Haemophilus influenzae Meningitis Direct Diagnosis by Metagenomic Next-Generation Sequencing: A Case Report
Abstract
1. Introduction
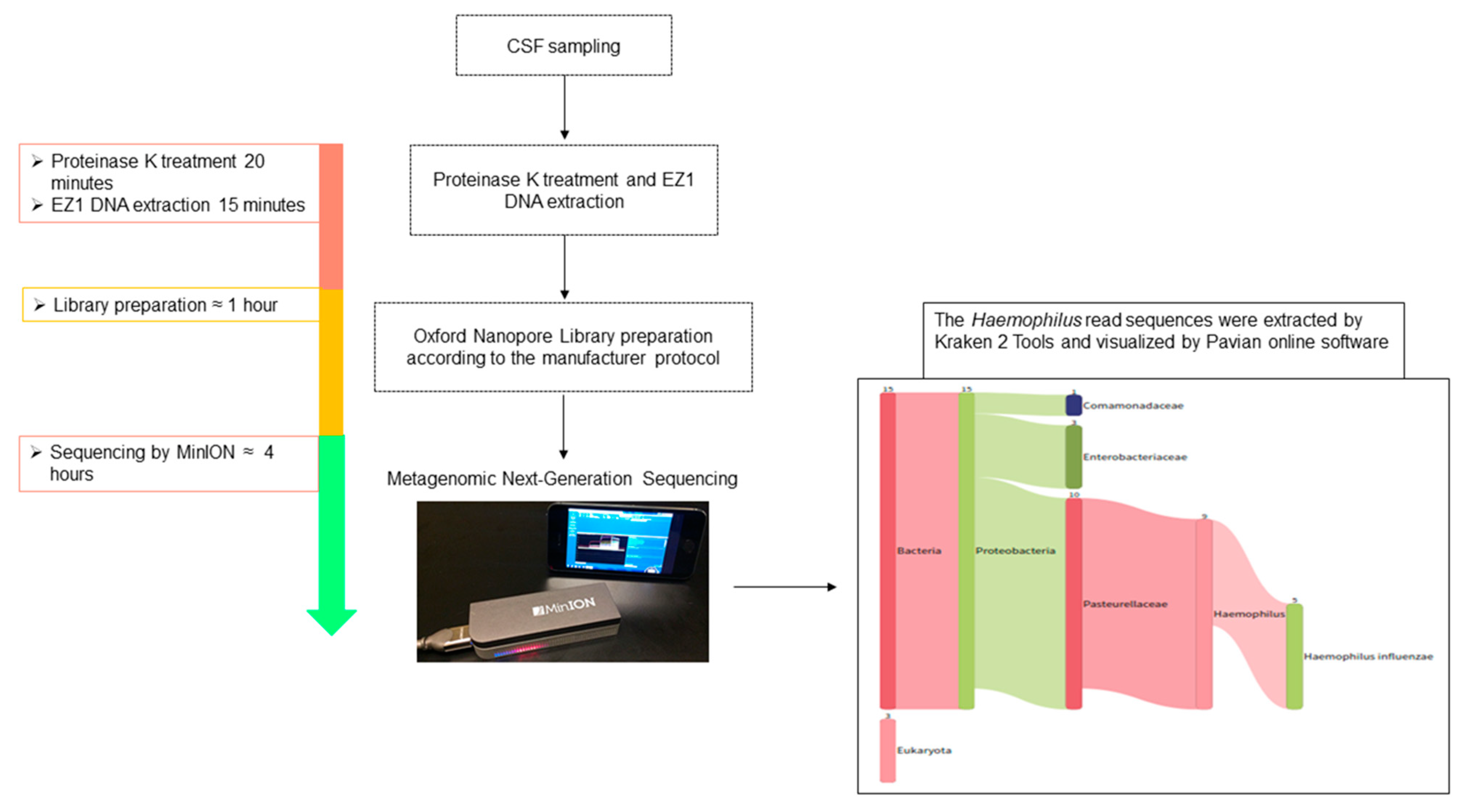
2. Case Report and Methods
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
List of Abbreviations
| dsDNA | double-strand desoxyribonucleic acid. |
| RT-PCR | real-time polymerase chain reaction. |
| POC | point-of-care. |
| CSF | cerebrospinal fluid. |
| mNGS | metagenomic next-generation sequencing. |
References
- Vincent, J.J.; Zandotti, C.; Baron, S.; Kandil, C.; Levy, P.Y.; Drancourt, M.; Raoult, D.; Ninove, L. Point-of-care multiplexed diagnosis of meningitis using the FilmArray® ME panel technology. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1573–1580. [Google Scholar] [CrossRef] [PubMed]
- Khumalo, J.; Nicol, M.; Hardie, D.; Muloiwa, R.; Mteshana, P.; Bamford, C. Diagnostic accuracy of two multiplex real-time polymerase chain reaction assays for the diagnosis of meningitis in children in a resource-limited setting. PLoS ONE 2017, 12, e0173948. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Hu, Z.; Sun, S.; Yao, F.; Yan, X.; Zhang, X.; Wu, B. Simultaneous detection and identification of bacteria and fungi in cerebrospinal fluid by TaqMan probe-based real-time PCR. Clin. Lab. 2014, 60, 1287–1293. [Google Scholar] [CrossRef] [PubMed]
- Fukasawa, L.O.; Gonçalves, M.G.; Higa, F.T.; Castilho, E.A.; Ibarz-Pavó, A.B.; Sacchi, C.T. Use of cerebrospinal fluid and serum samples impregnated on FTA TM Elute filter paper for the diagnosis of infections caused by Neisseria meningitidis, Streptococcus pneumoniae and Haemophilus influenzae. PLoS ONE 2017, 12, e0172794. [Google Scholar] [CrossRef] [PubMed]
- Paireau, J.; Chen, A.; Broutin, H.; Grenfell, B.; Basta, N.E.; Basta, N.E. Seasonal dynamics of bacterial meningitis: A time-series analysis. Lancet Glob. Health 2016, 4, e370–e377. [Google Scholar] [CrossRef]
- Hotomi, M.; Togawa, A.; Kono, M.; Sugita, G.; Sugita, R.; Fujimaki, Y.; Kamide, Y.; Uchizono, A.; Kanesada, K.; Sawada, S.; et al. An Application of Outer Membrane Protein P6-Specific Enzyme-Linked Immunosorbent Assay for Detection of Haemophilus influenzae in Middle Ear Fluids and Nasopharyngeal Secretions. PLoS ONE 2013, 8, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Moleres, J.; Fernández-Calvet, A.; Ehrlich, R.L.; Martí, S.; Pérez-Regidor, L.; Euba, B. Antagonistic pleiotropy in the bifunctional surface protein fadl (OmpP1) during adaptation of Haemophilus influenzae to chronic lung infection associated with chronic obstructive pulmonary disease. MBio 2018, 9, e01176-18. Available online: https://pubmed.ncbi.nlm.nih.gov/30254117/ (accessed on 5 April 2021). [CrossRef] [PubMed]
- Fleischmann, R.D.; Adams, M.D.; White, O.; Clayton, R.A.; Kirkness, E.F.; Kerlavage, A.R.; Bult, C.J.; Tomb, J.F.; Dougherty, B.A.; Merrick, J.M. Whole-genome random sequencing and assembly of Haemophilus influenzae. Science 1995, 269, 496–512. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morsli, M.; Kerharo, Q.; Delerce, J.; Roche, P.-H.; Troude, L.; Drancourt, M. Haemophilus influenzae Meningitis Direct Diagnosis by Metagenomic Next-Generation Sequencing: A Case Report. Pathogens 2021, 10, 461. https://doi.org/10.3390/pathogens10040461
Morsli M, Kerharo Q, Delerce J, Roche P-H, Troude L, Drancourt M. Haemophilus influenzae Meningitis Direct Diagnosis by Metagenomic Next-Generation Sequencing: A Case Report. Pathogens. 2021; 10(4):461. https://doi.org/10.3390/pathogens10040461
Chicago/Turabian StyleMorsli, Madjid, Quentin Kerharo, Jeremy Delerce, Pierre-Hugues Roche, Lucas Troude, and Michel Drancourt. 2021. "Haemophilus influenzae Meningitis Direct Diagnosis by Metagenomic Next-Generation Sequencing: A Case Report" Pathogens 10, no. 4: 461. https://doi.org/10.3390/pathogens10040461
APA StyleMorsli, M., Kerharo, Q., Delerce, J., Roche, P.-H., Troude, L., & Drancourt, M. (2021). Haemophilus influenzae Meningitis Direct Diagnosis by Metagenomic Next-Generation Sequencing: A Case Report. Pathogens, 10(4), 461. https://doi.org/10.3390/pathogens10040461





