Abstract
Ticks transmit many pathogens with public health and veterinary importance. Despite the wide distribution of tick-borne pathogens in Sudan, the information on the tick–pathogen relationship needs to be updated, particularly using modern molecular techniques. This cross-sectional study, conducted between September and November 2019, used morphology, PCR, and sequencing to confirm the identity of adult cattle ticks (male and female; n = 536) from Khartoum State (n = 417) and East Darfur State (n = 119). Moreover, the presence of Theileria annulata, Babesia bigemina, B. bovis, Anaplasma marginale, and Ehrlichia ruminantium was detected and confirmed in each tick using species-specific PCR or nested PCR and sequencing. The most economically important tick genera, Rhipicephalus, Hyalomma, and Amblyomma, were prevalent in the study area, and 13 different tick species were identified. The most prevalent tick species were Rhipicephalus evertsi evertsi (34.3%) and Hyalomma anatolicum (57.3%) in Khartoum State, and Rhipicephalus annulatus (27%), Rhipicephalus decoloratus (25%), and Hyalomma rufipes (29%) in East Darfur State. We detected all five pathogens in both states. To the best of our knowledge, this is the first study to report the presence of E. ruminantium, its vector Amblyomma variegatum, and B. bovis in Khartoum State. Further, this is the first report on most tick and pathogen species identified in East Darfur State. Our findings indicate the migration of some tick and pathogen species beyond their distribution areas in the country, and this consideration is necessary to develop future control strategies.
Keywords:
ticks; tick-borne pathogens; epidemiology; cattle; Khartoum State; East Darfur State; Sudan 1. Introduction
Ticks (Acari: Ixodoidea) are important ectoparasites infesting livestock and human populations around the globe [1]. Ticks carry and transmit a large number of pathogens, including bacteria, viruses, and protozoa. They are second to mosquitoes in importance as disease vectors, greatly impacting human and animal health [2]. Ticks are voracious blood suckers, causing heavy blood losses leading to anemia, and the injuries caused by their attachment damage hides and predispose animals to secondary bacterial infections that may lead to mastitis. These wounds may also attract myiasis-causing flies. Long-hypostome ticks may induce abscesses due to secondary bacterial infections [3]. Moreover, ticks can cause paralysis [4]. Ticks have a wide range of hosts, with cattle being the favorite for many tick species [5].
Sudan occupies an area of 1.886 million km2 in the northeast of Africa and is located at latitude 15°47′9.85″ N and longitude 30°11′58.48″ E. Sudan harbors one of the largest cattle populations in Africa; the cattle population was estimated at approximately 31 million heads, raised in almost all parts of the country [6]. This large cattle population is severely affected, and its productivity is highly hampered by different ticks and tick borne-diseases. During the late 1940s and early 1950s, Hoogstraal recorded 25 tick species in Sudan [4]. Since then, scattered studies on tick distribution, morphological identification, morphological abnormalities, host range, and role in disease transmission were carried out in the country. These studies reported 70 tick species, including Hyalomma anatolicum, H. impressum, H. dromedarii, H. impeltatum, H. rufipes, H. truncatum, Rhipicephalus evertsi evertsi, R. sanguineus, R. praetextatus, R. annulatus, R. decoloratus, Amblyomma lepidum, and Am. variegatum, as the most prevalent tick species in the country [7,8,9,10,11,12,13,14,15,16]. As mentioned, ticks are very important disease vectors for livestock in Sudan; however, owing to the limited resources to perform molecular-based surveys, very few reports on the molecular identification of ticks, with data on only R. sanguineus, are available [14]. Moreover, very few studies have identified tick-borne pathogens in ticks in Sudan, such as Ehrlichia ruminantium in Am. lepidum and Am. variegatum [17,18], Rickettsia aeschlimannii and R. africae in different Hyalomma spp. and Am. lepidum [14], Crimean-Congo hemorrhagic fever virus in H. impeltatum [19], and R. africae in Am. variegatum [20]. Sudan consists, on the north-south direction, of desert, a semi-desert and a savannah that exhibits unique flora and fauna, with five identified ecological zones of rainfall [21]. In addition to this, its unique location in Africa, connecting north Africa with sub-Saharan countries, makes Sudan a very important country prone to transboundary animal diseases, including ticks and tick-borne diseases. Moreover, many factors determine the dynamic changes in tick distribution in the country, including animal movement for trade, nomadism, or migration due to civil unrest, which have been discussed in detail by Hassan and Salih [21]. Therefore, intensive and continuous screening for ticks and tick-borne diseases in Sudan is essential. The present study investigated the distribution and species of ticks infesting cattle in Khartoum State, central Sudan, and East Darfur State, western Sudan. We further screened the collected ticks for the presence of the most economically important tick-borne pathogens, including Thielira annulata, Babesia bovis, B. bigemina, Anaplasma marginale, and Ehrlichia ruminantium.
2. Results
2.1. Tick Species Identification
A total of 536 ticks were examined in this study. All individual ticks were morphologically identified. We identified three tick genera: Rhipicephalus, Hyalomma, and Amblyomma. The genus Hyalomma, with a prevalence of 56.9% (305/536), was found to be the most prevalent, followed by the genus Rhipicephalus [40.9% (219/536)] and Amblyomma [2.6% (14/536)].
Within the three genera, 13 tick species were morphologically identified, namely, R. annulatus, R. decoloratus, R. e. evertsi, R. praetextatus, R. simpsoni, Hyalomma rufipes, H. anatolicum, H. excavatum, H. dromedarii, H. impeltatum, H. marginatum, Am. lepidum, and Am. variegatum (Table 1). In general, within the identified tick species, H. anatolicum [44.6% (239/517)] and R. e. evertsi [27% (144/536)] were the most prevalent in the study area (Table 1).

Table 1.
Prevalence of different tick species in Khartoum State and East Darfur State.
A comparison of the prevalence of the different tick species in Khartoum State and East Darfur State revealed that the prevalence of H. anatolicum [57.3% (239/417)] and R. e. evertsi [34.3% (143/417)] in Khartoum State was significantly higher (p < 0.05) than that in East Darfur State, which was calculated to be 0% (0/119) and 0.84% (1/119), respectively (Table 1). In contrast, in East Darfur State, the prevalence of R. annulatus [27% (32/119)], R. decoloratus [25% (30/119)], H. rufipes [29% (34/119)], H. impeltatum [7.6% (9/119)], and Am. lepidum [8.4% (10/119)] was significantly higher (p < 0.05) than that in Khartoum State which [0% (0/417), 0% (0/417), 1.2% (5/417), 0.24% (1/417), and 0% (0/417), respectively] (Table 1). No statistically significant difference (p > 0.05) was observed between the prevalence of the remaining tick species in the two states (Table 1).
One to 15 ticks from all tick species morphologically identified were randomly selected for further confirmation using mitochondrial 12S rDNA gene analysis. The lengths of the amplified tick mitochondrial 12S rDNA sequences varied from 306 bp to 349 bp. A BLASTn analysis of the sequences obtained in this study showed an identity ranging from 97.64% to 100% with reference sequences from GenBank. However, matching mitochondrial 12S rDNA reference sequences for R. simpsoni are not available in the GenBank. All sequencing data are provided in Supplementary Table S1.
2.2. Pathogens Detected in the Ticks and Infection Rates
All tick DNA samples (536) were examined by PCR for the presence of five pathogens: Theileria annulata, Babesia bigemina, Babesia bovis, Anaplasma marginale, and Ehrlichia ruminantium. The most frequently observed pathogens in the tick samples were T. annulata [37.3% (200/536)] and B. bigemina [18.7% (100/536)], followed by A. marginale [10.6% (57/536)], E. ruminantium [10.07% (54/536)], and B. bovis [5.6% (30/536)] (Table 2). In Khartoum State the prevalence of T. annulata [42.7% (178/417)], A. marginale [12.5% (52/417)], and E. ruminantium [12.2% (51/417)] was significantly higher (p < 0.05) than that in East Darfur State [18.5% (22/119), 4.2% (5/119), and 2.5% (3/119), respectively]. In contrast, the prevalence of B. bigemina [31.1% (37/119)] in East Darfur State was significantly higher (p < 0.05) than that recorded in Khartoum State [15.1% (63/417)]. The prevalence of B. bovis in ticks did not differ to a statistically significant extent between the two states (Table 2).

Table 2.
Prevalence of different tick-borne pathogens in ticks in Khartoum State and East Darfur State.
2.3. Distribution of Tick-Borne Pathogens within Different Tick Species
In general, out of 536 tick samples examined, 339 (63%), representing 11 tick species, were positive for at least one of the five pathogens detected, while only two tick species, H. dromedarii and R. simpsoni, were negative for all pathogens. On the other hand, R. e. evertsi, H. anatolicum, and H. marginatum were found to harbor all the five pathogens (Table 3).

Table 3.
Distribution of tick-borne pathogens within different tick species.
Theileria annulata, the most prevalent pathogen in the study samples, with a prevalence of 37.3% (200/536) (Table 2), was found mainly in two tick species, R. e. evertsi [27.5% (55/200)] and H. anatolicum [56.5% (113/200)] (Table 3). B. bigemina, the second most prevalent pathogen, with a prevalence of 18.7% (100/536) (Table 2), was found mainly in four tick species, H. anatolicum [44% (44/100)], R. decoloratus [18% (18/100)], R. e. evertsi [15% (15/100)], and R. annulatus [11% (11/100)] (Table 3). Other relatively less prevalent pathogens, including B. bovis, A. marginale, and E. ruminantium, were found harbored in R. e. evertsi with a prevalence of 23.3% (7/30), 35.1% (20/57), and 24.1% (13/54), respectively, and in H. anatolicum with a prevalence of 43.3% (13/30), 49.1% (28/57), and 55.5% (30/54), respectively (Table 3).
2.4. Comparative Sequence Analyses of tams-1, ama1, msp4, spb4, and pCS20 Genes
The percentage of nucleotide identity of all four T. annulata tams-1 gene sequences (LC611432–LC611435) ranged from 94.7% to 95.4% among themselves. These sequences showed a 99.6% and 97% nucleotide sequence identity with sequences from previous studies conducted in Mauritania (AF214823) and India (MH277619). Further, the percentage of identity of all five sequences of B. bigemina ama1 gene (LC611412–LC611416) ranged from 98.1% to 99.05% among themselves. These sequences showed a 99% nucleotide sequence identity with sequences from Egypt (AB917262) and South Africa (KF626596). On the other hand, the percentage of nucleotide identity of all seven B. bovis spb4 gene sequences (LC611417–LC611423) ranged from 97.9% to 99.7% among themselves. These sequences showed a 99.6% and 95% nucleotide identity with sequences from previous studies conducted in Indonesia (KY484532) and South Africa (KF626635). Furthermore, the percentage of nucleotide identity of nine A. marginale msp4 gene sequences (LC611403- LC611411) ranged from 99.4% to 100% among themselves. These sequences shared a 100% nucleotide sequence identity with a previously reported sequence from Nigeria (EU106082). Additionally, the percentage of nucleotide identity of seven E. ruminantium pCS20 gene sequences (LC611424–LC611430) ranged from 98.9% to 99.8%. However, these sequences shared a 100% and 99.3% nucleotide sequence identity with previously reported sequences from South Africa (DQ631925) and Sudan (AB218277).
2.5. Phylogenetic Analysis
In this study, phylogenetic trees of T. annulata, B. bigemina, B. bovis, A. marginale, and E. ruminantium were constructed based on tams-1, ama1, spb4, msp4, and pCS20 genes, respectively, using sequences from the NCBI GenBank. Four sequences (LC611432–LC611435) of T. annulata were analyzed. LC611431 clustered on one clade of the phylogenetic tree with sequences AB917287 and KJ021627 from Egypt, while the other three sequences (LC611433, LC611434, and LC611435) clustered together with AF214919 from Tunisia and KJ021626 and AB917288 from Egypt (Figure 1).
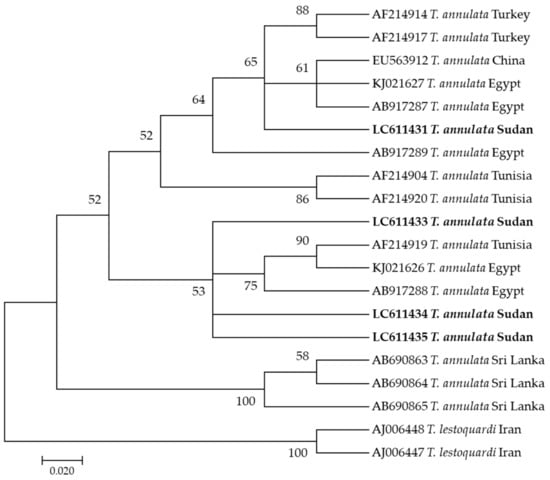
Figure 1.
Phylogenetic analysis of Theileria annulata identified in this study based on Tams-1 gene sequences using the maximum likelihood method. The number at the nodes represents the percentage occurrence of clade in a 1000 bootstrap replication of data. The best model was Tamura-Nei Model 92 with Gamma distribution. Sequences from this study are shown in bold font. The tree was constructed using the MEGA version X software program.
On the other hand, five sequences (LC611412–LC611416) of B. bigemina used in this study appeared in the same clade, that also involved sequences KF626596, KF626597, and KF626600 from South Africa as well as AB17262 from Egypt (Figure 2).
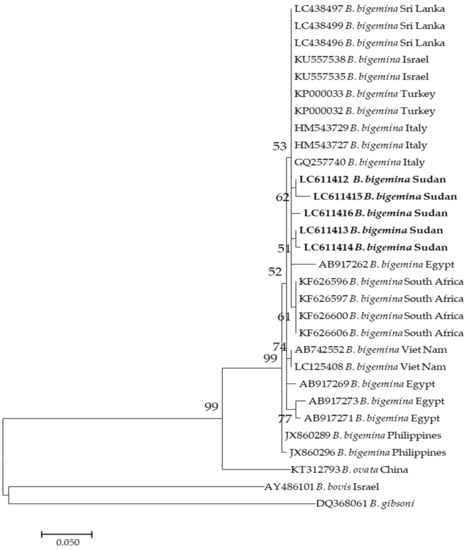
Figure 2.
Phylogenetic analysis of Babesia bigemina identified in this study based on ama1 gene sequences using the maximum likelihood method. The number at the nodes represents the percentage occurrence of clade in a 1000 bootstrap replication of data. The best model was the Kimura 2-parameter model. Sequences from this study are shown in bold font. The tree was constructed using the MEGA version X software program.
Moreover, seven sequences (LC611417–LC611423) of B. bovis generated in this study clustered together in the same clade with sequences KY484532 from Indonesia, KX685399 from Syria, and KX685399 from Egypt (Figure 3).
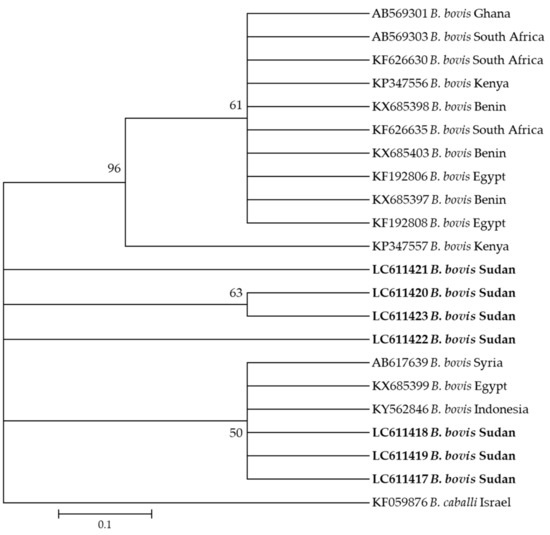
Figure 3.
Phylogenetic analysis of Babesia bovis identified in this study based on spb4 gene sequences using the maximum likelihood method. The number at the nodes represents the percentage occurrence of clade in a 1000 bootstrap replication of data. The best model was the Kimura 2-parameter model with invariant sites. Sequences from this study are shown in bold font. The tree was constructed using the MEGA version X software program.
Furthermore, the phylogenetic analysis revealed that the nine A. marginale sequences generated in this study (LC611403–LC611411) were in the same clade as sequences from Zimbabwe (AY666007, AY666011, and AY666006), from Kenya (AY666004), and from Nigeria (EU106082) (Figure 4).
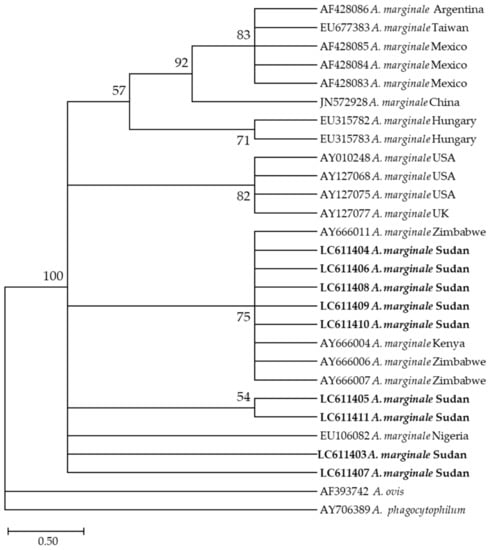
Figure 4.
Phylogenetic analysis of Anaplasma marginale identified in this study based on msp4 gene sequences using the maximum likelihood method. The number at the nodes represents the percentage occurrence of clade in a 1000 bootstrap replication of data. The best model was the Tamura 2-parameter model with invariant sites. Sequences from this study are shown in bold font. The tree was constructed using the MEGA version X software program.
Additionally, six E. ruminantium sequences (LC611424–LC611430) used in this study appeared in the same clade along with different sequences from different countries in Africa, such as those from South Africa (DQ631925, DQ631920, and AY236069) and a previously reported sequence from Sudan (AB218277) (Figure 5).
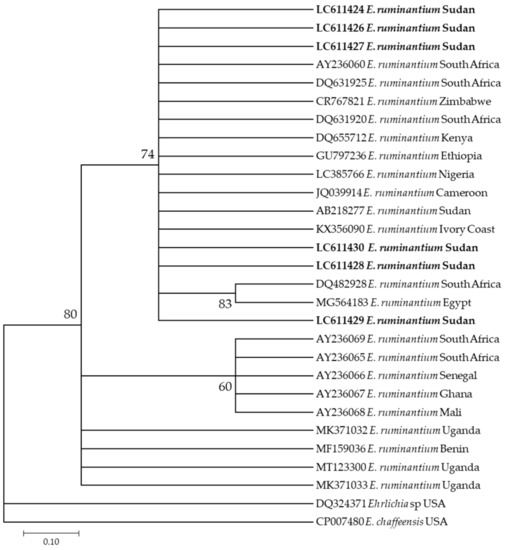
Figure 5.
Phylogenetic analysis of Ehrlichia ruminantium identified in this study based on pCS20 gene sequences using the maximum likelihood method. The number at the nodes represents the percentage occurrence of clade in 1000 a bootstrap replication of data. The best model was Tamura-Nei Model 92. Sequences from this study are shown in bold font. The tree was constructed using the MEGA version X software program.
3. Discussion
Tick infestations are common in Africa, including Sudan, where tick fauna comprises over 70 species prevalent in diverse ecological zones [21]. In this study, ticks were collected from cattle in two states—Khartoum State, in central Sudan, and East Darfur State, in western Sudan. The ticks were examined using morphology and molecular techniques (PCR, sequencing, and phylogenetics) to identify the species as well as to detect the microorganisms they carried. We morphologically identified 13 tick species of the three most economically important genera, namely, Rhipicephalus, Hyalomma, and Amblyomma, and confirmed their identity using molecular techniques. Within the genus Rhipicephalus, five different species were identified. R. annulatus and R. decoloratus, the main vectors of babesiosis [22], were detected only in East Darfur State. This is consistent with previous studies [4,16,23,24] that confirmed the presence of these two tick species in southern parts of the county, extending as far north as Wad Medani (14° 25′ N, 32° 75′ E), which lies to the south of Khartoum State, where these two ticks were not identified in this study. Hassan and Salih attributed the presence of these two ticks in the southern parts of the country, including East Darfur State, to humidity; Khartoum State is a dry area [21]. Interestingly, this is the first study to identify R. simpsoni tick species (the greater cane rat tick), which is considered native to the Republic of South Sudan, in Khartoum State [4,25,26]. The presence of R. simpsoni in Khartoum State, hundreds of kilometers north of the Republic of South Sudan, indicates that ticks in general are highly dynamic and can move with the animals migrating for different reasons, such as nomadism or civil unrest, especially after the separation of Sudan and the Republic of South Sudan in 2011, as discussed by Hassan and Salih [21]. We detected a significantly higher prevalence of R. e. evertsi in Khartoum Sate. With a prevalence of 34.3%, R. e. evertsi was the second most abundant tick species in Khartoum State after H. anatolicum. However, only one tick belonging to R. e. evertsi was recorded in East Darfur State, while all R. praetextatus ticks identified in this study were from Khartoum State. This data on R. e. evertsi and R. praetextatus is consistent with their distribution recorded in previous studies [27,28,29]. Within the genus Hyalomma, the prevalence of H. anatolicum, the most important tick species in Sudan and the main vector for T. annulata in the country [21], was found to be significantly higher in Khartoum State. This finding is consistent with previous studies [8,30]. H. anatolicum is a xerophilic species thriving in semi-desert conditions in northern Sudan, including Khartoum Sate [10]. Although in this study we did not detect H. anatolicum in East Darfur State, Abdalla and Hassan have identified it in Nyala, in South Darfur State [31], which borders East Darfur Sate. The absence of this tick species in East Darfur State in our study confirms that H. anatolicum is established in Nyala, South Darfur Sate, but has not yet spread to other localities of Darfur, as discussed previously [21]. We also reported a significantly higher prevalence of H. impeltatum and H. rufipes in East Darfur State that is consistent with the findings of previous studies [29,32] that reported their prevalence in South Darfur Sate, bordering East Darfur Sate. Two ticks of the genus Amblyomma were detected in this study. Am. lepidum, which is classified as a dry region tick abundant in the eastern part of Sudan [15,33], was detected in far western Sudan in East Darfur. Although this tick was previously reported in South Darfur State [31], the present study is the first report in East Darfur State. The presence of Am. lepidum in East Darfur State is alarming because it suggests the possible spread of this species, that was previously confined to eastern parts of the African continent [33], to West African countries in the near future. This would aggravate and complicate the heartwater incidence in West African countries, as discussed by Hassan and Salih [21]. On the other hand, within Amblyomma, only two female Am. variegatum ticks were detected in Khartoum State. The occurrence of Am. variegatum in Khartoum State is surprising because this tick species has not been previously identified in the area; however, it explains the presence of E. ruminantium, detected in this study for the first time in Khartoum State.
We investigated five main tick-borne pathogens, T. annulata, B. bigemina, B. bovis, A. marginale, and E. ruminantium, in Sudan. All pathogens detected in this study were further confirmed using sequencing and phylogenetic analyses by clustering them with those in the GenBank. In Khartoum State, endemic for T. annulata and H. anatolicum [34], the parasite was detected in 42.7% (178/417) of the ticks, and its main vector, H. anatolicum, showed a 57.3% (239/417) prevalence. Interestingly, although H. anatolicum was not detected in East Darfur State in this study, we detected T. annulata for the first time in this area, in 18.5% (22/119) of the ticks belonging to different species of the genus Hyalomma. These data indicate the presence of alternative vectors for T. annulata in East Darfur State, such as H. rufipes, H. impeltatum, and H. marginatum, that were previously shown to transmit the parasite [34,35,36,37]. Although T. annulata was not detected in H. dromedarii in this study, it can act as an alternative vector [35]. We identified two Babesia parasites, B. bigemina and B. bovis, in the present study. In East Darfur State, both these parasites and their classical vectors, R. annulatus and R. decoloratus [22], were found. Similarly, we confirmed the presence of both B. bigemina and B. bovis in ticks from Khartoum State. Supporting this, B. bigemina was previously detected and microscopically identified in two cows in Khartoum Sate [38]. Notably, R. annulatus and R. decoloratus were not found in Khartoum State in this study. This finding should alert the veterinary authorities to the fact that these two Babesia parasites are prevalent, and control measures should be implemented, especially considering the high prevalence of R. e. evertsi (34.3%) in Khartoum Sate, which can transmit these parasites as an alternative vector [22]. This study is the first to confirm the presence of E. ruminantium and its vector Am. variegatum in Khartoum State. Our finding indicates that the bacterium and its vector are well established in this area and may have been overlooked in previous studies. This is cause for alarm, and the authorities should begin an organized screening of the clinical cases to develop better control strategies. More studies investigating the biological aspects of Am. variegatum and its presence in Khartoum State, which was considered free of this tick species [4,25], are required. In the present study, the prevalence of E. ruminantium in East Darfur State was consistent with that of its vector, Am. lepidum. It is worth mentioning that this is the first report on the presence of both the pathogen and its vector in East Darfur State, although they were previously detected in South Darfur State [18,31]. In this study, the prevalence of A. marginale was confirmed in ticks from Khartoum State and East Darfur State. Like most of the pathogens investigated in this study, this is the first report on the prevalence of A. marginale in East Darfur State. A. marginale was previously reported in Khartoum State [38,39]. Further, Anaplasma marginale was proven to be transmitted by H. rufipes, R. annulatus, R. decoloratus, and R. e. evertsi [35]. All these tick species were identified in the two states in this study except for R. annulatus and R. decoloratus, that were only identified in East Darfur State.
Although all ticks assessed in this study were feeding ticks collected from the animals, and the pathogen source may be the infested animal itself, all identified tick species harboring the respective pathogens could be considered as potential vectors for these pathogens. This study provides basic information on the prevalence of the most economically important ticks and tick-borne pathogens in Khartoum State and East Darfur State. Our findings provide a better understanding of the tick–pathogen relationship in Sudan. Studies on the prevalence and distribution of ticks and tick-borne pathogens in Sudan are highly important to develop better control strategies.
4. Materials and Methods
4.1. Study Area
This cross-sectional study was carried out between September and November 2019 in Khartoum State, central Sudan (longitude 31.5° to 34° E and latitude 15° to 16° N), and East Darfur State, western Sudan (longitude 26° to 30° E and latitude 11° to 10.01° N) (Figure 6). Although Khartoum State is the smallest state in the country based on area (22,142 km2), it is considered a big market, and livestock is transported from different parts of the country to meet the consumer demand of meat [40]. Moreover, 17 dairy camps were established around Khartoum State, under the supervision of the Ministry of Agriculture, Animal Resources, and Irrigation of Khartoum State, under an intensive traditional farming system, to meet approximately 80% of the milk requirement of the population [41]. In contrast, in East Darfur State, livestock is owned by the traditional sector, mostly made up of nomads who migrate with their animals as far as beyond latitude 8° N in the south and 14° N in the north, in search for pastures throughout the year [21]. Considering the differences in their geographic location and animal husbandry practices, we selected these two states for tick screening in this study.
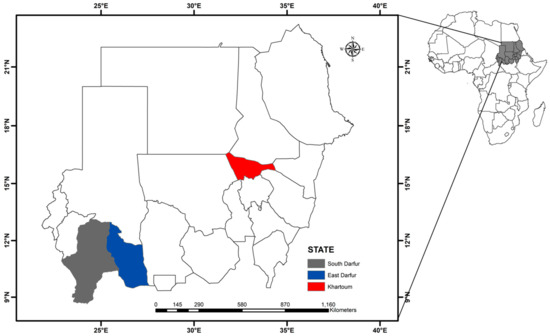
Figure 6.
A map of Sudan. Map created using the ArcMap 10.1 software program (Esri, Redlands, CA, USA).
4.2. Tick Samples
A total of 536 adult feeding ticks [male (n = 328) and female (n = 208)] were collected from 153 infested cattle (three to five ticks from each animal) in Khartoum State (n = 417) and East Darfur State (n = 119). The tick sampling was conducted with the consent of the livestock owners, and care was taken to minimize animal discomfort. All procedures of tick sampling and sample processing were performed in accordance with the ethical guidelines of the Obihiro University of Agriculture and Veterinary Medicine (approval No: 2019020 and 200049). Collected ticks were isolated in 1.5 mL tubes containing 70% ethanol, labeled with the location and date of collection, and stored at room temperature until processing. Prior to DNA extraction, identification and morphological classification of the tick species were carried out using a binocular microscope (Olympus SZX16, Tokyo, Japan) with previously established standard taxonomic keys [4,35].
4.3. DNA Extraction
Each tick was washed in three 70% ethanol baths, rinsed in double distilled water, air dried, and collected in sterile 1.5 mL tubes. The tick-containing tubes were plunged in liquid nitrogen for 3–5 min, and the frozen ticks were crushed using sterile pellet mixers. The tissues of crushed ticks were transferred aseptically to sterile 1.5 mL tubes and used for DNA extraction. DNA was extracted using NucleoSpin® Tissue Mini kit for DNA from cells and tissue (Macherey-Nagel, Dueren, Germany) according to the manufacturer’s protocol. Extracted DNA was stored at −30 °C until molecular analysis.
4.4. Molecular Characterization of Ticks
To confirm the species, tick DNA was amplified by PCR, and an approximately 360-bp fragment of tick mitochondrial 12S rDNA was sequenced. The amplification was performed with forward (F1; 5′-AAACTAGGATTAGATACCCT-3′) and reverse (R1; 5′-AATGAGAGCGACGGGCGATGT-3′) primers [42] under the following cycling conditions: initial denaturation at 98 °C for 2 min, 40 cycles of denaturation at 98 °C for 10 s, annealing at 55 °C for 15 s, and extension at 68 °C for 45 s. Each 10 µL PCR reaction method included 5 µL of 2× MightyAmp Buffer Ver.3, 1 µL of 10× Additive for High Specificity, 0.2 µL of MightyAmp DNA Polymerase Ver. 3 (Takara, Shiga, Japan), 0.5 µL each of 10 mM forward and reverse primers, 1.8 µL of double-distilled water, and 1 µL of the tick DNA template. The DNA of Amblyomma variegatum ticks collected in Benin [43] and distilled water were used as positive and negative controls, respectively. PCRs were conducted on a Veriti™ Thermal cycler (Thermo Fisher Scientific Inc., Waltham, MA, USA). The PCR products were electrophoresed on 1.5% agarose gels that were stained with ethidium bromide and visualized under UV light. Then, all amplicons were extracted from the gels using a QIAquick Gel Extraction Kit (QIAGEN, Hilden, Germany) and sequenced.
4.5. Detection and Characterization of Tick-Borne Pathogens
To detect the presence of T. annulata, B. bigemina, B. bovis, A. marginale, and E. ruminantium in the extracted DNA, individual tick DNA was used as template to amplify the T. annulata merozoite surface antigen (tams-1) [44], B. bigemina apical membrane antigen-1 (ama1) [44], B. bovis spherical body protein-4 (sbp4) [45], A. marginale major surface protein 4 (msp4) [46], and E. ruminantium pCS20 region [47]. The primer sequences used in all PCR assays are listed in Table 4. PCR cycling conditions for all pathogen assays were the same as documented in respective referenced publications, with some modifications. Briefly, the cycling conditions were as follows: initial denaturation at 98 °C for 2 min; 40 cycles of denaturation at 98 °C for 10 s, annealing at varying temperatures (indicated in Table 4) for 15 s, and extension at 68 °C for 45 s. The reaction mixtures included MightyAmpTM DNA Polymerase Ver. 3 (Takara, Shiga, Japan), and the amplified templates were observed as described in 4.4. Amplicons were purified using a QIAquick Gel Extraction Kit and used for sequencing.

Table 4.
Primer sequences used in this study.
4.6. Sequencing Analysis
The selected gel-extracted PCR amplicons were sequenced bi-directionally using the same PCR primers. Approximately 300 ng/µL of purified DNA was used for sequencing the PCR amplicons using a Big Dye Terminator kit. The PCR product was ethanol-precipitated and dissolved in 20 µL of a Hi-Di formamide solution prior to DNA sequencing. The sequencing procedure consisted of 30 cycles of denaturation at 96 °C for 1 min, annealing at 50 °C for 5 s, and extension at 60 °C for 2 min. The gene sequence was analyzed using an ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Carlsbad, CA, USA). The forward and reverse raw sequences were processed using sangeranalyseR [48] to generate consensus sequences. Low quality sequences were excluded and cleaned manually.
4.7. BLASTn Analysis, Sequence Alignment, and Phylogenetic Analyses
Pairwise distances among the sequences were evaluated using MEGA version X [49], and the percentage identity was determined by the Basic Local Alignment Search Tool (BLASTn) in NCBI GenBank. Reference sequences were downloaded from NCBI, and the Simulate PCR tool was used to extract amplicons with respective primers. The reference extracts and sequenced products were aligned using MAFFT online version 7 (https://mafft.cbrc.jp/alignment/software/(accessed date 15 February 2021)). The alignments were imported to MEGA version X, and modeling was performed to determine the substitution model for each alignment. The phylogenetic analysis was performed by Maximum Likelihood using the respective substitution models and 1000 bootstraps. The sequences generated in this study were deposited in the GenBank database under the accession numbers (LC612433–LC612524 for different tick species), (LC611403–LC611411 for A. marginale), (LC611412–LC611416 for B. bigemnia), (LC611417–LC611423 for B. bovis), (LC611424–LC611430 for E. ruminantium), and (LC611431–LC611435 for T. annulata).
4.8. Statistical Analysis
The prevalence (P) of different ticks and tick-borne pathogens was calculated using the following formula: P (%) = number of positive samples/total number of samples × 100. A Chi-squared test and a Fisher’s exact test were used to investigate the differences in the prevalence with different variables using GraphPad (GraphPad Software Inc., San Diago, CA, USA). p-values < 0.05 were considered to indicate statistically significant differences.
5. Conclusions
We documented the prevalence of 13 different tick species belonging the most economically important tick genera, Rhipicephalus, Hyalomma, and Amblyomma, in two states in Sudan, namely, Khartoum Sate and East Darfur State. Moreover, we documented five pathogens, namely, T. annulata, B. bigemina, A. marginale, B. bovis, and E. ruminantium, in the collected ticks. We confirmed the identity of all ticks and pathogens using molecular techniques and suggested the potential roles of different tick species in the transmission of the respective pathogens. To the best of our knowledge, this is the first study to detect the presence of E. ruminantium, its vector Am. variegatum, and B. bovis in Khartoum State. Moreover, as very few studies have been carried out in East Darfur State, this is the first report on the most identified tick and pathogen species in East Darfur State. These findings should alert the veterinary authorities to the need for an effective control strategy based on the distribution of ticks and pathogens in the study area as well as throughout the country. Our findings provide insights that could be used for the improvement of diagnostic protocols and the development of better treatment and control strategies of the detected pathogens. Further studies are required to identify the exact role of each tick species in the transmission of the corresponding pathogens. Further studies are anticipated to confirm the establishment of different tick species, especially in Khartoum State.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/pathogens10050580/s1, Table S1: Results of tick species identification based on mitochondrial 12S rDNA gene partial sequence.
Author Contributions
Conceptualization, E.M. and X.X.; data curation, E.M.; formal analysis, E.M., A.G., K.S., Y.O., B.S., and M.L.; funding acquisition, E.M., R.U.-S., and X.X.; investigation, E.M., Y.O.M., K.S., and R.U.-S.; methodology, E.M.; project administration, X.X.; resources, K.S., and R.U.-S.; supervision, X.X.; writing—original draft, E.M.; writing—review & editing, A.G., Y.O.M., K.S., R.U.-S., Y.O., B.S., M.L., and X.X. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by a grant from the Japanese Society for the Promotion of Science (JSPS) to EM (grant no. 19F19403). This study was also supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan as a project of the Joint Usage/Research Center. Additional funding was obtained from the Ministry of Higher Education and Scientific Research, Republic of Sudan (grant no. SRI-VS-2015–71933).
Institutional Review Board Statement
Permission for the sampling, DNA experiments (2019020), and animal handling (200049) was obtained from Obihiro University of Agriculture and Veterinary Medicine, Obihiro, Hokkaido, Japan.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sajid, M.S.; Kausar, A.; Iqbal, A.; Abbas, H.; Iqbal, Z.; Jones, M.K. An insight into the ecobiology, vector significance and control of Hyalomma ticks (Acari: Ixodidae): A review. Acta. Trop. 2018, 187, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Adjou Moumouni, P.F.; Thekisoe, O.; Gao, Y.; Liu, M.; Li, J.; Galon, E.M.; Efstratiou, A.; Wang, G.; Jirapattharasate, C.; et al. Genetic characterization of tick-borne pathogens in ticks infesting cattle and sheep from three South African provinces. Ticks Tick Borne Dis. 2019, 10, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Ambrose, N.; Lloyd, D.; Maillard, J.-C. Immune responses to Dermatophilus congolensis infections. Parasitol. Today 1999, 15, 295–300. [Google Scholar] [CrossRef]
- Hoogstraal, H. Ticks of the Sudan, (with Special Reference to Equatoria Province and with Preliminary Reviews of the Genera Boophilus, Margaropus, and Hyallomma); U.S. Government Printing Office: Washington, DC, USA, 1956.
- Moyo, B.; Masika, P. Tick control methods used by resource-limited farmers and the effect of ticks on cattle in rural areas of the Eastern Cape Province, South Africa. Trop. Anim. Health Prod. 2009, 41, 517–523. [Google Scholar] [CrossRef]
- FAO. Special Report—2019 FAO Crop and Food Supply Assessment Mission to the Sudan; FAO: Rome, Italy, 2020. [Google Scholar] [CrossRef]
- Elhaj, M.T.; Taha, K.M.; El Ghali, A.; Hassan, S.M.; Salih, D.A.; Ahmed, J.; Clausen, P.-H.; EL Hussein, A.R.M. Baseline survey of Ixodid ticks infesting cattle in Northern State, Sudan. J. Veterina. Sci. Res. 2019, 1, 37–46. [Google Scholar] [CrossRef]
- Salih, D.A.; Sharieff, O.E.; Lazarus, A.G.; Hassan, S.M.; El Hussein, A.M. Natural infection rates and transmission of Theileria annulata by Hyalomma anatolicum ticks in the Sudan. Onderstepoort J. Vet. Res. 2005, 72, 303–307. [Google Scholar] [CrossRef] [PubMed]
- Salih, D.A.; Hassan, S.M.; El Hussein, A.M.; Jongejan, F. Preliminary survey of ticks (Acari: Ixodidae) on cattle in northern Sudan. Onderstepoort J. Vet. Res. 2004, 71, 319–326. [Google Scholar] [CrossRef][Green Version]
- Jongejan, F.; Zivkovic, D.; Pegram, R.G.; Tatchell, R.J.; Fison, T.; Latif, A.A.; Paine, G. Ticks (Acari: Ixodidae) of the Blue and White Nile ecosystems in the Sudan with particular reference to the Rhipicephalus sanguineus group. Exp. Appl. Acarol. 1987, 3, 331–346. [Google Scholar] [CrossRef]
- Ahmed, B.M.; El Hussein, A.M.; El Khider, A.O. Some observations on ticks (Acari: Ixodidae) infesting sheep in river Nile Province of Northern Sudan. Onderstepoort J. Vet. Res. 2005, 72, 239–243. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shuaib, Y.A.; Isaa, M.H.; Ezz-Eldin, M.I.-E.; Abdalla, M.A.; Bakhiet, A.O.; Chitimia-Dobler, L. Morphological abnormalities in ticks (Acari: Ixodidae) collected from domestic animal species in Sudan. Exp. Appl. Acarol. 2020, 82, 161–169. [Google Scholar] [CrossRef]
- Hayati, M.A.; Hassan, S.M.; Ahmed, S.K.; Salih, D.A. Prevalence of ticks (Acari: Ixodidae) and Theileria annulata infection of cattle in Gezira State, Sudan. Parasite Epidemiol. Control 2020, 20, e00148. [Google Scholar] [CrossRef]
- Shuaib, Y.A.; Elhag, A.M.-A.W.; Brima, Y.A.; Abdalla, M.A.; Bakiet, A.O.; Mohmed-Noor, S.E.-T.; Lemhöfer, G.; Bestehorn, M.; Poppert, S.; Schaper, S.; et al. Ixodid tick species and two tick-borne pathogens in three areas in the Sudan. Parasitol. Res. 2020, 119, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Karrar, G.; Kaiser, M.N.; Hoogstraal, H. Ecology and host-relationships of ticks (Ixodoidea) infesting domestic animals in Kassala Province, Sudan, with special reference to Amblyomma lepidum Dönitz. 1. Bull. Entomol. Res. 1963, 54, 509–522. [Google Scholar] [CrossRef]
- Osman, O.; El Hussein, A.; Ahmed, N.; Abdulla, H. Ecological studies on ticks (Acarina, Ixodidae) of Kordofan Region, Sudan. Bull. Anim. Health. Prod. Afr. 1982, 30, 45–53. [Google Scholar]
- Muramatsu, Y.; Ukegawa, S.-y.; El Hussein, A.R.M.; Rahman, M.B.A.; Gabbar, K.M.A.A.; Chitambo, A.M.; Komiya, T.; Mwase, E.T.; Morita, C.; Tamura, Y. Ehrlichia ruminantium, Sudan. Emerg. Infect. Dis. 2005, 11, 1792–1793. [Google Scholar] [CrossRef] [PubMed]
- Sayed, M.; Hassan, S.M.; Elhussein, A.M.; Samir, E.; Mazahir, M.S.; Abdelrahman, M.B. Estimation of the prevalence of Ehrlichia ruminantium in Amblyomma spp. and blood of domestic ruminants in the Sudan by conventional PCR based on pCS20 gene region. Sud. J. Sci. Tech. 2013, 14, 15–27. [Google Scholar]
- Chitimia-Dobler, L.; Issa, M.H.; Ezalden, M.E.; Yagoub, I.A.; Abdalla, M.A.; Bakhiet, A.O.; Schaper, S.; Rieß, R.; Vollmar, P.; Grumbach, A.; et al. Crimean-Congo haemorrhagic fever virus in Hyalomma impeltatum ticks from North Kordofan, the Sudan. Int. J. Infect. Dis. 2019, 89, 81–83. [Google Scholar] [CrossRef]
- Nakao, R.; Qiu, Y.; Salim, B.; Hassan, S.M.; Sugimoto, C. Molecular detection of Rickettsia africae in Amblyomma variegatum collected from Sudan. Vector Borne Zoonotic Dis. 2015, 15, 323–325. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.; Salih, D. An overview of factors responsible for geographic distribution pattern of ixodid ticks in the Sudan. Sokoto J. Vet. Sci. 2013, 11, 1–9. [Google Scholar] [CrossRef][Green Version]
- Bock, R.; Jackson, L.; de Vos, A.; Jorgensen, W. Babesiosis of cattle. Parasitology 2004, 129, S247–S269. [Google Scholar] [CrossRef]
- ElImam, A. Ecological studies of ticks (Acari: Ixodidae) infesting cattle in Kosti Province, Sudan. Sud. J. Vet. Sci. Anim. Husb. 2003, 42, 62–71. [Google Scholar]
- Sowar, A. Epidemiology and Ecology of Ticks and Some Tick-Borne Diseases in (Kadogli and Dilling) Southern Kordofan State, Sudan. Master’s Thesis, Department of Preventive Medicine, University of Khartoum, Khartoum State, Sudan, 2002. [Google Scholar]
- ElGhali, A.A.; Hassan, S.M. Ticks infesting animals in the Sudan and southern Sudan: Past and current status. Onderstepoort J. Vet. Res. 2012, 79, E1–E6. [Google Scholar] [CrossRef]
- Walker, A. The genus Rhipicephalus (Acari, Ixodidae): A guide to the brown ticks of the world. In Tropical Animal Health and Production; Walker, J.B., Keirans, J.E., Horak, I.G., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2000; pp. 409–416. [Google Scholar]
- Gad Elrab, N. A Survey of Sheep Piroplasmosis in Khartoum Province, Central Sudan. Master’s Thesis, Department of Parasitology, University of Khartoum, Khartoum State, Sudan, 1986. [Google Scholar]
- Osman, I. Some Studies on Malignant Ovine Theileriosis in Northern Sudan. Master’s Thesis, Department of Parasitology, University of Khartoum, Khartoum State, Sudan, 1999. [Google Scholar]
- Ali, M.M. Studies on Ticks and Tick-Borne Diseases of Cattle in South Darfur State, Sudan. Master’s Thesis, Department of Parasitology, University of Khartoum, Khartoum State, Sudan, 2007. [Google Scholar]
- Walker, A.R.; Latif, A.A.; Morzaria, S.P.; Jongejan, F. Natural infection rates of Hyalomma anatolicum with Theileria in Sudan. Res. Vet. Sci. 1983, 35, 87–90. [Google Scholar] [CrossRef]
- Abdalla, M.M.; Hassan, S.M. Current status of distribution of ticks (Acari: Ixodidae) infesting cattle in South Darfur State, Sudan. Univ. Khartoum. J. Vet. Med. Anim. Prod. 2010, 1, 76–97. [Google Scholar]
- Abaker, I.A.; Salih, D.A.; El Haj, L.M.; Ahmed, R.E.; Osman, M.M.; Ali, A.M. Prevalence of Theileria annulata in dairy cattle in Nyala, South Darfur State, Sudan. Vet. World. 2017, 10, 1475–1480. [Google Scholar] [CrossRef]
- Walker, J.B.; Olwage, A. The tick vectors of Cowdria ruminantium (Ixodoidea, Ixodidae, genus Amblyomma) and their distribution. Onderstepoort J. Vet. Res. 1987, 54, 353–379. [Google Scholar]
- El Hussein, A.M.; Hassan, S.M.; Salih, D.A. Current situation of tropical theileriosis in the Sudan. Parasitol. Res. 2012, 111, 503–508. [Google Scholar] [CrossRef]
- Walker, A.; Bouattour, A.; Camicas, J.L.; Estrada-Peña, A.; Horak, I.; Latif, A.; Pegram, R.G.; Preston, P.M. Ticks of Domestic Animals in Africa: A Guide to Identification of Species; Bioscience Reports: Edinburgh, UK, 2003; pp. 98–114. [Google Scholar]
- Mustafa, U.E.H.; Jongejan, F.; Morzaria, S. Note on the transmission of Theileria annulata by Hyalomma ticks in the Sudan. Vet. Q. 1983, 5, 112–113. [Google Scholar] [CrossRef]
- Taha, K. Experimental Transmission of Theileria lestoquardi and Theileria annulata (Apicomplexa: Theileridae) and Their Cross Transmission between Cattle and Sheep. Ph.D. Thesis, Sudan Academy of Science, Khartoum, Sudan, 2009. [Google Scholar]
- Ibrahim, A.; Geysen, D.; Ismail, A.; Shadia, A.M. Haemoparasites identification in two dairy cattle farms in Khartoum State with reference of Babesia bigemina: Molecular confirmation. Sudan J. Vet. Res. 2010, 25, 37–42. [Google Scholar]
- Eisawi, N.M.; El Hussein, A.R.M.; Hassan, D.A.; Musa, A.B.; Hussien, M.O.; Enan, K.A.; Bakheit, M.A. A molecular prevalence survey on Anaplasma infection among domestic ruminants in Khartoum State, Sudan. Trop. Anim. Health Prod. 2020, 52, 1845–1852. [Google Scholar] [CrossRef]
- Karim, I.E.E.A.; Emam, A.A.; Gafar, S. Beef and mutton consumption in Khartum state: Demand analysis (2000–2004). J. Sc. Tech. 2009, 10, 12–26. [Google Scholar]
- El Zubeir, I.E.; Ahmed, G.M. An overview of the management practices and constrains at the dairy camps in Khartoum State, Sudan. Res. Opin. Anim. Vet. Sci. 2011, 1, 425–428. [Google Scholar]
- Beati, L.; Keirans, J.E. Analysis of the systematic relationships among ticks of the genera Rhipicephalus and Boophilus (Acari: Ixodidae) based on mitochondrial 12S ribosomal DNA gene sequences and morphological characters. J. Parasitol. 2001, 87, 32–48. [Google Scholar] [CrossRef]
- Adjou Moumouni, P.F.; Terkawi, M.A.; Jirapattharasate, C.; Cao, S.; Liu, M.; Nakao, R.; Umemiya-Shirafuji, R.; Yokoyama, N.; Sugimoto, C.; Fujisaki, K.; et al. Molecular detection of spotted fever group rickettsiae in Amblyomma variegatum ticks from Benin. Ticks Tick. Borne Dis. 2016, 7, 828–833. [Google Scholar] [CrossRef]
- Sivakumar, T.; Altangerel, K.; Battsetseg, B.; Battur, B.; AbouLaila, M.; Munkhjargal, T.; Yoshinari, T.; Yokoyama, N.; Igarashi, I. Genetic detection of Babesia bigemina from Mongolian cattle using apical membrane antigen-1 gene-based PCR assay. Vet. Parasitol. 2012, 187, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Terkawi, M.A.; Huyen, N.X.; Shinuo, C.; Inpankaew, T.; Maklon, K.; Aboulaila, M.; Ueno, A.; Goo, Y.-K.; Yokoyama, N.; Jittapalapong, S. Molecular and serological prevalence of Babesia bovis and Babesia bigemina in water buffaloes in the northeast region of Thailand. Vet. Parasitol. 2011, 178, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Torina, A.; Agnone, A.; Blanda, V.; Alongi, A.; D’Agostino, R.; Caracappa, S.; Marino, A.M.; Di Marco, V.; de la Fuente, J. Development and validation of two PCR tests for the detection of and differentiation between Anaplasma ovis and Anaplasma marginale. Ticks Tick Borne Dis. 2012, 3, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Mtshali, K.; Khumalo, Z.T.; Nakao, R.; Grab, D.J.; Sugimoto, C.; Thekisoe, O.M. Molecular detection of zoonotic tick-borne pathogens from ticks collected from ruminants in four South African provinces. J. Vet. Med. Sci. 2015, 77, 1573–1579. [Google Scholar] [CrossRef] [PubMed]
- Chao, K.-H.; Barton, K.; Palmer, S.; Lanfear, R. sangeranalyseR: Simple and interactive analysis of Sanger sequencing data in R. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).