36:3 | NA | 1.04 | NA | NA | 0.310 | 0.008 | 0.218 |
| 772.6201 | 4.73 | C44H86NO7P | PC O-36:2 | NA | 1.8 | NA | NA | 0.016 | 0.008 | 0.393 |
| 794.6051 | 4.42 | C46H84NO8P | PC O-38:5 | 0 | 0 | 1 | 5 | 0.056 | 0.008 | 0.481 |
| 796.6205 | 4.91 | C46H86NO7P | PC O-38:4 | NA | 1.26 | NA | NA | 0.151 | 0.008 | 0.105 |
| Dysregulated in both ear center and ear edge | ||||||||||
| 155.0498 | 0.85 | C11H6O | NA | NA | NA | NA | NA | 0.008 | 0.222 | 0.684 |
| m/z | RT (min) | Predicted Molecular Formula (SIRIUS) or Spectral Match Formula | Spectral Match on GNPS | Mass Difference | PPM Error | Cosine Score | Number of Matched Peaks for GNPS Spectral Match | p Values (Infected Center vs Uninfected Center) 1 | p Values (Infected Edge vs Uninfected Edge) | p Values (Infected vs Uninfected) |
|---|---|---|---|---|---|---|---|---|---|---|
| Dysregulated in ear center | ||||||||||
| 377.2679 | 3.53 | C11H34N10O3 ([M+Na]+) | NA 2 | NA | NA | NA | NA | 0.008 | 0.222 | 0.043 |
| 744.5848 | 5.59 | C42H82NO7P | LPC 34:2 or LPC O-34:3;O or PC O-34:2 | NA | 7.25 | NA | NA | 0.011 | 0.052 | 0.0003 |
| 768.5862 3 | 5.26 | C44H82NO7P | PC O-36:4 | NA | 5.2 | NA | NA | 0.008 | 0.008 | 1.08 × 10−5 |
| 770.6019 | 5.7 | C44H84NO7P | PC O-36:3 | NA | 5.06 | NA | NA | 0.008 | 0.095 | 0.0001 |
| 790.5424 | 5.43 | C45H76NO8P | PC 37:7 | NA | 5.44 | NA | NA | 0.548 | 0.691 | 0.631 |
| 792.5574 | 5.71 | C45H78NO8P | PC O-16:0/22:6 | 0.01 | 8 | 0.86 | 7 | 0.012 | 0.691 | 0.026 |
| 806.5682 | 5.42 | C46H80NO8P | PC 38:6 or PC O-38:7;O | 0.02 | 22 | 0.81 | 18 | 0.008 | 1 | 0.09 |
| 828.5516 | 5.44 | C48H78NO8P | PC O-40:10;O or PC 40:9 | NA | 1.2 | NA | NA | 0.008 | 0.691 | 0.143 |
| 834.5994 | 5.91 | C48H84NO8P | PC 40:6 or PC O-40:7;O | NA | 1.56 | NA | NA | 0.008 | 0.548 | 0.353 |
| 854.5676 | 5.36 | C38H79N9O10S | NA | NA | NA | NA | NA | 0.056 | 0.222 | 0.218 |
| 856.5826 | 5.87 | C43H73N11O7 | NA | NA | NA | NA | NA | 0.008 | 1 | 0.105 |
| 856.5826 | 5.9 | C43H73N11O7 | NA | NA | NA | NA | NA | 0.008 | 0.841 | 0.075 |
| 1017.687 | 3.04 | C45H84N20O7 | NA | NA | NA | NA | NA | 0.008 | 0.151 | 0.002 |
| Dysregulated in ear edge | ||||||||||
| 813.6845 | 5.27 | C46H92N4O5S | NA | NA | NA | NA | NA | 0.016 | 0.008 | 0.912 |
| Dysregulated in both ear center and ear edge | ||||||||||
| 332.6611 | 2.29 | no prediction in SIRIUS | NA | NA | NA | NA | NA | 0.008 | 0.222 | 0.0003 |
| m/z | RT (min) | Predicted Molecular Formula (SIRIUS) or Spectral Match Formula | Spectral Match on GNPS/LIPID MAPS/Molecular Networking | Mass Difference | PPM Error | Cosine Score | Number of Matched Peaks for GNPS Spectral Match | p Values 1 |
|---|---|---|---|---|---|---|---|---|
| 206.1067 | 4.51 | C7H16N3O2P | NA 2 | NA | NA | NA | NA | 0.008 |
| 210.1121 | 2.74 | C12H15N2 ([M+Na]+) | NA | NA | NA | NA | NA | 0.008 |
| 212.1651 | 2.75 | C12H21NO2 | NA | NA | NA | NA | NA | 0.008 |
| 230.1756 | 2.74 | C12H23NO3 | NA | NA | NA | NA | NA | 0.008 |
| 281.0052 | 2.55 | C9H13O4PS2 | NA | NA | NA | NA | NA | 0.008 |
| 303.2312 | 4.1 | C20H32O3 ([M+H-H2O]+) | 5,6-Epoxy-8Z,11Z,14Z-eicosatrienoic acid | 0 | 4 | 0.89 | 8 | 0.008 3 |
| 327.2325 | 4.05 | C22H30O2 | NA | NA | NA | NA | NA | 0.008 |
| 331.2638 | 4.31 | C22H34O2 | NA | NA | NA | NA | NA | 0.008 |
| 368.2591 | 4.06 | C24H33NO2 | NA | NA | NA | NA | NA | 0.008 |
| 377.1461 | 2.41 | C14H24N4O6S | NA | NA | NA | NA | NA | 0.012 |
| 377.2661 | 4.32 | C18H36N2O6 | NA | NA | NA | NA | NA | 0.008 |
| 425.3375 | 3.18 | C24H44N2O4 | NA | NA | NA | NA | NA | 0.008 |
| 508.3764 | 4.01 | C26H54NO6P | PC(P-18:0/0:0) | 0 | 2 | 0.91 | 10 | 0.008 |
| 522.2834 | 4.16 | C24H44NO9P | NA | NA | NA | NA | NA | 0.008 |
| 549.2233 | 2.49 | C22H36N4O10S | NA | NA | NA | NA | NA | 0.008 |
| m/z | RT (min) | Predicted Molecular Formula (SIRIUS) or Spectral Match Formula | Spectral Match on GNPS/LIPID MAPS/Molecular Networking | Mass Difference | PPM Error | Cosine Score | Number of Matched Peaks for GNPS Spectral Match | p Values 1 |
|---|---|---|---|---|---|---|---|---|
| 352.2937 | 4.71 | C16H40N4O2P | NA 2 | NA | NA | NA | NA | 0.008 3 |
| 480.3097 | 2.81 | C23H46NO7P | PE(18:1/0:0) | NA | 2 | NA | NA | 0.008 |
| 519.4891 | 3.79 | C29H65N3O2P | NA | NA | NA | NA | NA | 0.008 |
| 585.534 | 3.53 | C33H68N4O4 | NA | NA | NA | NA | NA | 0.016 |
| 703.5752 | 4.7 | C40H75N6O2P | NA | NA | NA | NA | NA | 0.008 |
| 720.5895 | 6.63 | C44H79N3OS ([M+Na]+) | NA | NA | NA | NA | NA | 0.008 |
| 722.4983 | 7.85 | C42H69NO7 ([M+Na]+) | NA | NA | NA | NA | NA | 0.095 |
| 744.5891 | 6.01 | C42H82NO7P | LPC 34:2 or LPC O-34:3;O or PC O-34:2 | NA | 2 | NA | NA | 0.008 |
| 768.5885 | 5.89 | C44H82NO7P | PC O-36:4 | NA | 2.21 | NA | NA | 0.008 |
| 794.6035 | 5.97 | C46H84NO7P | PC O-38:5 | 0 | 3 | 0.81 | 7 | 0.008 |
| 796.6135 | 6.64 | C44H83N7O3 ([M+K]+) | NA | NA | NA | NA | NA | 0.008 |
| 796.6182 | 6.59 | C46H86NO7P | PC O-38:4 | NA | 4 | NA | NA | 0.008 |
| 811.6686 | 6.55 | C47H91N2O6P | SM 42:3;O2 | NA | 0 | NA | NA | 0.008 |
| 813.6867 | 7.51 | C47H93N2O6P | SM 18:1;O2/24:1 | 0 | 4 | 0.91 | 6 | 0.008 |
| 828.5521 | 5.36 | C37H78N7O11P | NA | NA | NA | NA | NA | 0.008 |
| Ear Aqueous Extraction | |
| Start | 2% B |
| 1 min | 2% B |
| 1.5 min | 40% B |
| 4 min | 98% B |
| 5 min | 98% B |
| 6 min | 2% B |
| 7 min | 2% B |
| Ear Organic Extraction | |
| Start | 2% B |
| 1 min | 2% B |
| 1.5 min | 60% B |
| 5.5 min | 98% B |
| 7.5 min | 98% B |
| 8.5 min | 2% B |
| 10.5 min | 2% B |
| Footpad Aqueous Extraction | |
| Start | 2% B |
| 1 min | 2% B |
| 1.5 min | 40% B |
| 6 min | 98% B |
| 6.5 min | 98% B |
| 7 min | 2% B |
| Footpad Organic Extraction | |
| Start | 2% B |
| 1 min | 2% B |
| 1.5 min | 70% B |
| 7 min | 98% B |
| 8 min | 98% B |
| 9 min | 2% B |
| 10.5 min | 2% B |
| Detection Mode | Positive |
|---|---|
| Nebulizer gas pressure | 2 Bar |
| Capillary voltage | 4500 V |
| Ion source temperature | 200 °C |
| Dry gas flow | 9.0 L/min |
| Spectra rate acquisition | 3 spectra/s |
| Mass Detection | |
| MS level 1: Noise level | 1 × 103 |
| MS level 2: Noise level | 10 |
| Mass detector | Centroid |
| Chromatogram Builder | |
| Min time span | 0.06 min |
| Min peak height | 3 × 103 |
| m/z tolerance | 1 × 10−6 or 10 ppm |
| Chromatogram Deconvolution | |
| Algorithm | Baseline cutoff |
| Min peak height | 3 × 103 |
| Peak duration range (min) | 0.06–2 min (ear), 0.01–7 min (footpad) |
| Baseline level | 1 × 102 (ear), 1.5 × 103 (footpad) |
| m/z range for MS2 scan pairing (Da) | 0.01 |
| RT range for MS2 scan pairing (min) | 0.2 min |
| Isotopic Peaks Grouper | |
| m/z tolerance | 1 × 10−6 or 10 ppm |
| Retention time tolerance (absolute: min) | 0.05 min |
| Monotonic shape | Enabled |
| Maximum charge | 3 |
| Representative isotope | Most intense |
| Join Aligner | |
| m/z tolerance | 1 × 10−6 or 10 ppm |
| Weight for m/z | 7 |
| Retention time tolerance (absolute: min) | 0.5 min |
| Weight for RT | 3 |
| Manual Filtering | |
| Min number of peaks per row | 3 |
| RT range | 0.2–10.5 (ear organic and footpad), 0.2–6.9 (ear aqueous) |
| MS2 | required |
| Manual validation of peak shape | |
| Gap-Filing | |
| m/z tolerance | 1 × 10−6 or 10 ppm |
| RT tolerance | 0.5 min |
| Intensity tolerance | 30% |
| RT correction | Enabled |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parab, A.R.; Thomas, D.; Lostracco-Johnson, S.; Siqueira-Neto, J.L.; McKerrow, J.H.; Dorrestein, P.C.; McCall, L.-I. Dysregulation of Glycerophosphocholines in the Cutaneous Lesion Caused by Leishmania major in Experimental Murine Models. Pathogens 2021, 10, 593. https://doi.org/10.3390/pathogens10050593
Parab AR, Thomas D, Lostracco-Johnson S, Siqueira-Neto JL, McKerrow JH, Dorrestein PC, McCall L-I. Dysregulation of Glycerophosphocholines in the Cutaneous Lesion Caused by Leishmania major in Experimental Murine Models. Pathogens. 2021; 10(5):593. https://doi.org/10.3390/pathogens10050593
Chicago/Turabian StyleParab, Adwaita R., Diane Thomas, Sharon Lostracco-Johnson, Jair L. Siqueira-Neto, James H. McKerrow, Pieter C. Dorrestein, and Laura-Isobel McCall. 2021. "Dysregulation of Glycerophosphocholines in the Cutaneous Lesion Caused by Leishmania major in Experimental Murine Models" Pathogens 10, no. 5: 593. https://doi.org/10.3390/pathogens10050593
APA StyleParab, A. R., Thomas, D., Lostracco-Johnson, S., Siqueira-Neto, J. L., McKerrow, J. H., Dorrestein, P. C., & McCall, L.-I. (2021). Dysregulation of Glycerophosphocholines in the Cutaneous Lesion Caused by Leishmania major in Experimental Murine Models. Pathogens, 10(5), 593. https://doi.org/10.3390/pathogens10050593








