Inactivation of Human Coronavirus by FATHHOME’s Dry Sanitizer Device: Rapid and Eco-Friendly Ozone-Based Disinfection of SARS-CoV-2
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Human Coronavirus
2.3. RNA Extraction and qRT-PCR
2.4. Virus Infectivity through Immunofluorescence Assay
2.5. FATHHOME Device
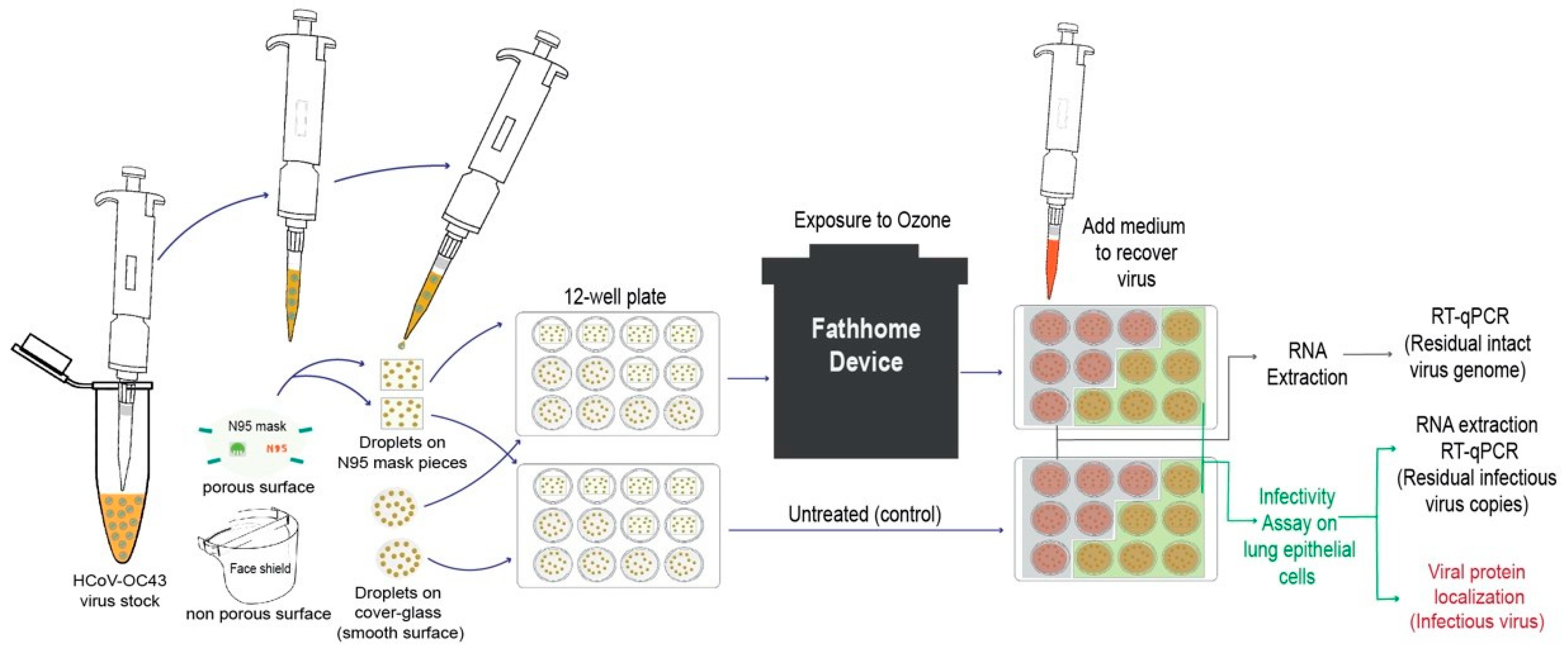
2.6. Experiment Set up
2.7. Statistical Analysis
3. Results
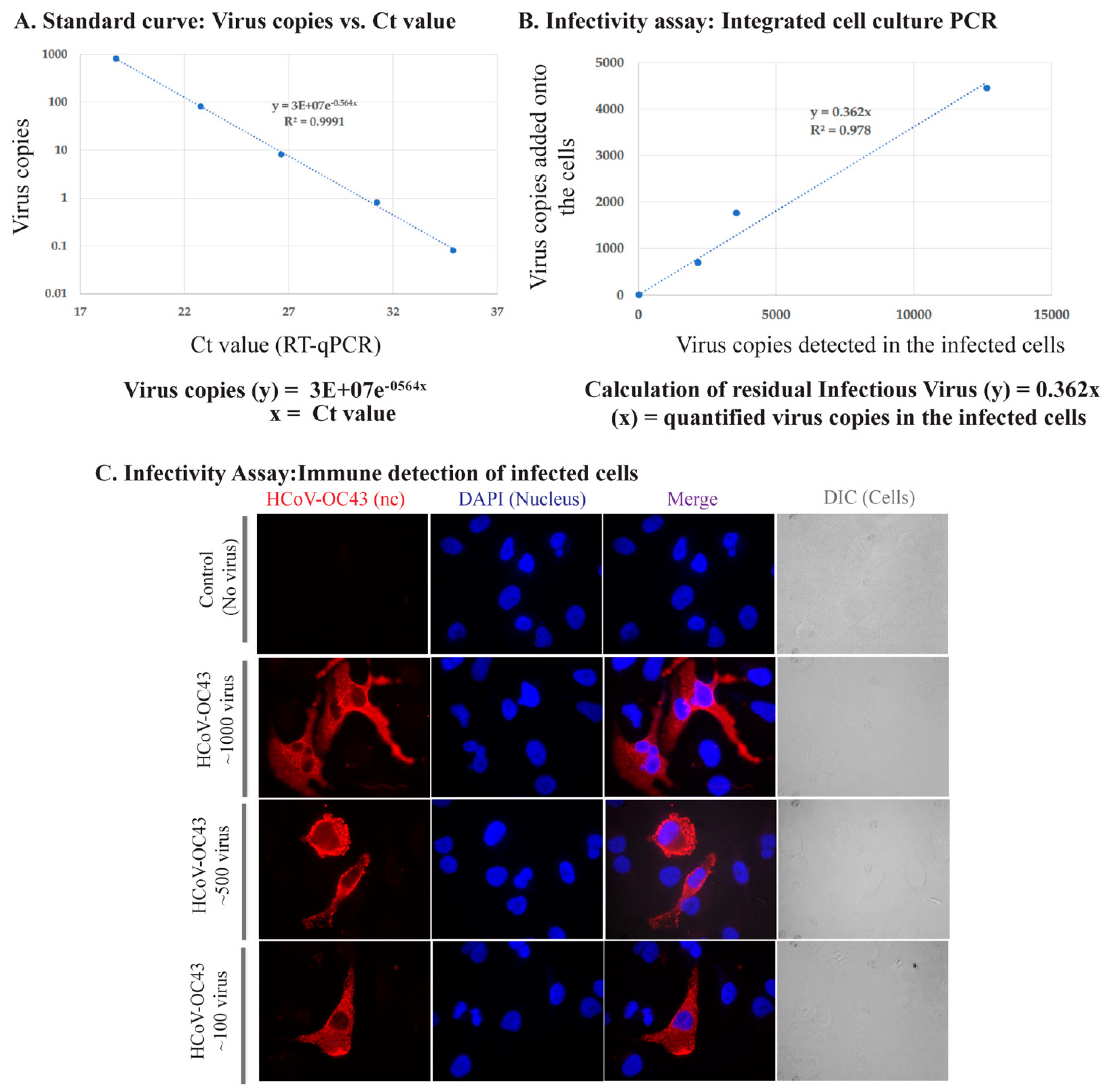
3.1. Assay Development to Quantify Infectious HCoV-OC43
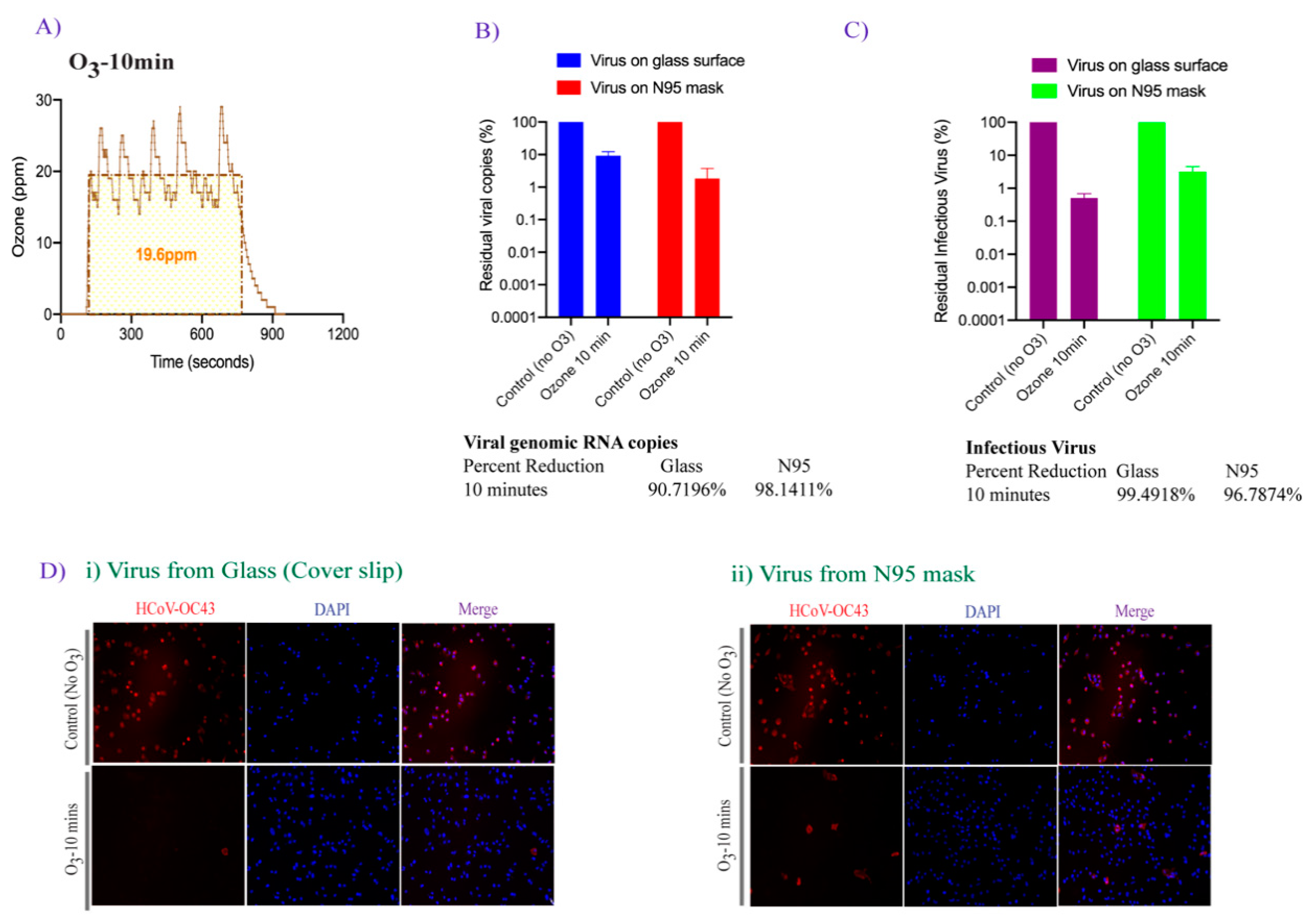
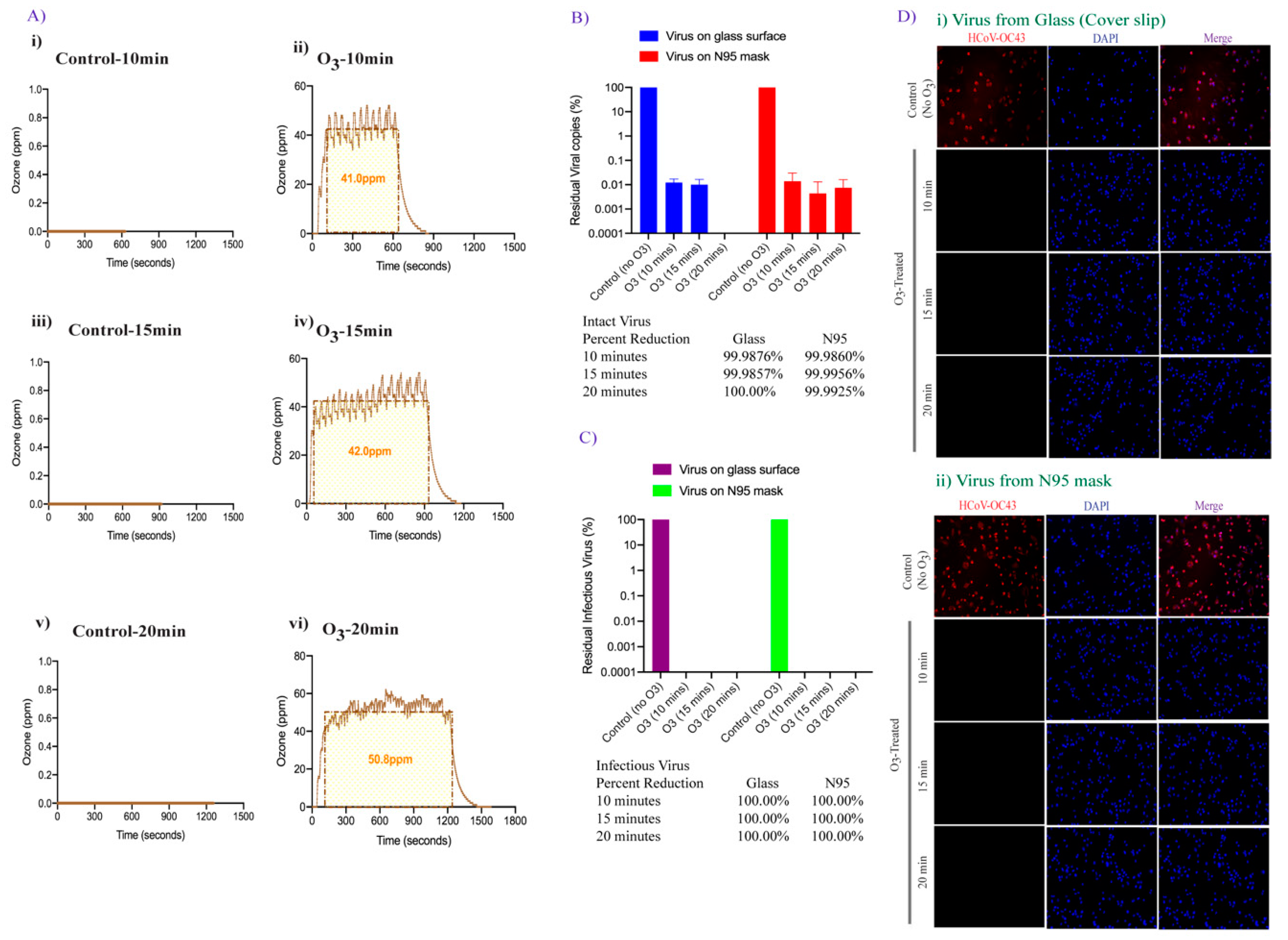
3.2. Effect of Ozone Exposure on HCoV-OC43 Virus Stability on Different Surfaces
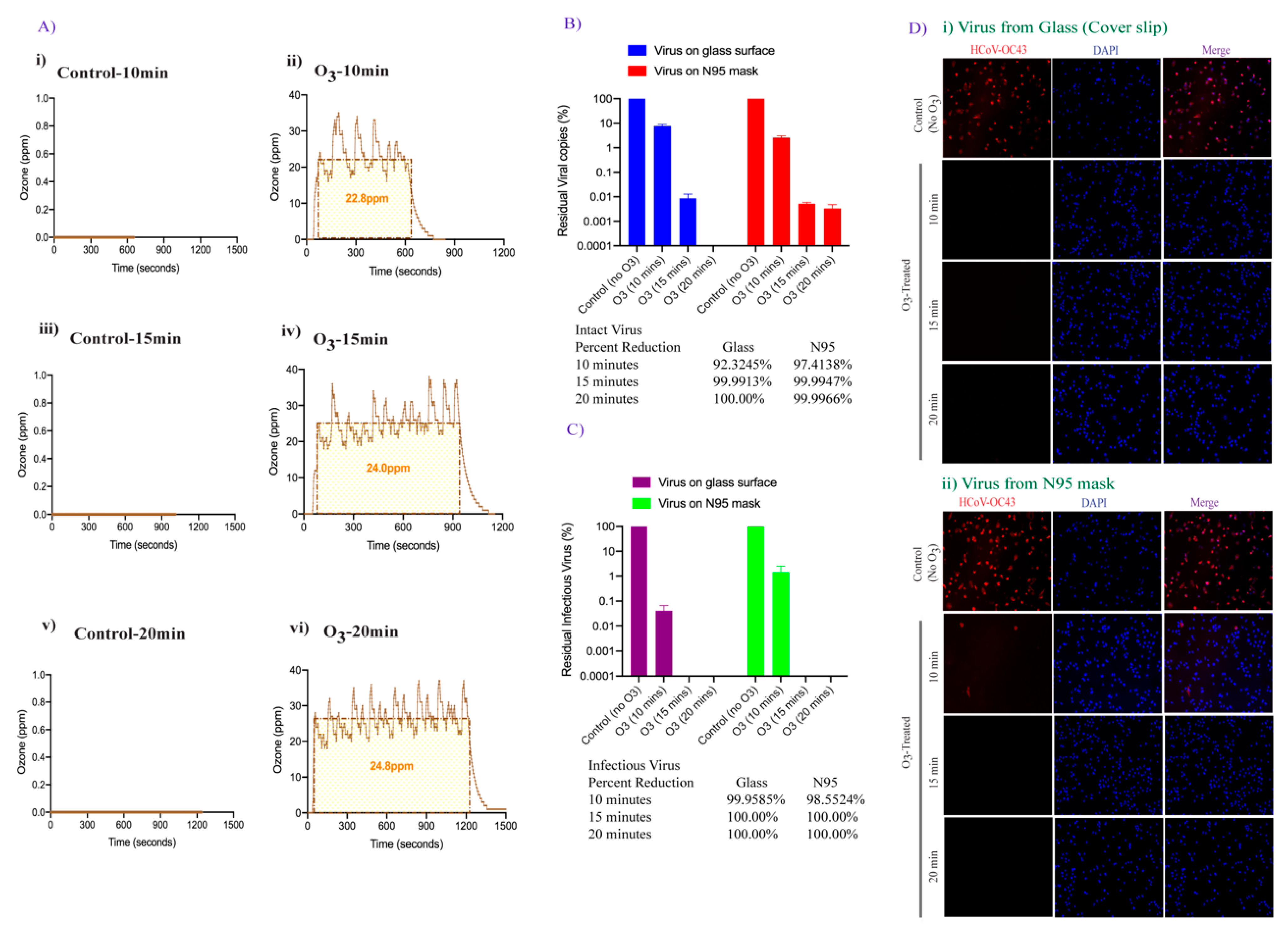
3.3. Effect of Increased Contact Time of Ozone Exposure on HCoV-OC43 Virus Inactivation
3.4. Doubling the Ozone Concentration Reduced Exposure Time to Achieve Faster Virus Inactivation
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ye, Z.-W.; Yuan, S.; Yuen, K.-S.; Fung, S.-Y.; Chan, C.-P.; Jin, D.-Y. Zoonotic Origins of Human Coronaviruses. Int. J. Biol. Sci. 2020, 16, 1686–1697. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef]
- Wei, W.E. Presymptomatic Transmission of SARS-CoV-2—Singapore, January 23 March 16, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 14. [Google Scholar] [CrossRef]
- Buitrago-Garcia, D.; Egli-Gany, D.; Counotte, M.J.; Hossmann, S.; Imeri, H.; Ipekci, A.M.; Salanti, G.; Low, N. Occurrence and Transmission Potential of Asymptomatic and Presymptomatic SARS-CoV-2 Infections: A Living Systematic Review and Meta-Analysis. PLOS Med. 2020, 17, e1003346. [Google Scholar] [CrossRef]
- COVID-19 Map. Available online: https://coronavirus.jhu.edu/map.html (accessed on 19 January 2021).
- Oran, D.P.; Topol, E.J. Prevalence of Asymptomatic SARS-CoV-2 Infection. Ann. Intern. Med. 2020, 173, 362–367. [Google Scholar] [CrossRef]
- Wiersinga, W.J.; Rhodes, A.; Cheng, A.C.; Peacock, S.J.; Prescott, H.C. Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19): A Review. JAMA 2020, 324, 782–793. [Google Scholar] [CrossRef]
- Gibson, P.G.; Qin, L.; Puah, S.H. COVID-19 Acute Respiratory Distress Syndrome (ARDS): Clinical Features and Differences from Typical Pre-COVID-19 ARDS. Med. J. Aust. 2020, 213, 54–56. [Google Scholar] [CrossRef] [PubMed]
- Archer Stephen, L.; Sharp Willard, W.; Weir, E. Kenneth Differentiating COVID-19 Pneumonia From Acute Respiratory Distress Syndrome and High Altitude Pulmonary Edema. Circulation 2020, 142, 101–104. [Google Scholar] [CrossRef] [PubMed]
- Badraoui, R.; Alrashedi, M.M.; El-May, M.V.; Bardakci, F. Acute Respiratory Distress Syndrome: A Life Threatening Associated Complication of SARS-CoV-2 Infection Inducing COVID-19. J. Biomol. Struct. Dyn. 2020, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Tojo, K.; Yamamoto, N.; Mihara, T.; Abe, M.; Goto, T. Characteristics of Changes in Circulating Markers of Alveolar Epithelial and Endothelial Injury in Acute Respiratory Distress Syndrome with COVID-19. MedRxiv 2021. [Google Scholar] [CrossRef]
- A Review of Current Interventions for COVID-19 Prevention-PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/32409144/ (accessed on 19 January 2021).
- Stability of SARS-CoV-2 in Different Environmental Conditions-PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/32835322/ (accessed on 19 January 2021).
- Kampf, G.; Todt, D.; Pfaender, S.; Steinmann, E. Persistence of Coronaviruses on Inanimate Surfaces and Their Inactivation with Biocidal Agents. J. Hosp. Infect. 2020, 104, 246–251. [Google Scholar] [CrossRef]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef]
- Tong, Y.; Bao, A.; Chen, H.; Huang, J.; Lv, Z.; Feng, L.; Cheng, Y.; Wang, Y.; Bai, L.; Rao, W.; et al. Necessity for Detection of SARS-CoV-2 RNA in Multiple Types of Specimens for the Discharge of the Patients with COVID-19. J. Transl. Med. 2020, 18, 411. [Google Scholar] [CrossRef]
- Chia, P.Y.; Coleman, K.K.; Tan, Y.K.; Ong, S.W.X.; Gum, M.; Lau, S.K.; Lim, X.F.; Lim, A.S.; Sutjipto, S.; Lee, P.H.; et al. Detection of Air and Surface Contamination by SARS-CoV-2 in Hospital Rooms of Infected Patients. Nat. Commun. 2020, 11, 2800. [Google Scholar] [CrossRef]
- Moore, G.; Rickard, H.; Stevenson, D.; Aranega-Bou, P.; Pitman, J.; Crook, A.; Davies, K.; Spencer, A.; Burton, C.; Easterbrook, L.; et al. Detection of SARS-CoV-2 within the Healthcare Environment: A Multi-Centre Study Conducted during the First Wave of the COVID-19 Outbreak in England. J. Hosp. Infect. 2021, 108, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Kampf, G. Potential Role of Inanimate Surfaces for the Spread of Coronaviruses and Their Inactivation with Disinfectant Agents. Infect. Prev. Pract. 2020, 2, 100044. [Google Scholar] [CrossRef]
- Kampf, G. Antiseptic Stewardship: Biocide Resistance and Clinical Implications; Springer International Publishing: Cham, Switzerland, 2018; ISBN 978-3-319-98784-2. [Google Scholar]
- Chen, H.; Wu, R.; Xing, Y.; Du, Q.; Xue, Z.; Xi, Y.; Yang, Y.; Deng, Y.; Han, Y.; Li, K.; et al. Influence of Different Inactivation Methods on Severe Acute Respiratory Syndrome Coronavirus 2 RNA Copy Number. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef]
- Grinshpun, S.A.; Yermakov, M.; Khodoun, M. Autoclave Sterilization and Ethanol Treatment of Re-Used Surgical Masks and N95 Respirators during COVID-19: Impact on Their Performance and Integrity. J. Hosp. Infect. 2020, 105, 608–614. [Google Scholar] [CrossRef]
- Patterson, E.I.; Prince, T.; Anderson, E.R.; Casas-Sanchez, A.; Smith, S.L.; Cansado-Utrilla, C.; Solomon, T.; Griffiths, M.J.; Acosta-Serrano, Á.; Turtle, L.; et al. Methods of Inactivation of SARS-CoV-2 for Downstream Biological Assays. J. Infect. Dis. 2020, 222, 1462–1467. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.T.; Lien, C.Z.; Liu, S.; Selvaraj, P. Effective Heat Inactivation of SARS-CoV-2. MedRxiv 2020. [Google Scholar] [CrossRef]
- Ratnesar-Shumate, S.; Williams, G.; Green, B.; Krause, M.; Holland, B.; Wood, S.; Bohannon, J.; Boydston, J.; Freeburger, D.; Hooper, I.; et al. Simulated Sunlight Rapidly Inactivates SARS-CoV-2 on Surfaces. J. Infect. Dis. 2020, 222, 214–222. [Google Scholar] [CrossRef]
- Kratzel, A.; Todt, D.; V’kovski, P.; Steiner, S.; Gultom, M.; Thao, T.T.N.; Ebert, N.; Holwerda, M.; Steinmann, J.; Niemeyer, D.; et al. Inactivation of Severe Acute Respiratory Syndrome Coronavirus 2 by WHO-Recommended Hand Rub Formulations and Alcohols. Emerg. Infect. Dis. 2020, 26, 1592–1595. [Google Scholar] [CrossRef] [PubMed]
- Meyers, C.; Kass, R.; Goldenberg, D.; Milici, J.; Alam, S.; Robison, R. Ethanol and Isopropanol Inactivation of Human Coronavirus on Hard Surfaces. J. Hosp. Infect. 2021, 107, 45–49. [Google Scholar] [CrossRef]
- Pascoe, M.J.; Robertson, A.; Crayford, A.; Durand, E.; Steer, J.; Castelli, A.; Wesgate, R.; Evans, S.L.; Porch, A.; Maillard, J.-Y. Dry Heat and Microwave-Generated Steam Protocols for the Rapid Decontamination of Respiratory Personal Protective Equipment in Response to COVID-19-Related Shortages. J. Hosp. Infect. 2020, 106, 10–19. [Google Scholar] [CrossRef]
- Inagaki, H.; Saito, A.; Sudaryatma, P.E.; Sugiyama, H.; Okabayashi, T.; Fujimoto, S. Rapid Inactivation of SARS-CoV-2 with Ozone Water. BioRxiv 2020. [Google Scholar] [CrossRef]
- Saini, V.; Sikri, K.; Batra, S.D.; Kalra, P.; Gautam, K. Development of a Highly Effective Low-Cost Vaporized Hydrogen Peroxide-Based Method for Disinfection of Personal Protective Equipment for Their Selective Reuse during Pandemics. Gut Pathog. 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Solar Ultraviolet Radiation Sensitivity of SARS-CoV-2–The Lancet Microbe. Available online: https://www.thelancet.com/journals/lanmic/article/PIIS2666-5247(20)30013-6/fulltext (accessed on 19 January 2021).
- Heilingloh, C.S.; Aufderhorst, U.W.; Schipper, L.; Dittmer, U.; Witzke, O.; Yang, D.; Zheng, X.; Sutter, K.; Trilling, M.; Alt, M.; et al. Susceptibility of SARS-CoV-2 to UV Irradiation. Am. J. Infect. Control 2020, 48, 1273–1275. [Google Scholar] [CrossRef]
- Viscusi, D.J.; Bergman, M.S.; Eimer, B.C.; Shaffer, R.E. Evaluation of Five Decontamination Methods for Filtering Facepiece Respirators. Ann. Occup. Hyg. 2009, 53, 815–827. [Google Scholar] [CrossRef] [PubMed]
- Siddharta, A.; Pfaender, S.; Vielle, N.J.; Dijkman, R.; Friesland, M.; Becker, B.; Yang, J.; Engelmann, M.; Todt, D.; Windisch, M.P.; et al. Virucidal Activity of World Health Organization–Recommended Formulations Against Enveloped Viruses, Including Zika, Ebola, and Emerging Coronaviruses. J. Infect. Dis. 2017, 215, 902–906. [Google Scholar] [CrossRef] [PubMed]
- Chou, R.; Dana, T.; Buckley, D.I.; Selph, S.; Fu, R.; Totten, A.M. Epidemiology of and Risk Factors for Coronavirus Infection in Health Care Workers: A Living Rapid Review. Ann. Intern. Med. 2020, 173, 120–136. [Google Scholar] [CrossRef]
- Phua, J.; Weng, L.; Ling, L.; Egi, M.; Lim, C.-M.; Divatia, J.V.; Shrestha, B.R.; Arabi, Y.M.; Ng, J.; Gomersall, C.D.; et al. Intensive Care Management of Coronavirus Disease 2019 (COVID-19): Challenges and Recommendations. Lancet Respir. Med. 2020, 8, 506–517. [Google Scholar] [CrossRef]
- de Perio, M.A.; Dowell, C.H.; Delaney, L.J.; Radonovich, L.J.; Kuhar, D.T.; Gupta, N.; Patel, A.; Pillai, S.K.; D’Alessandro, M. Strategies for Optimizing the Supply of N95 Filtering Facepiece Respirators During the Coronavirus Disease 2019 (COVID-19) Pandemic. Disaster Med. Public Health Prep. 2020, 14, 658–669. [Google Scholar] [CrossRef] [PubMed]
- Strategies for Optimizing the Supply of N95 Respirators: COVID-19 | CDC. Available online: https://www.cdc.gov/coronavirus/2019-ncov/hcp/respirators-strategy/index.html (accessed on 31 January 2021).
- Mills, D.; Harnish, D.A.; Lawrence, C.; Sandoval-Powers, M.; Heimbuch, B.K. Ultraviolet Germicidal Irradiation of Influenza-Contaminated N95 Filtering Facepiece Respirators. Am. J. Infect. Control 2018, 46, e49–e55. [Google Scholar] [CrossRef] [PubMed]
- Heimbuch, B.K.; Wallace, W.H.; Kinney, K.; Lumley, A.E.; Wu, C.-Y.; Woo, M.-H.; Wander, J.D. A Pandemic Influenza Preparedness Study: Use of Energetic Methods to Decontaminate Filtering Facepiece Respirators Contaminated with H1N1 Aerosols and Droplets. Am. J. Infect. Control 2011, 39, e1–e9. [Google Scholar] [CrossRef]
- Lore, M.B.; Heimbuch, B.K.; Brown, T.L.; Wander, J.D.; Hinrichs, S.H. Effectiveness of Three Decontamination Treatments against Influenza Virus Applied to Filtering Facepiece Respirators. Ann. Occup. Hyg. 2012, 56, 92–101. [Google Scholar] [CrossRef]
- Bergman, M.S.; Viscusi, D.J.; Heimbuch, B.K.; Wander, J.D.; Sambol, A.R.; Ronald, S.; Shaffer, R.E. Evaluation of Multiple (3-Cycle) Decontamination Processing for Filtering Facepiece Respirators. J. Eng. Fibers Fabr. 2010, 5. Available online: https://journals.sagepub.com/doi/abs/10.1177/155892501000500405 (accessed on 19 January 2021). [CrossRef]
- Seresirikachorn, K.; Phoophiboon, V.; Chobarporn, T.; Tiankanon, K.; Aeumjaturapat, S.; Chusakul, S.; Snidvongs, K. Decontamination and Reuse of Surgical Masks and N95 Filtering Facepiece Respirators during the COVID-19 Pandemic: A Systematic Review. Infect. Control Hosp. Epidemiol. 2021, 42, 25–30. [Google Scholar] [CrossRef]
- Hydrogen Peroxide Vapor Sterilization of N95 Respirators for Reuse. MedRxiv 2020. Available online: https://www.medrxiv.org/content/10.1101/2020.03.24.20041087v1 (accessed on 19 January 2021).
- FDA, Commissioner, O. of the Investigating Decontamination and Reuse of Respirators in Public Health Emergencies. Available online: https://www.fda.gov/emergency-preparedness-and-response/mcm-regulatory-science/investigating-decontamination-and-reuse-respirators-public-health-emergencies (accessed on 19 January 2021).
- Dennis, R.; Pourdeyhimi, B.; Cashion, A.; Emanuel, S.; Hubbard, D. Durability of Disposable N95 Mask Material When Exposed to Improvised Ozone Gas Disinfection. J. Sci. Med. 2020, 2. [Google Scholar] [CrossRef]
- Ran, L.; Chen, X.; Wang, Y.; Wu, W.; Zhang, L.; Tan, X. Risk Factors of Healthcare Workers with Coronavirus Disease 2019: A Retrospective Cohort Study in a Designated Hospital of Wuhan in China. Clin. Infect. Dis. 2020, 71, 2218–2221. [Google Scholar] [CrossRef]
- Zhu, Z.; Lian, X.; Su, X.; Wu, W.; Marraro, G.A.; Zeng, Y. From SARS and MERS to COVID-19: A Brief Summary and Comparison of Severe Acute Respiratory Infections Caused by Three Highly Pathogenic Human Coronaviruses. Respir. Res. 2020, 21, 224. [Google Scholar] [CrossRef]
- Human Coronavirus OC43—An Overview. ScienceDirect. Available online: https://www.sciencedirect.com/topics/veterinary-science-and-veterinary-medicine/human-coronavirus-oc43 (accessed on 19 January 2021).
- Khazaieli, A.; Rowe, A. Vacuum-Based Method and Apparatus for Cleaning Soiled Articles. US 10,772,477 B2, 15 September 2020. [Google Scholar]
- Llanes, A.; Restrepo, C.M.; Caballero, Z.; Rajeev, S.; Kennedy, M.A.; Lleonart, R. Betacoronavirus Genomes: How Genomic Information Has Been Used to Deal with Past Outbreaks and the COVID-19 Pandemic. Int. J. Mol. Sci. 2020, 21, 4546. [Google Scholar] [CrossRef] [PubMed]
- Zamorano, C.N.; Grandvaux, N. ACE2: Evidence of Role as Entry Receptor for SARS-CoV-2 and Implications in Comorbidities. Elife 2020, 9, e61390. [Google Scholar] [CrossRef] [PubMed]
- Wijdicks, E.F.M. Historical Lessons from Twentieth-Century Pandemics Due to Respiratory Viruses. Neurocrit. Care 2020, 33, 591–596. [Google Scholar] [CrossRef]
- Morens, D.M.; Daszak, P.; Taubenberger, J.K. Escaping Pandora’s Box—Another Novel Coronavirus. N. Engl. J. Med. 2020, 382, 1293–1295. [Google Scholar] [CrossRef]
- Transmission of SARS-CoV-2: Implications for Infection Prevention Precautions. Available online: https://www.who.int/news-room/commentaries/detail/transmission-of-sars-cov-2-implications-for-infection-prevention-precautions (accessed on 19 January 2021).
- Elvis, A.M.; Ekta, J.S. Ozone Therapy: A Clinical Review. J. Nat. Sci. Biol. Med. 2011, 2, 66–70. [Google Scholar] [CrossRef]
- Photodissociation–an Overview. ScienceDirect. Available online: https://www.sciencedirect.com/topics/earth-and-planetary-sciences/photodissociation (accessed on 19 January 2021).
- Kim, C.K.; Gentile, D.M.; Sproul, O.J. Mechanism of Ozone Inactivation of Bacteriophage F2. Appl. Environ. Microbiol. 1980, 39, 210–218. [Google Scholar] [CrossRef]
- MacNair, S.D.B.; Lesher, E.C. Proceedings of the Pathological Society of Great Britain and Ireland. J. Pathol. Bacteriol. 1963, 85, 567–568. [Google Scholar] [CrossRef]
- Roy, D.; Wong, P.K.; Engelbrecht, R.S.; Chian, E.S. Mechanism of Enteroviral Inactivation by Ozone. Appl. Environ. Microbiol. 1981, 41, 718–723. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Wananabe, Y.; Miyata, H. Virucidal Effect of Ozone Treatment of Laboratory Animal Viruses. Exp. Anim. 1990, 39, 223–229. [Google Scholar] [CrossRef]
- Tseng, C.; Li, C. Inactivation of surface viruses by gaseous ozone. J. Environ. Health 2008, 70, 56–62. Available online: https://pubmed.ncbi.nlm.nih.gov/18561570/ (accessed on 19 January 2021).
- Hudson, J.B.; Sharma, M.; Petric, M. Inactivation of Norovirus by Ozone Gas in Conditions Relevant to Healthcare. J. Hosp. Infect. 2007, 66, 40–45. [Google Scholar] [CrossRef]
- Dubuis, M.-E.; Dumont-Leblond, N.; Laliberté, C.; Veillette, M.; Turgeon, N.; Jean, J.; Duchaine, C. Ozone Efficacy for the Control of Airborne Viruses: Bacteriophage and Norovirus Models. PLoS ONE 2020, 15, e0231164. [Google Scholar] [CrossRef]
- Manjunath, S.N.; Sakar, M.; Katapadi, M.; Balakrishna, R.G. Recent case studies on the use of ozone to combat coronavirus: Problems and perspectives. Environ.Technol. & Innov. 2021, 21, 101313. [Google Scholar]
- Jia-min, Z.; Chong-yi, Z.; Geng-fu, X.; Yuan-quan, Z.; Rong, G. Examination of the efficacy of ozone solution disinfectant in in activating SARS virus. Chin. J. Disinfect. 2004, 01. Available online: https://en.cnki.com.cn/Article_en/CJFDTotal-ZGXD200401010.htm (accessed on 19 January 2021).
- Fontes, B.; Cattani Heimbecker, A.M.; de Souza Brito, G.; Costa, S.F.; van der Heijden, I.M.; Levin, A.S.; Rasslan, S. Effect of Low-Dose Gaseous Ozone on Pathogenic Bacteria. BMC Infect. Dis. 2012, 12, 358. [Google Scholar] [CrossRef]
- Ozone: A Potential Oxidant for COVID-19 Virus (SARS-CoV-2). Available online: https://www.tandfonline.com/doi/full/10.1080/01919512.2020.1795614 (accessed on 19 January 2021).
- Virion Disruption by Ozone-Mediated Reactive Oxygen Species-PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/18598719/ (accessed on 19 January 2021).
- Environmental and Decontamination Issues for Human Coronaviruses and Their Potential Surrogates. J. Med. Virol. 2020, 92, 2498–2510. Available online: https://onlinelibrary.wiley.com/doi/10.1002/jmv.26170 (accessed on 19 January 2021). [CrossRef]
- Full Article: Development of a Practical Method for Using Ozone Gas as a Virus Decontaminating Agent. Available online: https://www.tandfonline.com/doi/full/10.1080/01919510902747969 (accessed on 19 January 2021).
- Beaudry, M.S.; Frederick, J.C.; Lott, M.E.J.; Norfolk, W.A.; Glenn, T.C.; Lipp, E.K. Effectiveness of an Ozone Disinfecting and Sanitizing Cabinet to Decontaminate a Surrogate Virus for SARS-CoV-2 on N-95 Masks. MedRxiv 2020. [Google Scholar] [CrossRef]
- Blanchard, E.L.; Lawrence, J.D.; Noble, J.A.; Xu, M.; Joo, T.; Ng, N.L.; Schmidt, B.E.; Santangelo, P.J.; Finn, M.G. Enveloped Virus Inactivation on Personal Protective Equipment by Exposure to Ozone; Infectious Diseases (except HIV/AIDS). MedRxiv 2020. [Google Scholar] [CrossRef]
- Disinfection of N95 Respirators with Ozone. MedRxiv. Available online: https://www.medrxiv.org/content/10.1101/2020.05.28.20097402v1 (accessed on 28 February 2021).
- FDA, Health, C. for D. and R. Decontamination Systems for Personal Protective Equipment EUAs. Available online: https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/decontamination-systems-personal-protective-equipment-euas (accessed on 28 February 2021).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uppal, T.; Khazaieli, A.; Snijders, A.M.; Verma, S.C. Inactivation of Human Coronavirus by FATHHOME’s Dry Sanitizer Device: Rapid and Eco-Friendly Ozone-Based Disinfection of SARS-CoV-2. Pathogens 2021, 10, 339. https://doi.org/10.3390/pathogens10030339
Uppal T, Khazaieli A, Snijders AM, Verma SC. Inactivation of Human Coronavirus by FATHHOME’s Dry Sanitizer Device: Rapid and Eco-Friendly Ozone-Based Disinfection of SARS-CoV-2. Pathogens. 2021; 10(3):339. https://doi.org/10.3390/pathogens10030339
Chicago/Turabian StyleUppal, Timsy, Amir Khazaieli, Antoine M. Snijders, and Subhash C. Verma. 2021. "Inactivation of Human Coronavirus by FATHHOME’s Dry Sanitizer Device: Rapid and Eco-Friendly Ozone-Based Disinfection of SARS-CoV-2" Pathogens 10, no. 3: 339. https://doi.org/10.3390/pathogens10030339
APA StyleUppal, T., Khazaieli, A., Snijders, A. M., & Verma, S. C. (2021). Inactivation of Human Coronavirus by FATHHOME’s Dry Sanitizer Device: Rapid and Eco-Friendly Ozone-Based Disinfection of SARS-CoV-2. Pathogens, 10(3), 339. https://doi.org/10.3390/pathogens10030339






