Microeukaryotic Communities on the Fruit of Gardenia thunbergia Thunb. with a Focus on Pathogenic Fungi
Abstract
1. Introduction
2. Results
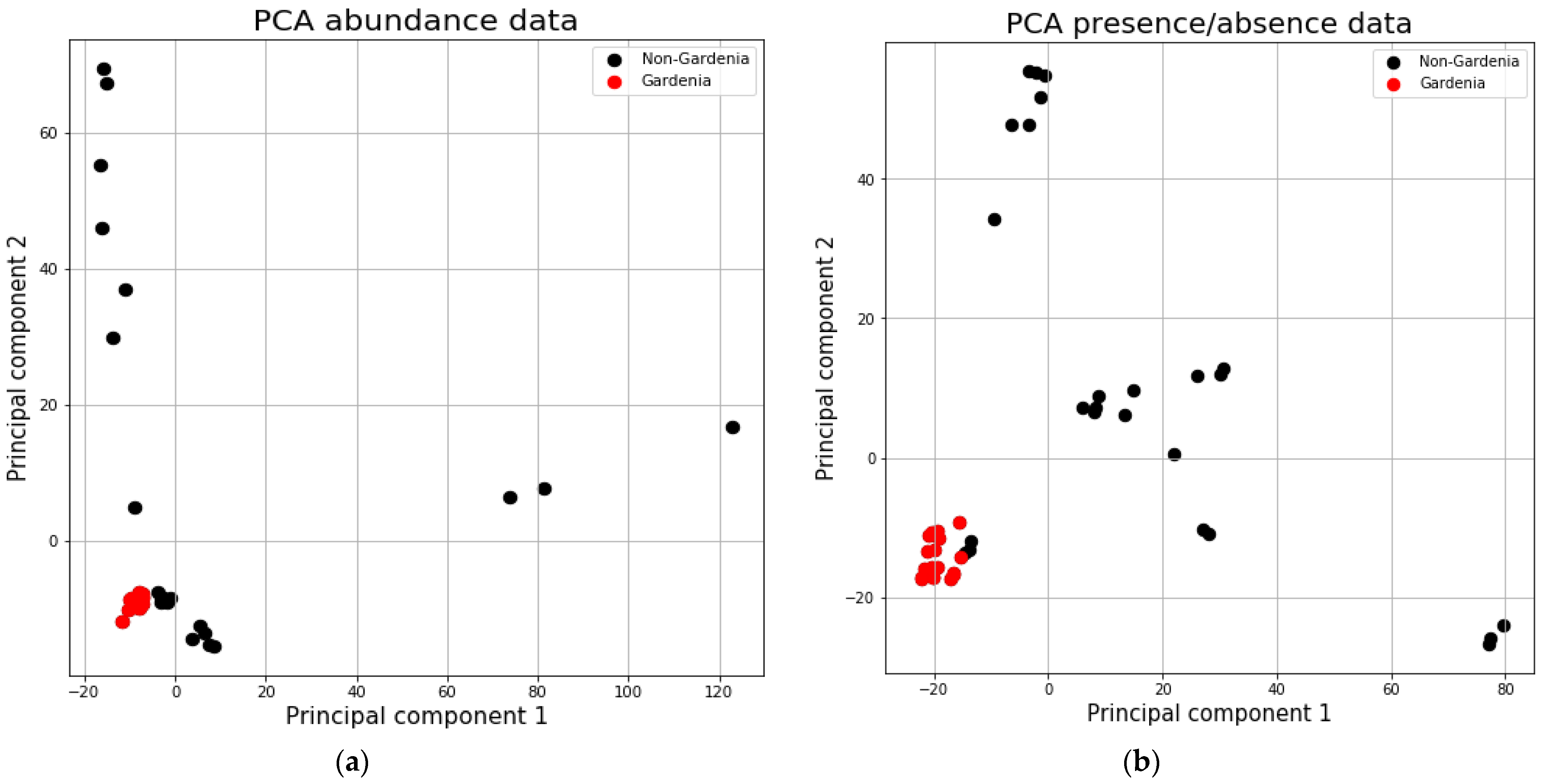
2.1. General Overview on the Microeukaryote Communities of All Samples
2.2. Abundances of the Microbial Communities
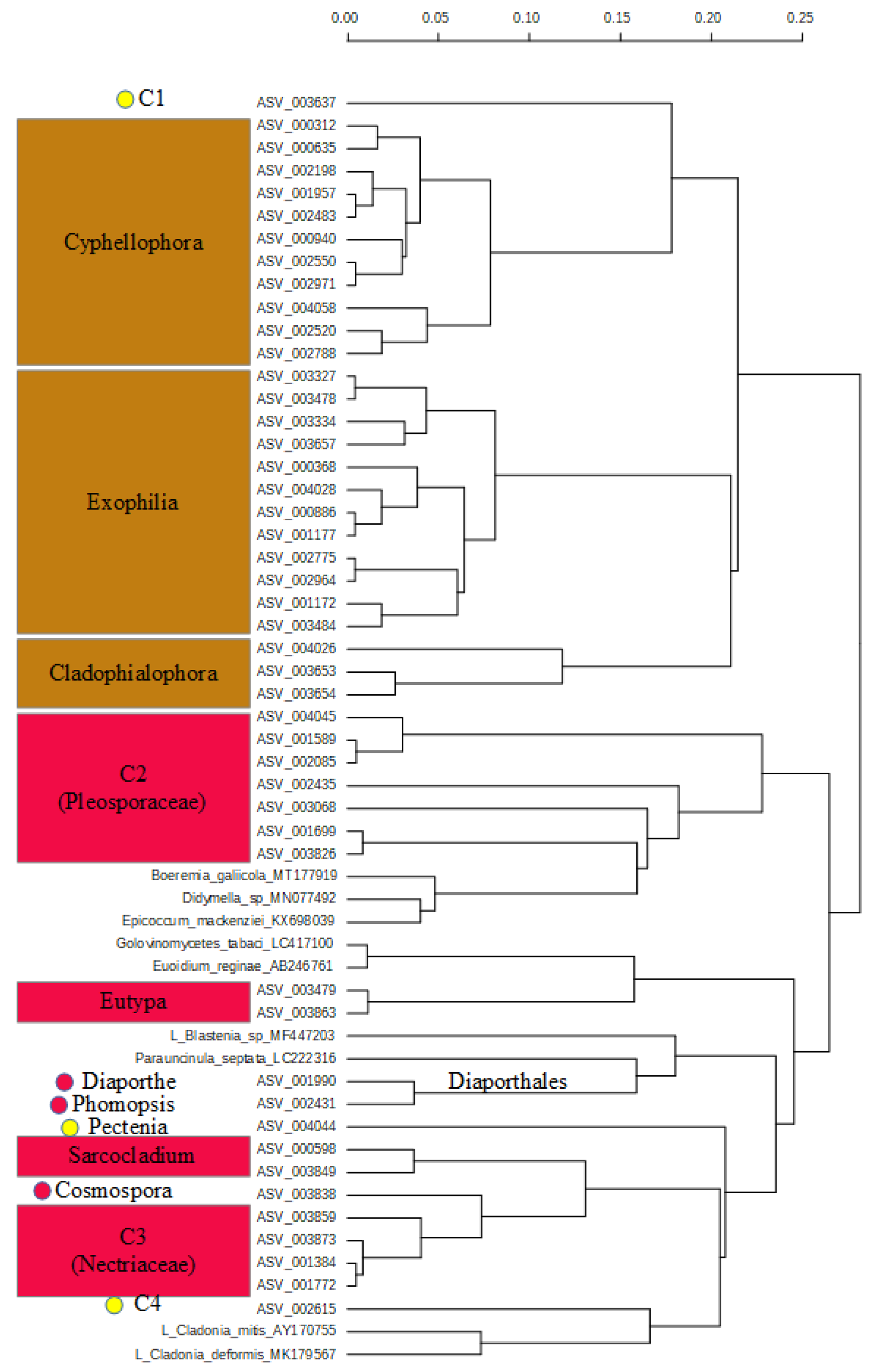
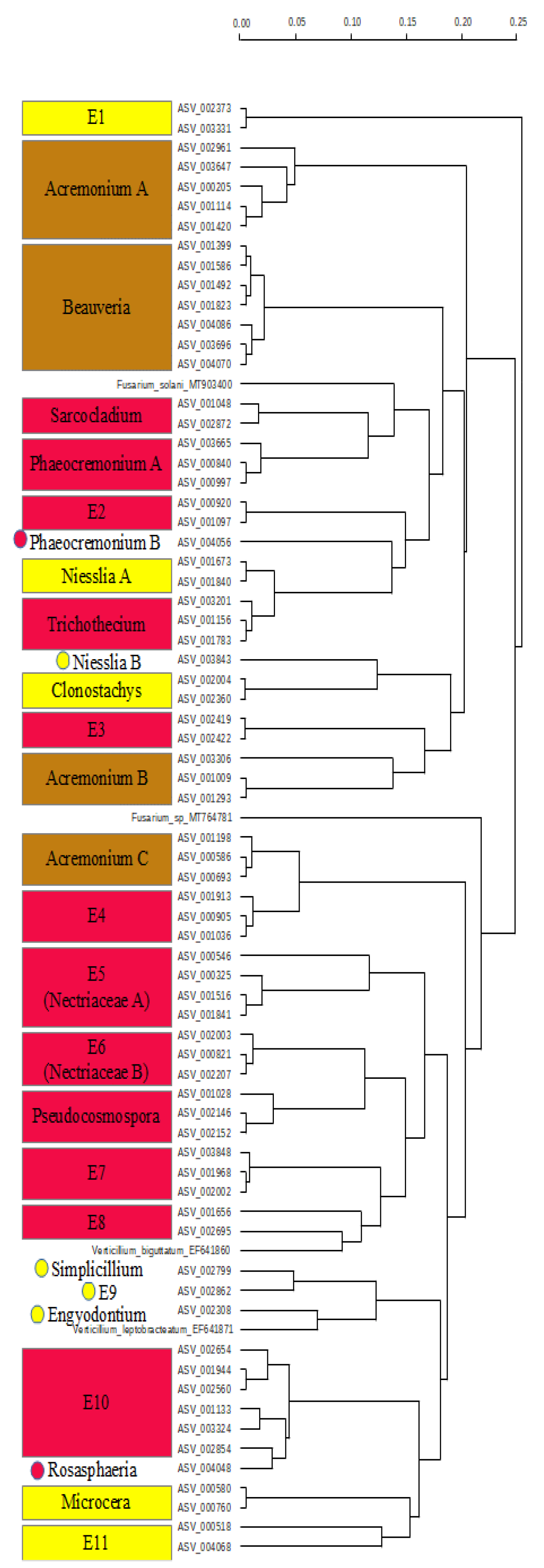
2.3. Diversity and Composition of Fungi on Fruit of Gardenia Thunbergia
3. Discussion
3.1. Overall Microeukaryote Communities
3.2. Pathogen Fungi on Gardenia thunbergia Fruit
3.3. Lichen on Gardenia thunbergia Fruit
4. Materials and Methods
4.1. Methods Overview
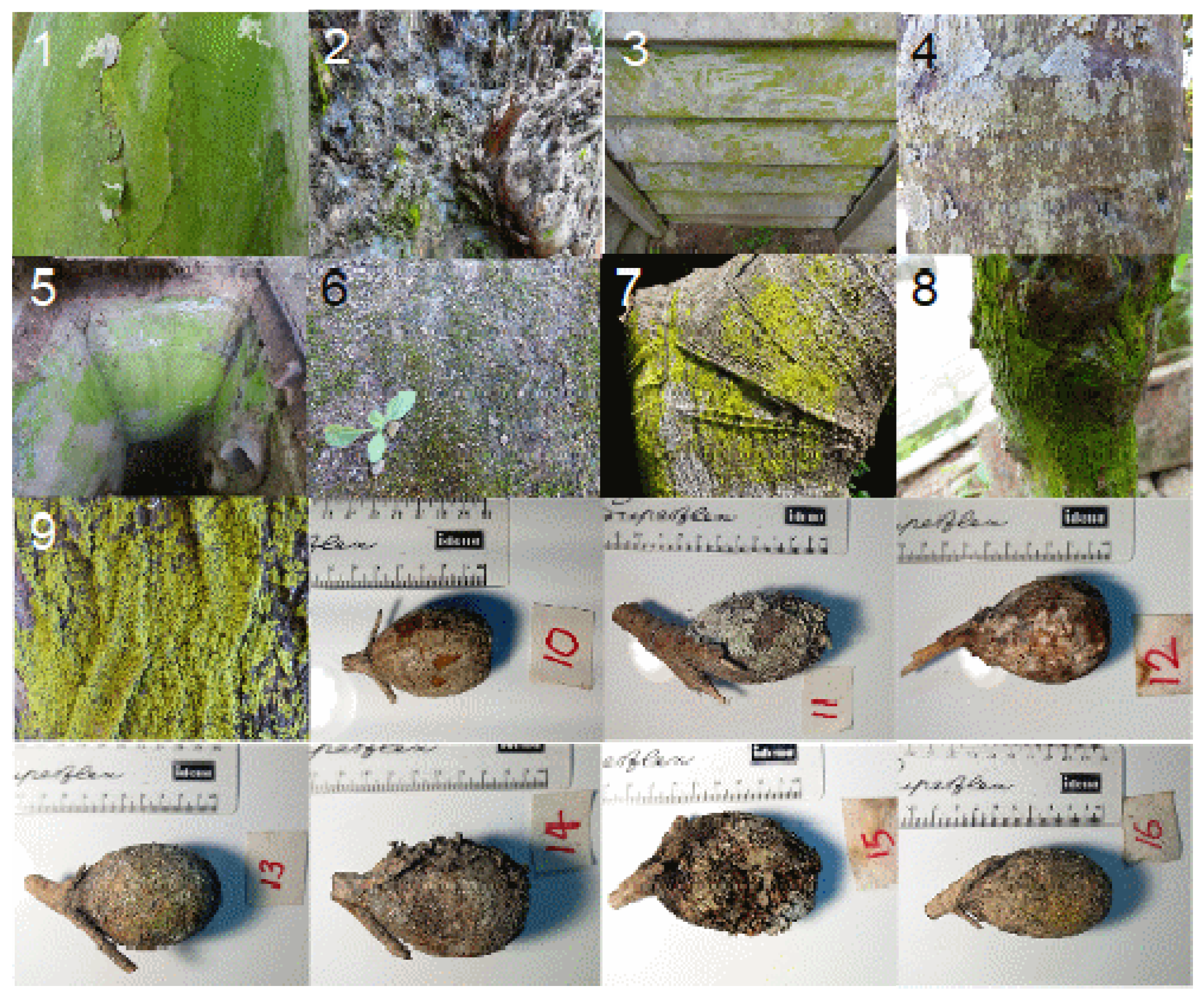
4.2. Study Region and Sample Collection
4.3. DNA Preparation and PCR
4.4. Data Processing and Selection of Sequences
4.5. Data Records
4.6. Statistics
4.7. Construction of Dendrograms
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement.
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Richardson, D.M.; Pysek, P.; Rejmanek, M.; Barbour, M.G.; Panetta, F.D.; West, C.J. Naturalization and invasion of alien plants: Concepts and definitions. Divers. Distrib. 2000, 6, 93–107. [Google Scholar] [CrossRef]
- Frenot, Y.; Chown, S.L.; Whinam, J.; Selkirk, P.M.; Convey, P.; Skotnicki, M.; Bergstrom, D.M. Biological invasions in the Antarctic: Extent, impacts and implications. Biol. Rev. 2005, 80, 45–72. [Google Scholar] [CrossRef]
- Bandyopadhyay, R.; Frederiksen, R.A. Contemporary global movement of emerging plant diseases. Ann. N. Y. Acad. Sci. 1999, 894, 28–36. [Google Scholar] [CrossRef] [PubMed]
- MacLeod, A.; Pautasso, M.; Jeger, M.J.; Haines-Young, R. Evolution of the international regulation of plant pests and challenges for fu-ture plant health. Food Secur. 2010, 2, 49–70. [Google Scholar] [CrossRef]
- Santini, A.; Ghelardini, L.; De Pace, C.; Desprez-Loustau, M.L.; Capretti, P.; Chandelier, A.; Cech, T.; Chira, D.; Diamandis, S.; Gaitniekis, T.; et al. Biogeographical patterns and determinants of invasion by forest pathogens in Europe. New Phytol. 2013, 197, 238–250. [Google Scholar] [CrossRef]
- O’Malley, M.A. ‘Everything is everywhere: But the environment selects’: Ubiquitous distribution and ecological determinism in microbial biogeography. Stud. Hist. Philos. Sci. Part. C Stud. Hist. Philos. Biol. Biomed. Sci. 2008, 39, 314–325. [Google Scholar] [CrossRef]
- Xu, X.; Wang, N.; Lipson, D.; Sinsabaugh, R.; Schimel, J.; He, L.; Soudzilovskaia, N.A.; Tedersoo, L. Microbial macroecology: In search of mechanisms governing microbial biogeographic patterns. Glob. Ecol. Biogeogr. 2020, 29, 1870–1886. [Google Scholar] [CrossRef]
- Miller, S.A.; Beed, F.D.; Harmon, C.L. Plant Disease Diagnostic Capabilities and Networks. Annu. Rev. Phytopathol. 2009, 47, 15–38. [Google Scholar] [CrossRef]
- Parnell, S.; Bosch, F.V.D.; Gottwald, T.; Gilligan, C.A. Surveillance to Inform Control of Emerging Plant Diseases: An Epidemiological Perspective. Annu. Rev. Phytopathol. 2017, 55, 591–610. [Google Scholar] [CrossRef]
- Rimbaud, L.; Dallot, S.; Bruchou, C.; Thoyer, S.; Jacquot, E.; Soubeyrand, S.; Thébaud, G. Improving Management Strategies of Plant Diseases Using Sequential Sensitivity Analyses. Phytopathology 2019, 109, 1184–1197. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.-H.; Su, J.-H.; Shang, J.-J.; Wu, Y.-Y.; Li, Y.; Bao, D.-P.; Yao, Y.-J. Evaluation of the ribosomal DNA internal transcribed spacer (ITS), specifically ITS1 and ITS2, for the analysis of fungal diversity by deep sequencing. PLoS ONE 2018, 13, e0206428. [Google Scholar] [CrossRef]
- Fan, S.; Miao, L.; Li, H.; Lin, A.; Song, F.; Zhang, P. Illumina-based analysis yields new insights into the diversity and composition of endophytic fungi in cultivated Huperzia serrata. PLoS ONE 2020, 15, e0242258. [Google Scholar] [CrossRef]
- Berbee, M.L. The phylogeny of plant and animal pathogens in the Ascomycota. Physiol. Mol. Plant. Pathol. 2001, 59, 165–187. [Google Scholar] [CrossRef]
- Van Der Does, H.C.; Rep, M. Virulence Genes and the Evolution of Host Specificity in Plant-Pathogenic Fungi. Mol. Plant.-Microbe Interact. 2007, 20, 1175–1182. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Kadooka, C.; Uchida, J.Y. Fusarium species as pathogen on orchids. Microbiol. Res. 2018, 207, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Sexton, A.C.; Howlett, B.J. Parallels in Fungal Pathogenesis on Plant and Animal Hosts. Eukaryot. Cell 2006, 5, 1941–1949. [Google Scholar] [CrossRef]
- Kim, W.G.; Lee, B.D.; Kim, W.S.; Cho, W.D. Root rot of moth orchid caused by Fusarium spp. Plant. Pathol. J. 2002, 18, 225–227. [Google Scholar] [CrossRef]
- Almanza-Álvarez, J.; Garibay-Orijel, R.; Salgado-Garciglia, R.; Fernández-Pavía, S.P.; Lappe-Oliveras, P.; Arellano-Torres, E.; Ávila-Díaz, I. Identification and control of pathogenic fungi in neotropical valued orchids (Laelia spp.). Trop. Plant. Pathol. 2017, 42, 339–351. [Google Scholar] [CrossRef]
- Sarsaiya, S.; Jia, Q.; Fan, X.; Jain, A.; Shu, F.; Chen, J.; Lu, Y.; Shi, J. First Report of Leaf Black Circular Spots on Dendrobium nobile Caused by Trichoderma longibrachiatum in Guizhou Province, China. Plant. Dis. 2019, 103, 3275. [Google Scholar] [CrossRef]
- Fernández-Herrera, E.; Rentería-Martínez, M.E.; Ramírez-Bustos, I.I.; Moreno-Salazar, S.F.; Ochoa-Meza, A.; Guillén-Sánchez, D. Colletotrichum karstii: Causal agent of anthracnose of Dendrobium nobile in Mexico. Can. J. Plant. Pathol. 2020, 42, 514–519. [Google Scholar] [CrossRef]
- Gebru, S.T.; Mammel, M.K.; Gangiredla, J.; Tournas, V.H.; Lampel, K.A.; Tartera, C. Draft Genome Sequences of 12 Isolates from 3 Fusarium Species Recovered from Moldy Peanuts. Microbiol. Resour. Announc. 2019, 8, e01642-18. [Google Scholar] [CrossRef]
- Liew, E.C.Y.; Laurence, M.H.; Pearce, C.A.; Shivas, R.G.; Johnson, G.I.; Tan, Y.; Edwards, J.; Perry, S.; Cooke, A.; Summerell, B.A. Review of Fusarium species isolated in association with mango malformation in Australia. Australas. Plant. Pathol. 2016, 45, 547–559. [Google Scholar] [CrossRef]
- Nhung, N.P.; Thu, P.Q.; Dell, B.; Chi, N.M. First report of canker disease in Dalbergia tonkinensis caused by Fusarium lateritium and Fusarium decemcellulare. Australas. Plant. Pathol. 2018, 47, 317–323. [Google Scholar] [CrossRef]
- Walsh, T.J.; Groll, A.; Hiemenz, J.; Fleming, R.; Roilides, E.; Anaissie, E. Infections due to emerging and uncommon medically im-portant fungal pathogens. Clin. Microbiol. Inf. 2004, 10, 48–66. [Google Scholar] [CrossRef] [PubMed]
- Guitard, J.; Degulys, A.; Buot, G.; Aline-Fardin, A.; Dannaoui, E.; Rio, B.; Marie, J.P.; Lapusan, S.; Hennequin, C. Acremonium scleroti-genum-Acremonium egyptiacum: A multi-resistant fungal pathogen complicating the course of aplastic anaemia. Clin. Microbiol. Inf. 2014, 20, O30–O32. [Google Scholar] [CrossRef]
- Jones, D.R.; Baker, R.H.A. Introductions of non-native plant pathogens into Great Britain, 1970–2004. Plant. Pathol. 2007, 56, 891–910. [Google Scholar] [CrossRef]
- Moralejo, E.; Pérez-Sierra, A.M.; Álvarez, L.A.; Belbahri, L.; Lefort, F.; Descals, E. Multiple alien Phytophthora taxa discovered on diseased ornamental plants in Spain. Plant. Pathol. 2009, 58, 100–110. [Google Scholar] [CrossRef]
- Jung, T.; Orlikowski, L.; Henricot, B.; Abad-Campos, P.; Aday, A.G.; Aguín Casal, O.; Bakonyi, J.; Cacciola, S.O.; Cech, T.; Chavar-riaga, D.; et al. Widespread Phytophthora infestations in European nurseries put forest, semi-natural and horticultural ecosystems at high risk of Phy-tophthora diseases. For. Pathol. 2016, 46, 134–163. [Google Scholar] [CrossRef]
- Palgrave, K.C. Trees of Southern Africa, 3rd ed.; Struik Nature: Cape Town, South Africa, 2002. [Google Scholar]
- Hilal, A.A. New diseases of ornamentals in Egypt: V: Cut flower plant: Gardenia. Egypt. J. Phytopathol. 2004, 32, 145–146. [Google Scholar]
- Ohashi, H.; Tsurushima, T.; Ueno, T.; Fukami, H. Cerbinal, a pseudoazulene iridoid, as a potent antifungal compound isolated from Gardenia jasminoides Ellis. Agric. Biol. Chem. 1986, 50, 2655–2657. [Google Scholar] [CrossRef]
- Kafua, L.; Kritzinger, Q.; Hussein, A. Antifungal activity of Gardenia brighamii leaf extracts. S. Afr. J. Bot. 2010, 76, 411. [Google Scholar] [CrossRef][Green Version]
- Hitokoto, H.; Morozumi, S.; Wauke, T.; Sakai, S.; Kurata, H. Fungal contamination and mycotoxin detection of powdered herbal drugs. Appl. Environ. Microbiol. 1978, 36, 252–256. [Google Scholar] [CrossRef] [PubMed]
- Feau, N.; Vialle, A.; Allaire, M.; Tanguay, P.; Joly, D.L.; Frey, P.; Callan, B.E.; Hamelin, R.C. Fungal pathogen (mis-) identifications: A case study with DNA barcodes on Melampsora rusts of aspen and white poplar. Mycol. Res. 2009, 113, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Toju, H.; Tanabe, A.S.; Yamamoto, S.; Sato, H. High-coverage ITS primers for the DNA-based identification of ascomycetes and basid-iomycetes in environmental samples. PLoS ONE 2012, 7, e40863. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M., Gelfand, D., Shinsky, J., White, T., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Schneider, D.; Thürmer, A.; Gollnow, K.; Lugert, R.; Gunka, K.; Groß, U.; Daniel, R. Gut bacterial communities of diarrheic patients with indications of Clostridioides difficile infection. Sci. Data 2017, 4, 170152. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ. 2016, 4, e2584. [Google Scholar] [CrossRef]
- Tsuji, S.; Miya, M.; Ushio, M.; Sato, H.; Minamoto, T.; Yamanaka, H. Evaluating intraspecific genetic diversity using environmental DNA and denoising approach: A case study using tank water. Environ. DNA 2019, 2, 42–52. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef] [PubMed]
- Tipping, M.E.; Bishop, C.M. Probabilistic Principal Component Analysis. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 1999, 61, 611–622. [Google Scholar] [CrossRef]
- Wilkinson, S.P. kmer: An R Package for Fast Alignment-Free Clustering of Biological Sequences. Version 1.1.2.. 2019. Available online: https://shaunpwilkinson.github.io/post/kmer-vignette/ (accessed on 6 January 2020).
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2020. [Google Scholar]
| Sequence ID | Family | Genus | Cluster | No. seq. | No. samp. | Age |
|---|---|---|---|---|---|---|
| ASV_004045 | Pleosporaceae | ? | C | 21 | 1 | y |
| ASV_001589 | Pleosporaceae | ? | C | 94 | 2 | y |
| ASV_002085 | Pleosporaceae | ? | C | 55 | 1 | y |
| ASV_002435 | Pleosporaceae | ? | C | 133 | 5 | all |
| ASV_003068 | Pleosporaceae | ? | C | 33 | 1 | o |
| ASV_001699 | Pleosporaceae | ? | C | 57 | 1 | y |
| ASV_003826 | Pleosporaceae | ? | C | 10 | 1 | y |
| ASV_003479 | Diatrypaceae | Eutypa | C | 31 | 1 | o |
| ASV_003863 | Diatrypaceae | Eutypa | C | 18 | 1 | o |
| ASV_001990 | ? | ? | C | 365 | 1 | m |
| ASV_002431 | ? | ? | C | 309 | 1 | m |
| ASV_000598 | incertae sedis | Sarcocladium | C | 737 | 3 | y/o |
| ASV_003849 | incertae sedis | Sarcocladium | C | 26 | 1 | y |
| ASV_003838 | Nectriaceae | Cosmospora | C | 2445 | 7 | all |
| ASV_003859 | Nectriaceae | ? | C | 160 | 1 | o |
| ASV_003873 | Nectriaceae | ? | C | 23 | 1 | o |
| ASV_001384 | Nectriaceae | ? | C | 176 | 1 | o |
| ASV_001772 | Nectriaceae | ? | C | 144 | 1 | o |
| ASV_001048 | incertae sedis | Sarcocladium | E | 435 | 1 | o |
| ASV_002872 | incertae sedis | Sarcocladium | E | 51 | 1 | o |
| ASV_003665 | Togniniaceae | Phaeocremonium | E | 66 | 1 | o |
| ASV_000840 | Togniniaceae | Phaeocremonium | E | 240 | 1 | o |
| ASV_000997 | Togniniaceae | Phaeocremonium | E | 168 | 1 | o |
| ASV_000920 | ? | ? | E | 184 | 1 | y |
| ASV_001097 | ? | ? | E | 148 | 1 | y |
| ASV_004056 | Togniniaceae | Phaeocremonium | E | 48 | 1 | m |
| ASV_003201 | incertae sedis | Trichothecium | E | 28 | 2 | y/m |
| ASV_001156 | incertae sedis | Trichothecium | E | 146 | 2 | y/m |
| ASV_001783 | incertae sedis | Trichothecium | E | 87 | 2 | y/m |
| ASV_002419 | ? | ? | E | 39 | 3 | y/o |
| ASV_002422 | ? | ? | E | 34 | 3 | y/o |
| ASV_001913 | ? | ? | E | 147 | 2 | y/o |
| ASV_000905 | ? | ? | E | 164 | 3 | all |
| ASV_001036 | ? | ? | E | 190 | 4 | y/o |
| ASV_000546 | Nectriaceae | ? | E | 662 | 1 | m |
| ASV_000325 | Nectriaceae | ? | E | 769 | 1 | o |
| ASV_001516 | Nectriaceae | ? | E | 111 | 2 | m/o |
| ASV_001841 | Nectriaceae | ? | E | 83 | 1 | o |
| ASV_002003 | Nectriaceae | ? | E | 56 | 2 | y/m |
| ASV_000821 | Nectriaceae | ? | E | 236 | 1 | m |
| ASV_002207 | Nectriaceae | ? | E | 37 | 1 | m |
| ASV_001028 | Nectriaceae | Pseudocosmophora | E | 512 | 1 | m |
| ASV_002146 | Nectriaceae | Pseudocosmophora | E | 461 | 1 | m |
| ASV_002152 | Nectriaceae | Pseudocosmophora | E | 189 | 1 | m |
| ASV_003848 | ? | ? | E | 17 | 2 | y/o |
| ASV_001968 | ? | ? | E | 60 | 1 | o |
| ASV_002002 | ? | ? | E | 55 | 1 | o |
| ASV_001656 | ? | ? | E | 88 | 2 | o |
| ASV_002695 | ? | ? | E | 56 | 2 | y/o |
| ASV_002654 | ? | ? | E | 178 | 4 | all |
| ASV_001944 | ? | ? | E | 414 | 4 | m/o |
| ASV_002560 | ? | ? | E | 128 | 4 | all |
| ASV_001133 | ? | ? | E | 386 | 3 | all |
| ASV_003324 | ? | ? | E | 469 | 5 | all |
| ASV_002854 | ? | ? | E | 273 | 2 | m/o |
| ASV_004048 | Niessliaceae | Rosasphaeria | E | 37 | 5 | all |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Steudel, B.; Baijnath, H.; Schwedt, T.; Schmitt, A.O. Microeukaryotic Communities on the Fruit of Gardenia thunbergia Thunb. with a Focus on Pathogenic Fungi. Pathogens 2021, 10, 555. https://doi.org/10.3390/pathogens10050555
Steudel B, Baijnath H, Schwedt T, Schmitt AO. Microeukaryotic Communities on the Fruit of Gardenia thunbergia Thunb. with a Focus on Pathogenic Fungi. Pathogens. 2021; 10(5):555. https://doi.org/10.3390/pathogens10050555
Chicago/Turabian StyleSteudel, Bastian, Himansu Baijnath, Thorben Schwedt, and Armin Otto Schmitt. 2021. "Microeukaryotic Communities on the Fruit of Gardenia thunbergia Thunb. with a Focus on Pathogenic Fungi" Pathogens 10, no. 5: 555. https://doi.org/10.3390/pathogens10050555
APA StyleSteudel, B., Baijnath, H., Schwedt, T., & Schmitt, A. O. (2021). Microeukaryotic Communities on the Fruit of Gardenia thunbergia Thunb. with a Focus on Pathogenic Fungi. Pathogens, 10(5), 555. https://doi.org/10.3390/pathogens10050555






