Yersiniosis in New Zealand
Abstract
1. Introduction
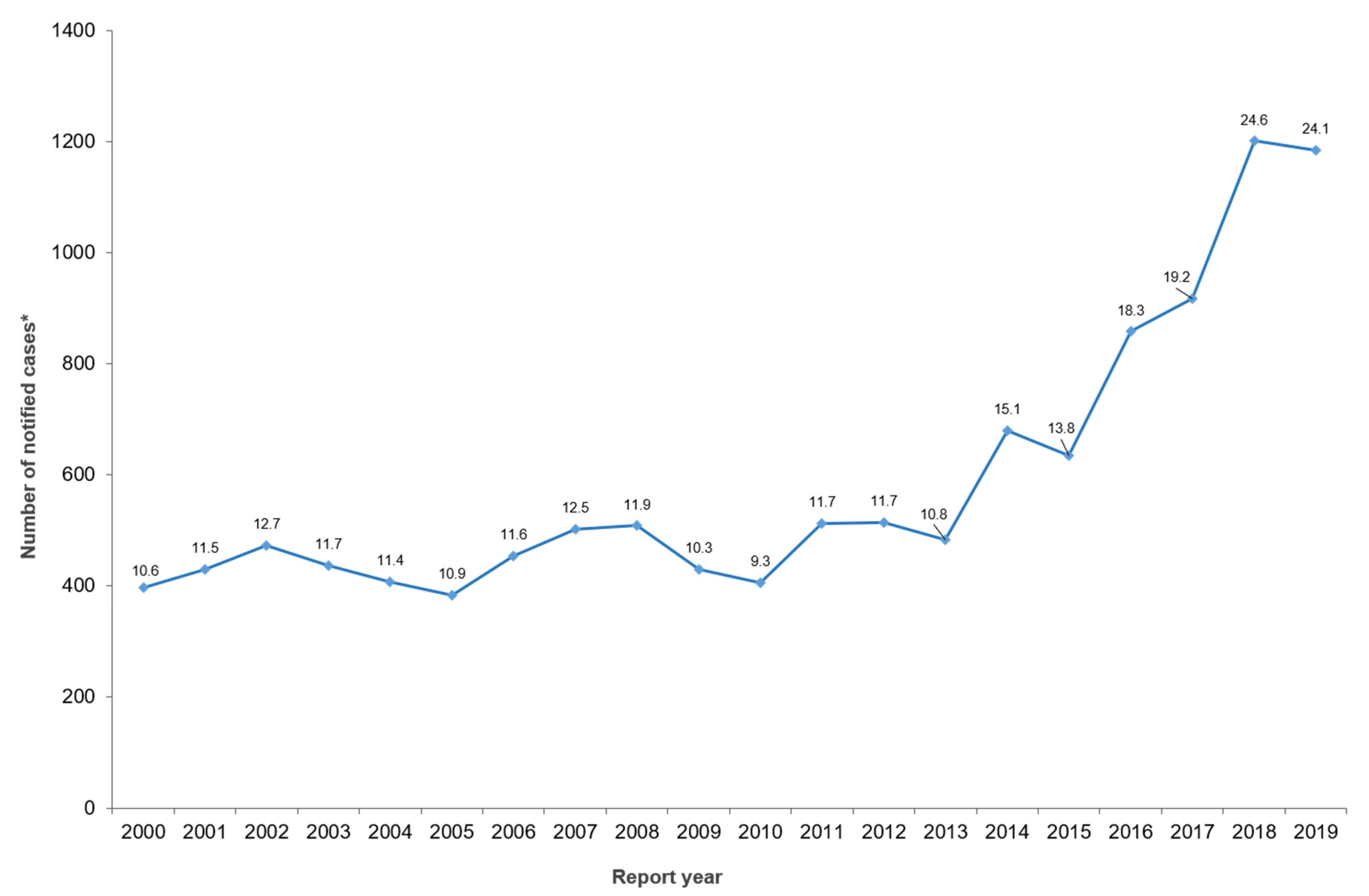
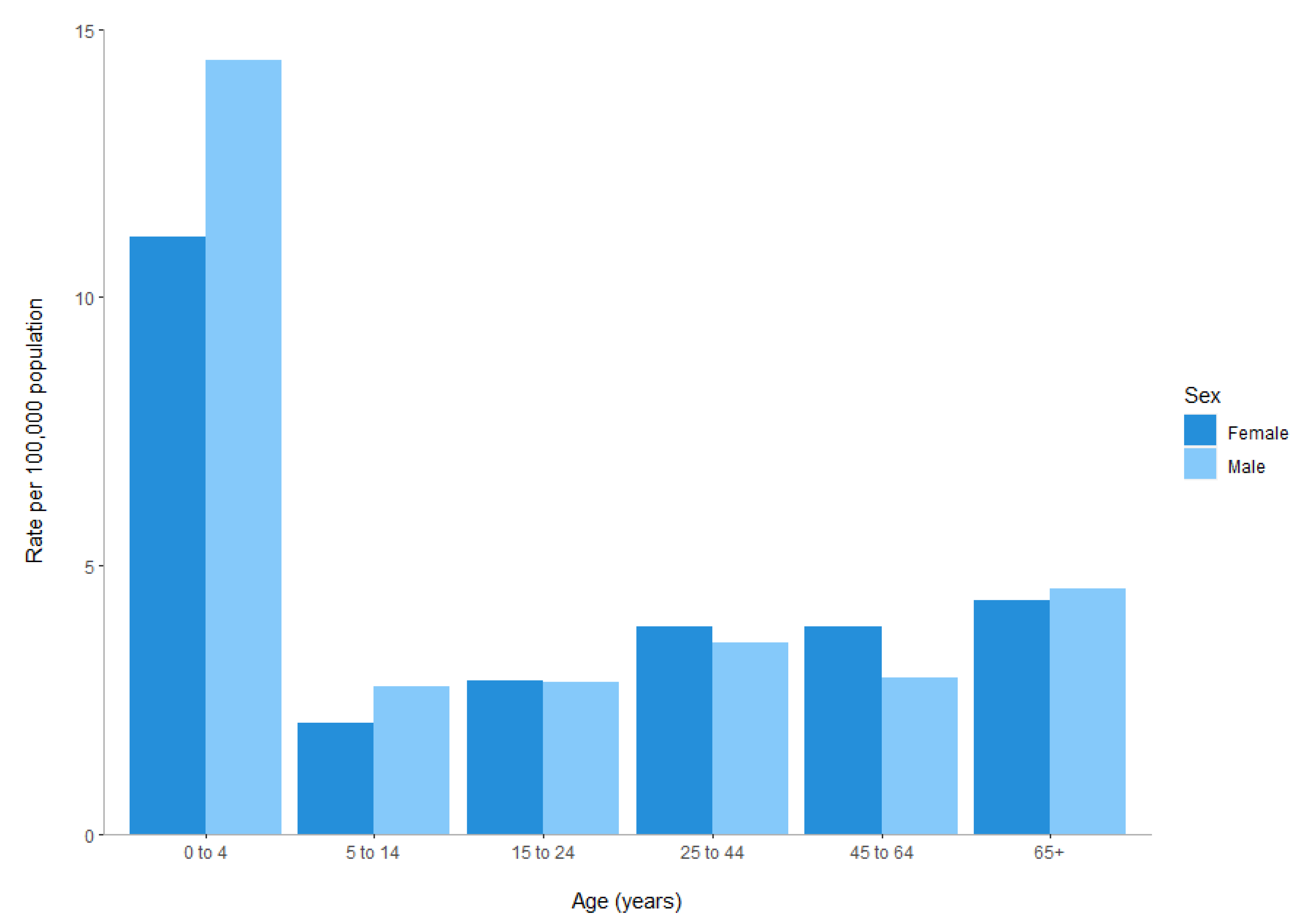
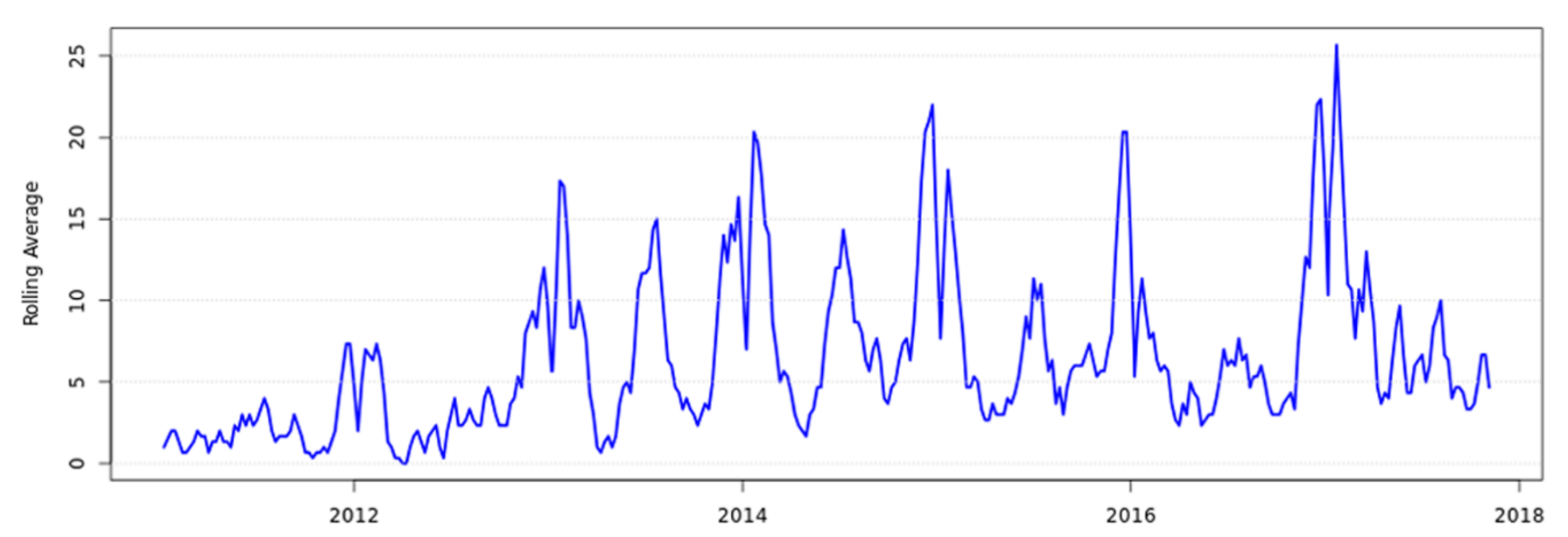
2. Human Clinical Yersiniosis Is Increasing in New Zealand
3. Has the Introduction of Culture-Independent Diagnostics Testing Influenced the Increase in Notified Cases?
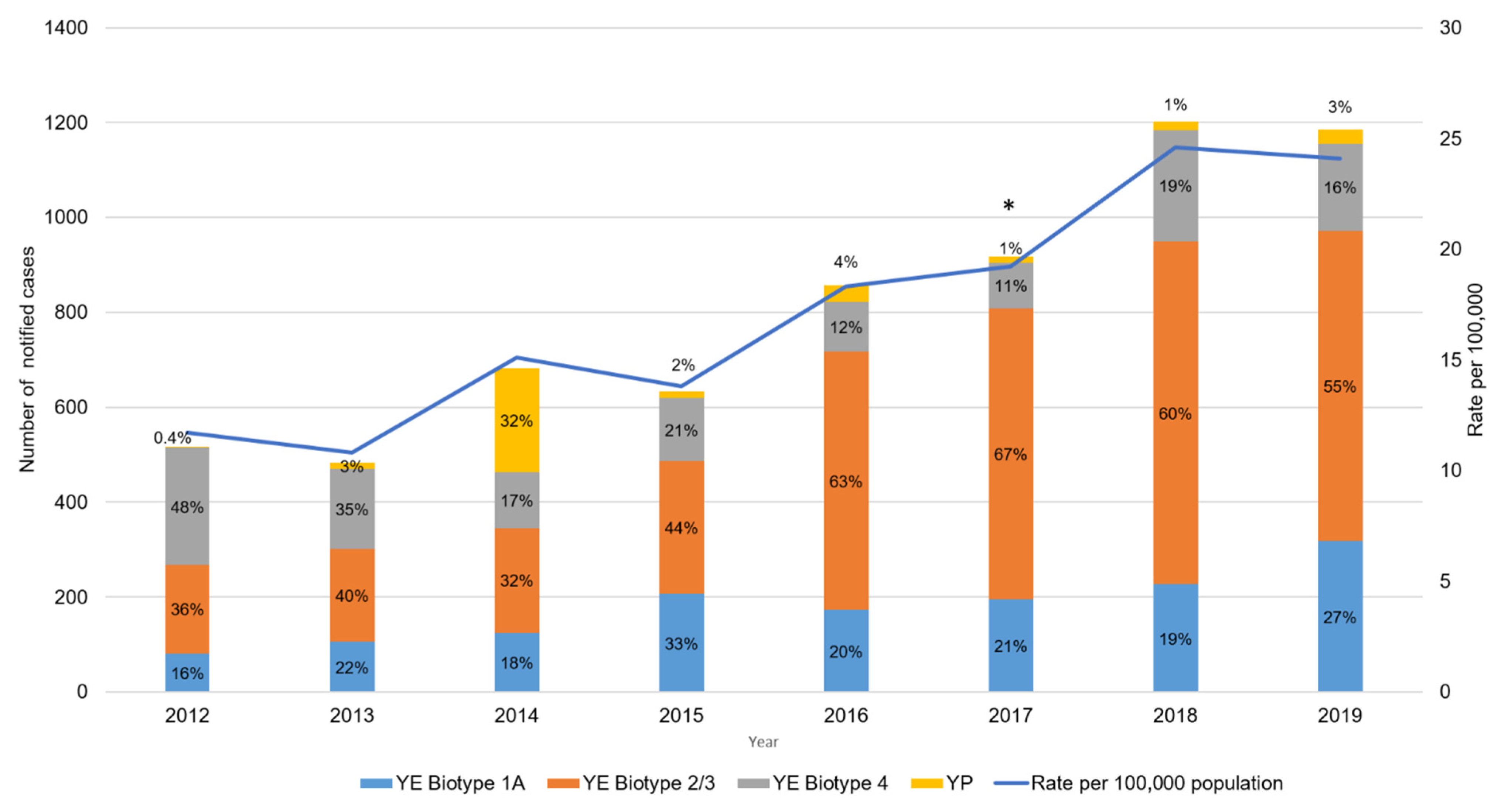
4. The Types of Yersinia enterocolitica Causing Human Yersiniosis Have Changed in New Zealand
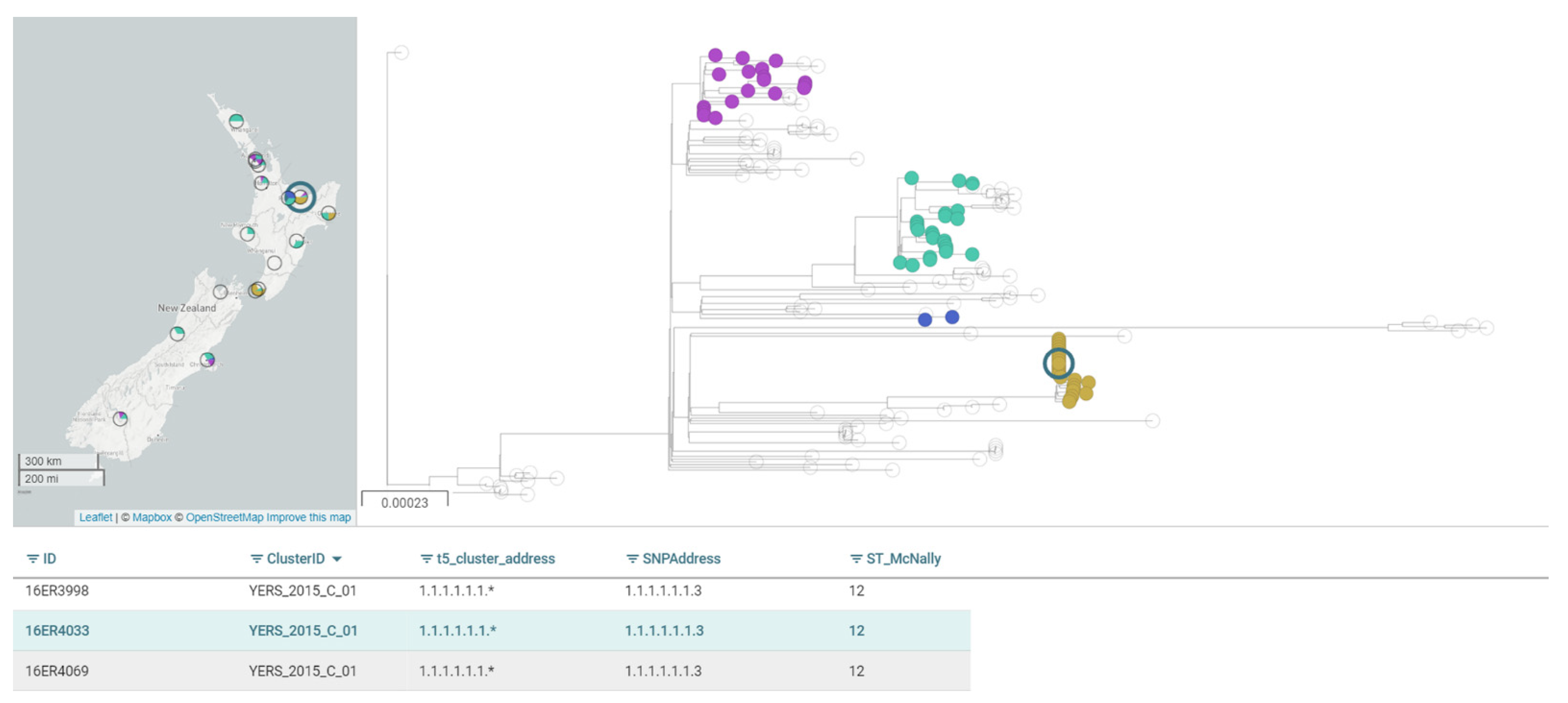
5. Whole-Genome Sequencing Is Providing Unprecedented Discriminatory Typing Power
6. Key Reservoirs, Sources and Transmission Routes of Yersinia in New Zealand Require Further Exploration
6.1. Foodborne Transmission
6.2. Possible Transmission Routes from Other Animal Species
6.3. Waterborne Transmission
6.4. Human-to-Human Transmission
7. Yersiniosis Is Also Increasing in Animals in New Zealand
8. Isolating Yersinia from Foods Can Be Challenging
9. Antimicrobial Susceptibly Data for Yersinia in New Zealand Is Currently Limited
10. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References and Notes
- Centre for Disease Control and Prevention (CDC). Yersinia enterocolitica (Yersiniosis). Available online: https://www.cdc.gov/yersinia/healthcare.html (accessed on 14 January 2021).
- New Zealand Ministry of Health. Yersiniosis. Available online: https://www.health.govt.nz/our-work/diseases-and-conditions/communicable-disease-control-manual/yersiniosis#_ftn2 (accessed on 14 January 2021).
- Fredriksson-Ahomaa, M.; Korkeala, H. Low occurrence of pathogenic Yersinia enterocolitica in clinical, food, and environmental samples: A methodological problem. Clin. Microbiol. Rev. 2003, 16, 220–229. [Google Scholar] [CrossRef] [PubMed]
- The Institute of Environmental Science and Research. Public Health Surveillance—Annual Surveillance Summary. Available online: https://surv.esr.cri.nz/surveillance/annual_surveillance.php (accessed on 11 December 2020).
- Williamson, D.A.; Baines, S.L.; Carter, G.P.; da Silva, A.G.; Ren, X.; Sherwood, J.; Dufour, M.; Schultz, M.B.; French, N.P.; Seemann, T.; et al. Genomic insights into a sustained national outbreak of Yersinia pseudotuberculosis. Genome Biol. Evol. 2016, 8, 3806–3814. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Yersiniosis; ECDC: Stockholm. 2019. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/AER_for_2018-yersiniosis-corrected.pdf (accessed on 14 January 2021).
- Tack, D.M.; Ray, L.; Griffin, P.M.; Cieslak, P.R.; Dunn, J.; Rissman, T.; Jervis, R.; Lathrop, S.; Muse, A.; Duwell, M.; et al. Preliminary incidence and trends of infections with pathogens transmitted commonly through food—foodborne diseases active surveillance network, 10 U.S. Sites, 2016–2019. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 509–514. [Google Scholar] [CrossRef] [PubMed]
- OzFoodNet Working Group. Reported foodborne illness and gastroenteritis in Australia: Annual report of the OzFoodNet network, 2004. Commun. Dis. Intell. 2005, 29, 164–190. [Google Scholar]
- Stats NZ. 2018 Census. Available online: https://www.stats.govt.nz/2018-census/ (accessed on 14 January 2021).
- Stats NZ. Ethnicity. Available online: https://www.stats.govt.nz/topics/ethnicity (accessed on 14 January 2021).
- The Institute of Environmental Science and Research Ltd. Notifiable Diseases in New Zealand: Annual Report 2018; The Institute of Environmental Science and Research Ltd.: Poirua, New Zealand, 2020; Available online: https://surv.esr.cri.nz/PDF_surveillance/AnnualRpt/AnnualSurv/2018/2018AnnualNDReport_FINAL.pdf (accessed on 14 January 2021).
- Chakraborty, A.; Komatsu, K.; Roberts, M.; Collins, J.; Beggs, J.; Turabelidze, G.; Safranek, T.; Maillard, J.M.; Bell, L.J.; Young, D.; et al. The descriptive epidemiology of yersiniosis: A multistate study, 2005–2011. Public Health Rep. 2015, 130, 269–277. [Google Scholar] [CrossRef]
- Long, C.; Jones, T.F.; Vugia, D.J.; Scheftel, J.; Strockbine, N.; Ryan, P.; Shiferaw, B.; Tauxe, R.V.; Gould, L.H. Yersinia pseudotuberculosis and Y. enterocolitica infections, FoodNet, 1996–2007. Emerg. Infect. Dis. 2010, 16, 566–567. [Google Scholar] [CrossRef]
- Lake, R.J.; Cressey, P.J.; Campbell, D.M.; Oakley, E. Risk ranking for foodborne microbial hazards in New Zealand: Burden of disease estimates. Risk Anal. 2010, 30, 743–752. [Google Scholar] [CrossRef]
- Porter, C.K.; Choi, D.; Cash, B.; Pimentel, M.; Murray, J.; May, L.; Riddle, M.S. Pathogen-specific risk of chronic gastrointestinal disorders following bacterial causes of foodborne illness. BMC Gastroenterol. 2013, 13, 46. [Google Scholar] [CrossRef]
- Highton, J.; Priest, D. Reactive arthritis: Characteristics in southern New Zealand. N. Z. Med. J. 1996, 109, 93–95. [Google Scholar]
- Rosner, B.M.; Werber, D.; Höhle, M.; Stark, K. Clinical aspects and self-reported symptoms of sequelae of Yersinia enterocolitica infections in a population-based study, Germany 2009–2010. BMC Infect. Dis. 2013, 13, 236. [Google Scholar] [CrossRef]
- Hannu, T.; Mattila, L.; Nuorti, J.P.; Ruutu, P.; Mikkola, J.; Siitonen, A.; Leirisalo-Repo, M. Reactive arthritis after an outbreak of Yersinia pseudotuberculosis serotype O:3 infection. Ann. Rheum. Dis. 2003, 62, 866–869. [Google Scholar] [CrossRef] [PubMed]
- Stolk-Engelaar, V.M.; Hoogkamp-Korstanje, J.A. Clinical presentation and diagnosis of gastrointestinal infections by Yersinia enterocolitica in 261 Dutch patients. Scand. J. Infect. Dis. 1996, 28, 571–575. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Inman, R.D.; Chiu, B.; Johnston, M.E.; Falk, J. Molecular mimicry in Reiter’s syndrome: Cytotoxicity and ELISA studies of HLA-microbial relationships. Immunology 1986, 58, 501–506. [Google Scholar] [PubMed]
- Sheehan, N.J. The ramifications of HLA-B27. J. R. Soc. Med. 2004, 97, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.L.; Wallace, M.C.; Jones, G.T.; van Rij, A.M.; Merriman, T.R.; Harrison, A.; White, D.; Stamp, L.K.; Ching, D.; Highton, J.; et al. Prevalence of HLA-B27 in the New Zealand population: Effect of age and ethnicity. Arthritis Res. Ther. 2013, 15, R158. [Google Scholar] [CrossRef]
- Bancerz-Kisiel, A.; Szweda, W. Yersiniosis—a zoonotic foodborne disease of relevance to public health. Ann. Agric. Environ. Med. 2015, 22, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Hugot, J.P.; Dumay, A.; Barreau, F.; Meinzer, U. Crohn’s disease: Is the cold chain hypothesis still hot? J. Crohns Colitis 2020. [Google Scholar] [CrossRef] [PubMed]
- Le Baut, G.; O’Brien, C.; Pavli, P.; Roy, M.; Seksik, P.; Tréton, X.; Nancey, S.; Barnich, N.; Bezault, M.; Auzolle, C.; et al. Prevalence of Yersinia species in the ileum of Crohn’s disease patients and controls. Front. Cell Infect. MicroBiol. 2018, 8, 336. [Google Scholar] [CrossRef]
- Effraimidis, G.; Tijssen, J.G.P.; Strieder, T.G.A.; Wiersinga, W.M. No causal relationship between Yersinia enterocolitica infection and autoimmune thyroid disease: Evidence from a prospective study. Clin. Exp. Immunol. 2011, 165, 38–43. [Google Scholar] [CrossRef]
- Bottone, E.J. Yersinia enterocolitica: The charisma continues. Clin. MicroBiol. Rev. 1997, 10, 257–276. [Google Scholar] [CrossRef]
- Horinouchi, T.; Nozu, K.; Hamahira, K.; Inaguma, Y.; Abe, J.; Nakajima, H.; Kugo, M.; Iijima, K. Yersinia pseudotuberculosis infection in Kawasaki disease and its clinical characteristics. BMC Pediatr. 2015, 15, 177. [Google Scholar] [CrossRef] [PubMed]
- Majowicz, S.E.; Panagiotoglou, D.; Taylor, M.; Gohari, M.R.; Kaplan, G.G.; Chaurasia, A.; Leatherdale, S.T.; Cook, R.J.; Patrick, D.M.; Ethelberg, S.; et al. Determining the long-term health burden and risk of sequelae for 14 foodborne infections in British Columbia, Canada: Protocol for a retrospective population-based cohort study. BMJ Open 2020, 10, e036560. [Google Scholar] [CrossRef] [PubMed]
- Pogreba-Brown, K.; Austhof, E.; Armstrong, A.; Schaefer, K.; Villa Zapata, L.; McClelland, D.J.; Batz, M.B.; Kuecken, M.; Riddle, M.; Porter, C.K.; et al. Chronic gastrointestinal and joint-related sequelae associated with common foodborne illnesses: A scoping review. Foodborne Pathog. Dis. 2020, 17, 67–86. [Google Scholar] [CrossRef]
- Nicol, C.; King, N.; Pirie, R.; Dufour, M. Diagnostic and Public Health Management Practices of Foodborne Bacterial Diseases; Institute of Environmental Science and Research: Wellington, New Zealand, 2010. Available online: https://www.mpi.govt.nz/dmsdocument/22312/direct (accessed on 14 January 2021).
- Addidle, M. Clinical Microbiologist, New Zealand Microbiology Network Liaison., Email Communication with J. Wright, December 2020
- International Organization for Standardization (ISO). ISO15189—Medical Laboratories—Requirements for Quality and Competence. 2012. Available online: https://www.iso.org/obp/ui/#iso:std:iso:15189:ed-3:v2:en (accessed on 14 January 2021).
- King, G. An Outbreak of Yersiniosis in Tauranga during October and November 2016; Institute of Environmental Science and Research: Wellington, New Zealand, 2017; pp. 6–7. Available online: https://surv.esr.cri.nz/PDF_surveillance/NZPHSR/2017/NZPHSRSeptember2017.pdf (accessed on 14 January 2021).
- Paixão, R.; Moreno, L.Z.; Sena de Gobbi, D.D.; Raimundo, D.C.; Hofer, E.; Matté, M.H.; Ferreira, T.S.P.; de Moura Gomes, V.T.; Costa, B.L.P.; Moreno, A.M. Characterization of Yersinia enterocolitica biotype 1A strains isolated from swine slaughterhouses and markets. Sci. World J. 2013, 2013, 769097. [Google Scholar] [CrossRef]
- Carniel, E.; Guilvout, I.; Prentice, M. Characterization of a large chromosomal “high-pathogenicity island” in biotype 1B Yersinia enterocolitica. J. Bacteriol. 1996, 178, 6743–6751. [Google Scholar] [CrossRef] [PubMed]
- Wauters, G.; Kandolo, K.; Janssens, M. Revised biogrouping scheme of Yersinia enterocolitica. Contrib. MicroBiol. Immunol. 1987, 9, 14–21. [Google Scholar] [PubMed]
- Hall, M.; Chattaway, M.A.; Reuter, S.; Savin, C.; Strauch, E.; Carniel, E.; Connor, T.; Van Damme, I.; Rajakaruna, L.; Rajendram, D.; et al. Use of whole-genus genome sequence data to develop a multilocus sequence typing tool that accurately identifies Yersinia isolates to the species and subspecies levels. J. Clin. MicroBiol. 2015, 53, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Strydom, H.; Wang, J.; Paine, S.; Dyet, K.; Cullen, K.; Wright, J. Evaluating sub-typing methods for pathogenic Yersinia enterocolitica to support outbreak investigations in New Zealand. Epidemiol. Infect. 2019, 147, e186. [Google Scholar] [CrossRef] [PubMed]
- Reuter, S.; Connor, T.R.; Barquist, L.; Walker, D.; Feltwell, T.; Harris, S.R.; Fookes, M.; Hall, M.E.; Petty, N.K.; Fuchs, T.M.; et al. Parallel independent evolution of pathogenicity within the genus Yersinia. Proc. Natl. Acad. Sci. USA 2014, 111, 6768–6773. [Google Scholar] [CrossRef] [PubMed]
- Eksić, S.; Steigerwalt, A.G.; Bockemühl, J.; Huntley-Carter, G.P.; Brenner, D.J. Yersinia rohdei sp. nov. isolated from human and dog feces and surface water. Int. J. Syst. Evol. 1987, 37, 327–332. [Google Scholar] [CrossRef]
- Hilbink, F.; Fenwick, S.; Thompson, E.J.; Penrose, M.; Ross, G.P. Non-specific seroreactions against Brucella abortus in ruminants in New Zealand and the presence of Yersinia enterocolitica O:9. N. Z. Vet. J. 1995, 43, 175–178. [Google Scholar] [CrossRef]
- Fenwick, S. Domestic animals as potential sources of human Yersinia infection. Surveillance 1997, 24, 3–4. [Google Scholar]
- Staples, M. Senior Scientist, Reference Laboratory, Public Health Microbiology, Forensic and Scientific Services, Queensland Health, Australia November 2020, Email Communication with J. Wright, December 2020
- Campioni, F.; Falcao, J.P. Genotyping of Yersinia enterocolitica biotype 1A strains from clinical and nonclinical origins by pulsed-field gel electrophoresis. Can. J. MicroBiol. 2014, 60, 419–424. [Google Scholar] [CrossRef]
- Sihvonen, L.M.; Jalkanen, K.; Huovinen, E.; Toivonen, S.; Corander, J.; Kuusi, M.; Skurnik, M.; Siitonen, A.; Haukka, K. Clinical isolates of Yersinia enterocolitica biotype 1A represent two phylogenetic lineages with differing pathogenicity-related properties. BMC MicroBiol. 2012, 12, 208. [Google Scholar] [CrossRef]
- Tennant, S.M.; Grant, T.H.; Robins-Browne, R.M. Pathogenicity of Yersinia enterocolitica biotype 1A. FEMS Immunol. Med. MicroBiol. 2003, 38, 127–137. [Google Scholar] [CrossRef]
- Tuompo, R.; Hannu, T.; Huovinen, E.; Sihvonen, L.; Siitonen, A.; Leirisalo-Repo, M. Yersinia enterocolitica biotype 1A: A possible new trigger of reactive arthritis. Rheumatol.Int. 2017, 37, 1863–1869. [Google Scholar] [CrossRef]
- Grant, T.; Bennett-Wood, V.; Robins-Browne, R.M. Identification of virulence-associated characteristics in clinical isolates of Yersinia enterocolitica lacking classical virulence markers. Infect. Immun. 1998, 66, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Ramamurthy, T.; Yoshino, K.; Huang, X.; Balakrish Nair, G.; Carniel, E.; Maruyama, T.; Fukushima, H.; Takeda, T. The novel heat-stable enterotoxin subtype gene (ystB) of Yersinia enterocolitica: Nucleotide sequence and distribution of the yst genes. Microb. Pathog. 1997, 23, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Stephan, R.; Joutsen, S.; Hofer, E.; Säde, E.; Björkroth, J.; Ziegler, D.; Fredriksson-Ahomaa, M. Characteristics of Yersinia enterocolitica biotype 1A strains isolated from patients and asymptomatic carriers. Eur. J. Clin. MicroBiol. Infect. Dis. 2013, 32, 869–875. [Google Scholar] [CrossRef]
- Wojciech, Ł.; Staroniewicz, Z.; Jakubczak, A.; Ugorski, M. Typing of Yersinia enterocolitica isolates by ITS profiling, REP- and ERIC-PCR. J. Vet. Med. B 2004, 51, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Gilpin, B.J.; Robson, B.; Lin, S.; Hudson, J.A.; Weaver, L.; Dufour, M.; Strydom, H. The limitations of pulsed-field gel electrophoresis for analysis of Yersinia enterocolitica isolates. Zoonoses Public Health 2014, 61, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Gierczyński, R.; Golubov, A.; Neubauer, H.; Pham, J.N.; Rakin, A. Development of multiple-locus variable-number tandem-repeat analysis for Yersinia enterocolitica subsp. palearctica and its application to bioserogroup 4/O3 subtyping. J. Clin. MicroBiol. 2007, 45, 2508–2515. [Google Scholar] [CrossRef] [PubMed]
- Gulati, P.; Varshney, R.K.; Virdi, J.S. Multilocus variable number tandem repeat analysis as a tool to discern genetic relationships among strains of Yersinia enterocolitica biovar 1A. J. Appl. MicroBiol. 2009, 107, 875–884. [Google Scholar] [CrossRef]
- Besser, J.M.; Carleton, H.A.; Trees, E.; Stroika, S.G.; Hise, K.; Wise, M.; Gerner-Smidt, P. Interpretation of whole-genome sequencing for enteric disease surveillance and outbreak investigation. Foodborne Pathog. Dis. 2019, 16, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Inns, T.; Flanagan, S.; Greig, D.R.; Jenkins, C.; Seddon, K.; Chin, T.; Cartwright, J. First use of whole-genome sequencing to investigate a cluster of Yersinia enterocolitica, Liverpool, United Kingdom, 2017. J. Med. MicroBiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization of the United Nations. Technical Background Paper: Applications of Whole Genome Sequencing in Food Safety Management; Food and Agriculture Organization of the United Nations: Rome, Italy, 2016. Available online: http://www.fao.org/3/a-i5619e.pdf (accessed on 14 January 2021).
- Seecharran, T.; Kalin-Mänttäri, L.; Koskela, K.A.; Nikkari, S.; Dickins, B.; Corander, J.; Skurnik, M.; McNally, A. Phylogeographic separation and formation of sexually discrete lineages in a global population of Yersinia pseudotuberculosis. bioRxiv 2017, 149468. [Google Scholar] [CrossRef]
- Savin, C.; Criscuolo, A.; Guglielmini, J.; Le Guern, A.S.; Carniel, E.; Pizarro-Cerdá, J.; Brisse, S. Genus-wide Yersinia core-genome multilocus sequence typing for species identification and strain characterization. Microb. Genom. 2019, 5. [Google Scholar] [CrossRef]
- Zhou, Z.; Charlesworth, J.; Achtman, M. HierCC: A multi-level clustering scheme for population assignments based on core genome MLST. bioRxiv 2020. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.-F.; Mohamed, K.; Fan, Y.; Agama Study, G.; Achtman, M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef]
- Seemann, T.; Goncalves da Silva, A.; Bulach, D.M.; Schultz, M.B.; Kwong, J.C.; Howden, B.P. Nullarbor GitHub. Available online: https://github.com/tseemann/nullarbor (accessed on 6 September 2019).
- Laukkanen-Ninios, R.; Didelot, X.; Jolley, K.A.; Morelli, G.; Sangal, V.; Kristo, P.; Brehony, C.; Imori, P.F.M.; Fukushima, H.; Siitonen, A.; et al. Population structure of the Yersinia pseudotuberculosis complex according to multilocus sequence typing. Environ. MicroBiol. 2011, 13, 3114–3127. [Google Scholar] [CrossRef]
- Seemann, T.; Goncalves da Silva, A.; Bulach, D.M.; Schultz, M.B.; Kwong, J.C.; Howden, B.P. Snippy, Rapid Bacterial SNP Calling and Core Genome Alignments. Available online: https://github.com/tseemann/snippy (accessed on 13 December 2020).
- Dallman, T.; Ashton, P.; Schafer, U.; Jironkin, A.; Painset, A.; Shaaban, S.; Hartman, H.; Myers, R.; Underwood, A.; Jenkins, C.; et al. SnapperDB: A database solution for routine sequencing analysis of bacterial isolates. Bioinformatics 2018, 34, 3028–3029. [Google Scholar] [CrossRef] [PubMed]
- Argimón, S.; Abudahab, K.; Goater, R.J.E.; Fedosejev, A.; Bhai, J.; Glasner, C.; Feil, E.J.; Holden, M.T.G.; Yeats, C.A.; Grundmann, H.; et al. Microreact: Visualizing and sharing data for genomic epidemiology and phylogeography. Microb. Genom. 2016, 2, e000093. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2017, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Lake, R.; Hudson, A.; Cressey, P. Risk Profile: Yersinia enterocolitica in Pork; Institute of Environmental Science and Research: Wellington, New Zealand, 2004. Available online: https://www.agriculture.govt.nz/dmsdocument/26192/direct (accessed on 14 January 2021).
- Satterthwaite, P.; Pritchard, K.; Floyd, D.; Law, B. A case control study of Yersinia enterocolitica infections in Auckland. Aust. N. Z. J. Publ. Health 1999, 23, 482–485. [Google Scholar] [CrossRef]
- Wright, J. Gastrointestinal Infection in a New Zealand Community: A One Year Study; Massey University: Palmerston North, New Zealand, 1996; Available online: https://mro.massey.ac.nz/handle/10179/10745 (accessed on 14 January 2021).
- New Zealand Ministry of Health. Appendix 2: Enteric Disease. Available online: https://www.health.govt.nz/our-work/diseases-and-conditions/communicable-disease-control-manual/appendix-2-enteric-disease (accessed on 12 January 2021).
- Cressey, P.J.; Lake, R.J.; Thornley, C.; Campbell, D. Expert elicitation for estimation of the proportion foodborne for selected microbial pathogens in New Zealand. Foodborne Pathog. Dis. 2019, 16, 543–549. [Google Scholar] [CrossRef]
- Zanabria, R.; Racicot, M.; Leroux, A.; Xucen, L.; Cormier, M.; Ferrouillet, C.; Arsenault, J.; Mackay, A.; Griffiths, M.; Holley, R.; et al. Source attribution at the food sub-product level for the development of the Canadian Food Inspection Agency risk assessment model. Int. J. Food MicroBiol. 2019, 305, 108241. [Google Scholar] [CrossRef] [PubMed]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.-A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States--major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Butler, A.J.; Thomas, M.K.; Pintar, K.D. Expert elicitation as a means to attribute 28 enteric pathogens to foodborne, waterborne, animal contact, and person-to-person transmission routes in Canada. Foodborne Pathog. Dis. 2015, 12, 335–344. [Google Scholar] [CrossRef]
- Adak, G.K.; Long, S.M.; O’Brien, S.J. Trends in indigenous foodborne disease and deaths, England and Wales: 1992 to 2000. Gut 2002, 51, 832–841. [Google Scholar] [CrossRef]
- New Zealand Ministry for Primary Industries. Outbreak Source Investigation: Yersinia pseudotuberculosis 2014; MPI: Wellington, New Zealand, 2014. [Google Scholar]
- Espenhain, L.; Riess, M.; Muller, L.; Colombe, S.; Ethelberg, S.; Litrup, E.; Jernberg, C.; Kuhlmann-Berenzon, S.; Lindblad, M.; Hove, N.K.; et al. Cross-border outbreak of Yersinia enterocolitica O:3 associated with imported fresh spinach, Sweden and Denmark, March 2019. Euro Surveill 2019, 24. [Google Scholar] [CrossRef] [PubMed]
- Minami, K.; Yasuda, R.; Terakawa, R.; Koike, Y.; Takeuchi, K.; Higuchi, T.; Horiuchi, A.; Kubota, N.; Hidaka, E.; Kawakami, Y. Four sporadic pediatric cases of Yersinia enterocolitica O:8 infection in a rural area of Japan. Jpn. J. Infect. Dis. 2017, 70, 192–194. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, E.; Einoder-Moreno, M.; Borgen, K.; Thorstensen Brandal, L.; Diab, L.; Fossli, O.; Guzman Herrador, B.; Hassan, A.A.; Johannessen, G.S.; Johansen, E.J.; et al. National outbreak of Yersinia enterocolitica infections in military and civilian populations associated with consumption of mixed salad, Norway, 2014. Euro Surveill. 2016, 21, 30321. [Google Scholar] [CrossRef]
- MacDonald, E.; Heier, B.T.; Stalheim, T.; Cudjoe, K.S.; Skjerdal, T.; Wester, A.; Lindstedt, B.A.; Vold, L. Yersinia enterocolitica O:9 infections associated with bagged salad mix in Norway, February to April 2011. Euro Surveill. 2011, 16, 19866. [Google Scholar] [PubMed]
- Center for Disease Control and Prevention (CDC). Notes from the field: Yersinia enterocolitica infections associated with pasteurized milk—southwestern Pennsylvania, March-August, 2011. MMWR Morb. Mortal. Mortal. Wkly. Rep. 2011, 60, 1428. [Google Scholar]
- OzFoodNet. Communicable Disease Intelligence-Quarterly Report; OzFoodNet: Canberra, Australia, 2009. Available online: https://www1.health.gov.au/internet/main/publishing.nsf/content/F5DACD23D760D604CA257BF0001CFE6B/$File/CDI3304.pdf (accessed on 14 January 2021).
- Grahek-Ogden, D.; Schimmer, B.; Cudjoe, K.S.; Nygard, K.; Kapperud, G. Outbreak of Yersinia enterocolitica serogroup O:9 infection and processed pork, Norway. Emerg. Infect. Dis. 2007, 13, 754–756. [Google Scholar] [CrossRef]
- Sakai, T.; Nakayama, A.; Hashida, M.; Yamamoto, Y.; Takebe, H.; Imai, S. Outbreak of food poisoning by Yersinia enterocolitica serotype O8 in Nara prefecture: The first case report in Japan. Jpn. J. Infect. Dis. 2005, 58, 257–258. [Google Scholar]
- Centre for Disease Control and Prevention (CDC). Yersinia enterocolitica gastroenteritis among infants exposed to chitterlings—Chicago, Illinois, 2002. MMWR Morb. Mortal. Mortal. Wkly. Rep. 2003, 52, 956–958. [Google Scholar]
- Pärn, T.; Hallanvuo, S.; Salmenlinna, S.; Pihlajasaari, A.; Heikkinen, S.; Telkki-Nykänen, H.; Hakkinen, M.; Ollgren, J.; Huusko, S.; Rimhanen-Finne, R. Outbreak of Yersinia pseudotuberculosis O:1 infection associated with raw milk consumption, Finland, spring 2014. Euro Surveill. 2015, 20. [Google Scholar] [CrossRef]
- Rimhanen-Finne, R.; Niskanen, T.; Hallanvuo, S.; Makary, P.; Haukka, K.; Pajunen, S.; Siitonen, A.; Ristolainen, R.; Poyry, H.; Ollgren, J.; et al. Yersinia pseudotuberculosis causing a large outbreak associated with carrots in Finland, 2006. Epidemiol. Infect. 2008, 137, 342–347. [Google Scholar] [CrossRef]
- Kangas, S.; Takkinen, J.; Hakkinen, M.; Nakari, U.M.; Johansson, T.; Henttonen, H.; Virtaluoto, L.; Siitonen, A.; Ollgren, J.; Kuusi, M. Yersinia pseudotuberculosis O:1 traced to raw carrots, Finland. Emerg. Infect. Dis. 2008, 14, 1959–1961. [Google Scholar] [CrossRef]
- Jalava, K.; Hakkinen, M.; Valkonen, M.; Nakari, U.M.; Palo, T.; Hallanvuo, S.; Ollgren, J.; Siitonen, A.; Nuorti, J.P. An outbreak of gastrointestinal illness and erythema nodosum from grated carrots contaminated with Yersinia pseudotuberculosis. J. Infect. Dis. 2006, 194, 1209–1216. [Google Scholar] [CrossRef] [PubMed]
- Boqvist, S.; Pettersson, H.; Svensson, A.; Andersson, Y. Sources of sporadic Yersinia enterocolitica infection in children in Sweden, 2004: A case-control study. Epidemiol. Infect. 2009, 137, 897–905. [Google Scholar] [CrossRef] [PubMed]
- Ostroff, S.M.; Kapperud, G.; Hutwagner, L.C.; Nesbakken, T.; Bean, N.H.; Lassen, J.; Tauxe, R.V. Sources of sporadic Yersinia enterocolitica infections in Norway: A prospective case-control study. Epidemiol. Infect. 1994, 112, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Jones, T.F.; Buckingham, S.C.; Bopp, C.A.; Ribot, E.; Schaffner, W. From pig to pacifier: Chitterling-associated yersiniosis outbreak among black infants. Emerg. Infect. Dis. 2003, 9, 1007–1009. [Google Scholar] [CrossRef]
- Guillier, L.; Fravalo, P.; Leclercq, A.; Thébault, A.; Kooh, P.; Cadavez, V.; Gonzales-Barron, U. Risk factors for sporadic Yersinia enterocolitica infections: A systematic review and meta-analysis. Microbial. Risk Anal. 2020. [Google Scholar] [CrossRef]
- MacDonald, E.; Heier, B.T.; Nygard, K.; Stalheim, T.; Cudjoe, K.S.; Skjerdal, T.; Wester, A.L.; Lindstedt, B.A.; Stavnes, T.L.; Vold, L. Yersinia enterocolitica outbreak associated with ready-to-eat salad mix, Norway, 2011. Emerg. Infect. Dis. 2012, 18, 1496–1499. [Google Scholar] [CrossRef]
- Tauxe, R.V.; Wauters, G.; Goossens, V.; Noyen, R.V.; Vandepitte, J.; Martin, S.M.; Mol, P.D.; Thiers, G. Yersinia enterocolitica infections and pork: The missing link. Lancet 1987, 329, 1129–1132. [Google Scholar] [CrossRef]
- Huovinen, E.; Sihvonen, L.M.; Virtanen, M.J.; Haukka, K.; Siitonen, A.; Kuusi, M. Symptoms and sources of Yersinia enterocolitica-infection: A case-control study. BMC Infect. Dis. 2010, 10, 122. [Google Scholar] [CrossRef]
- Rosner, B.M.; Stark, K.; Höhle, M.; Werber, D. Risk factors for sporadic Yersinia enterocolitica infections, Germany 2009–2010. Epidemiol. Infect. 2012, 140, 1738–1747. [Google Scholar] [CrossRef]
- Nuorti, J.P.; Niskanen, T.; Hallanvuo, S.; Mikkola, J.; Kela, E.; Hatakka, M.; Fredriksson-Ahomaa, M.; Lyytikainen, O.; Siitonen, A.; Korkeala, H.; et al. A widespread outbreak of Yersinia pseudotuberculosis O:3 infection from iceberg lettuce. J. Infect. Dis. 2004, 189, 766–774. [Google Scholar] [CrossRef] [PubMed]
- Jalava, K.; Hallanvuo, S.; Nakari, U.M.; Ruutu, P.; Kela, E.; Heinasmaki, T.; Siitonen, A.; Nuorti, J.P. Multiple outbreaks of Yersinia pseudotuberculosis infections in Finland. J. Clin. MicroBiol. 2004, 42, 2789–2791. [Google Scholar] [CrossRef]
- Fenwick, S. Yersinia enterocolitica Infections in People and Other Animals—A New Zealand Study; Massey University: Palmerston North, New Zealand, 1997; Available online: https://mro.massey.ac.nz/handle/10179/2732 (accessed on 14 January 2021).
- Hudson, J.A.; Mott, S.J.; Delacy, K.M.; Edridge, A.L. Incidence and coincidence of Listeria spp., motile aeromonads and Yersinia enterocolitica on ready-to-eat fleshfoods. Int. J. Food Microbiol. 1992, 16, 99–108. [Google Scholar] [CrossRef]
- Laukkanen-Ninios, R.; Fredriksson-Ahomaa, M.; Korkeala, H. Enteropathogenic Yersinia in the pork production chain: Challenges for control. Compr. Rev. Food Sci. Food Saf. 2014, 13, 1165–1191. [Google Scholar] [CrossRef]
- Drummond, N.; Murphy, B.P.; Ringwood, T.; Prentice, M.B.; Buckley, J.F.; Fanning, S. Yersinia enterocolitica: A brief review of the issues relating to the zoonotic pathogen, public health challenges, and the pork production chain. Foodborne Pathog. Dis. 2012, 9, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Messelhausser, U.; Kampf, P.; Colditz, J.; Bauer, H.; Schreiner, H.; Holler, C.; Busch, U. Qualitative and quantitative detection of human pathogenic Yersinia enterocolitica in different food matrices at retail level in Bavaria. Foodborne Pathog. Dis. 2011, 8, 39–44. [Google Scholar] [CrossRef]
- Fredriksson-Ahomaa, M.; Hielm, S.; Korkeala, H. High prevalence of yadA-positive Yersinia enterocolitica in pig tongues and minced meat at the retail level in Finland. J. Food Prot. 1999, 62, 123–127. [Google Scholar] [CrossRef][Green Version]
- Fredriksson-Ahomaa, M.; Lyhs, U.; Korte, T.; Korkeala, H. Prevalence of pathogenic Yersinia enterocolitica in food samples at retail level in Finland. Archiv. Lebensm. 2001, 52, 66–68. [Google Scholar]
- Arrausi-Subiza, M.; Ibabe, J.C.; Atxaerandio, R.; Juste, R.A.; Barral, M. Evaluation of different enrichment methods for pathogenic Yersinia species detection by real time PCR. BMC Vet. Res. 2014, 10, 192. [Google Scholar] [CrossRef]
- Van Damme, I.; Berkvens, D.; Vanantwerpen, G.; Bare, J.; Houf, K.; Wauters, G.; De Zutter, L. Contamination of freshly slaughtered pig carcasses with enteropathogenic Yersinia spp.: Distribution, quantification and identification of risk factors. Int. J. Food MicroBiol. 2015, 204, 33–40. [Google Scholar] [CrossRef]
- Nesbakken, T.; Eckner, K.; Røtterud, O.-J. The effect of blast chilling on occurrence of human pathogenic Yersinia enterocolitica compared to Campylobacter spp. and numbers of hygienic indicators on pig carcasses. Int. J. Food MicroBiol. 2008, 123, 130–133. [Google Scholar] [CrossRef]
- Van Damme, I.; De Zutter, L.; Jacxsens, L.; Nauta, M.J. Control of human pathogenic Yersinia enterocolitica in minced meat: Comparative analysis of different interventions using a risk assessment approach. Food MicroBiol. 2017, 64, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Martins, B.T.F.; Botelho, C.V.; Silva, D.A.L.; Lanna, F.; Grossi, J.L.; Campos-Galvão, M.E.M.; Yamatogi, R.S.; Falcão, J.P.; Bersot, L.D.S.; Nero, L.A. Yersinia enterocolitica in a Brazilian pork production chain: Tracking of contamination routes, virulence and antimicrobial resistance. Int. J. Food MicroBiol. 2018, 276, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Wang, X.; Xiao, Y.; Cui, Z.; Xia, S.; Hao, Q.; Yang, J.; Luo, L.; Wang, S.; Li, K.; et al. Prevalence of Yersinia enterocolitica in pigs slaughtered in Chinese abattoirs. Appl. Environ. MicroBiol. 2012, 78, 2949–2956. [Google Scholar] [CrossRef]
- Rahikainen Ibañez, T.; Laukkanen-Ninios, R.; Hakkinen, M.; Johansson, T.; Vilar, M.; Korkeala, H. Prevalence of pathogenic Yersinia enterocolitica in Finnish slaughter pigs. J. Food Prot. 2016, 79, 677–681. [Google Scholar] [CrossRef] [PubMed]
- Fondrevez, M.; Minvielle, B.; Labbé, A.; Houdayer, C.; Rose, N.; Esnault, E.; Denis, M. Prevalence of pathogenic Yersinia enterocolitica in slaughter-aged pigs during a one-year survey, 2010-2011, France. Int. J. Food MicroBiol. 2014, 174, 56–62. [Google Scholar] [CrossRef]
- Gürtler, M.; Alter, K.; Kasimir, S.; Linnebur, M.; Fahlhaber, K. Prevalence of Yersinia enterocolitica in fattening pigs. J. Food Prot. 2005, 68, 850–854. [Google Scholar] [CrossRef]
- Bonardi, S.; Bassi, L.; Brindani, F.; D’Incau, M.; Barco, L.; Carra, E.; Pongolini, S. Prevalence, characterization and antimicrobial susceptibility of Salmonella enterica and Yersinia enterocolitica in pigs at slaughter in Italy. Int. J. Food MicroBiol. 2013, 163, 248–257. [Google Scholar] [CrossRef]
- Bonardi, S.; Bruini, I.; D’Incau, M.; Van Damme, I.; Carniel, E.; Brémont, S.; Cavallini, P.; Tagliabue, S.; Brindani, F. Detection, seroprevalence and antimicrobial resistance of Yersinia enterocolitica and Yersinia pseudotuberculosis in pig tonsils in Northern Italy. Int. J. Food MicroBiol. 2016, 235, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Fois, F.; Piras, F.; Torpdahl, M.; Mazza, R.; Ladu, D.; Consolati, S.G.; Spanu, C.; Scarano, C.; De Santis, E.P.L. Prevalence, bioserotyping and antibiotic resistance of pathogenic Yersinia enterocolitica detected in pigs at slaughter in Sardinia. Int. J. Food MicroBiol. 2018, 283, 1–6. [Google Scholar] [CrossRef]
- Bolton, D.J.; Ivory, C.; McDowell, D. A small study of Yersinia enterocolitica in pigs from birth to carcass and characterisation of porcine and human strains. Food Control. 2013, 33, 521–524. [Google Scholar] [CrossRef]
- Choi, Y.M.; Park, H.J.; Jang, H.I.; Kim, S.A.; Imm, J.Y.; Hwang, I.G.; Rhee, M.S. Changes in microbial contamination levels of porcine carcasses and fresh pork in slaughterhouses, processing lines, retail outlets, and local markets by commercial distribution. Res. Vet. Sci. 2013, 94, 413–418. [Google Scholar] [CrossRef] [PubMed]
- Råsbäck, T.; Rosendal, T.; Stampe, M.; Sannö, A.; Aspán, A.; Järnevi, K.; Lahti, E.T. Prevalence of human pathogenic Yersinia enterocolitica in Swedish pig farms. Acta Vet. Scand. 2018, 60, 39. [Google Scholar] [CrossRef]
- Syczyło, K.; Platt-Samoraj, A.; Bancerz-Kisiel, A.; Szczerba-Turek, A.; Pajdak-Czaus, J.; Łabuć, S.; Procajło, Z.; Socha, P.; Chuzhebayeva, G.; Szweda, W. The prevalence of Yersinia enterocolitica in game animals in Poland. PLoS ONE 2018, 13, e0195136. [Google Scholar] [CrossRef]
- Sannö, A.; Aspán, A.; Hestvik, G.; Jacobson, M. Presence of Salmonella spp., Yersinia enterocolitica, Yersinia pseudotuberculosis and Escherichia coli O157:H7 in wild boars. Epidemiol. Infect. 2014, 142, 2542–2547. [Google Scholar] [CrossRef] [PubMed]
- Backhans, A.; Fellström, C.; Lambertz, S.T. Occurrence of pathogenic Yersinia enterocolitica and Yersinia pseudotuberculosis in small wild rodents. Epidemiol. Infect. 2011, 139, 1230–1238. [Google Scholar] [CrossRef]
- Reinhardt, M.; Hammerl, J.A.; Kunz, K.; Barac, A.; Nöckler, K.; Hertwig, S. Yersinia pseudotuberculosis prevalence and diversity in wild boars in Northeast Germany. Appl. Environ. MicroBiol. 2018, 84. [Google Scholar] [CrossRef] [PubMed]
- Niskanen, T.; Waldenström, J.; Fredriksson-Ahomaa, M.; Olsen, B.; Korkeala, H. virF-positive Yersinia pseudotuberculosis and Yersinia enterocolitica found in migratory birds in Sweden. Appl. Environ. MicroBiol. 2003, 69, 4670–4675. [Google Scholar] [CrossRef]
- Arrausi-Subiza, M.; Gerrikagoitia, X.; Alvarez, V.; Ibabe, J.C.; Barral, M. Prevalence of Yersinia enterocolitica and Yersinia pseudotuberculosis in wild boars in the Basque Country, northern Spain. Acta Vet. Scand. 2016, 58, 4. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson-Ahomaa, M.; Wacheck, S.; Bonke, R.; Stephan, R. Different enteropathogenic Yersinia strains found in wild boars and domestic pigs. Foodborne Pathog. Dis. 2011, 8, 733–737. [Google Scholar] [CrossRef]
- Niskanen, T.; Fredriksson-Ahomaa, M.; Korkeala, H. Occurence of Yersinia pseudotuberculosis in iceberg lettuce and environment. In The Genus Yersinia: Entering the Functional Genomic Era; Skurnik, M., Bengoechea, J.A., Granfors, K., Eds.; Springer: Boston, MA, USA, 2003; pp. 383–386. [Google Scholar] [CrossRef]
- Pattis, I.; Moriarty, E.; Billington, C.; Gilpin, B.; Hodson, R.; Ward, N. Concentrations of Campylobacter spp., Escherichia coli, Enterococci, and Yersinia spp. in the feces of farmed red deer in New Zealand. J. Environ. Qual. 2017, 46, 819–827. [Google Scholar] [CrossRef]
- Wang, X.; Cui, Z.; Wang, H.; Tang, L.; Yang, J.; Gu, L.; Jin, D.; Luo, L.; Qiu, H.; Xiao, Y.; et al. Pathogenic strains of Yersinia enterocolitica isolated from domestic dogs (Canis familiaris) belonging to farmers are of the same subtype as pathogenic Y. enterocolitica strains isolated from humans and may be a source of human infection in Jiangsu Province, China. J. Clin. MicroBiol. 2010, 48, 1604–1610. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson-Ahomaa, M.; Korte, T.; Korkeala, H. Transmission of Yersinia enterocolitica 4/O:3 to pets via contaminated pork. Lett. Appl. MicroBiol. 2001, 32, 375–378. [Google Scholar] [CrossRef]
- Le Guern, A.-S.; Martin, L.; Savin, C.; Carniel, E. Yersiniosis in France: Overview and potential sources of infection. Int. J. Infect. Dis. 2016, 46, 1–7. [Google Scholar] [CrossRef]
- Isobe, J.; Kimata, K.; Shimizu, M.; Kanatani, J.; Sata, T.; Watahiki, M. Water-borne outbreak of Yersinia enterocolitica O8 due to a small scale water system. Kansenshogaku Zasshi 2014, 88, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Eden, K.V.; Rosenberg, M.L.; Stoopler, M.; Wood, B.T.; Highsmith, A.K.; Skaliy, P.; Wells, J.G.; Feeley, J.C. Waterborne gastrointestinal illness at a ski resort. —Isolation of Yersinia enterocolitica from drinking water. Public Health Rep. 1977, 92, 245–250. [Google Scholar]
- New Zealand Ministry of Health. Drinking-Water Standards for New Zealand 2005 (Revised 2018); New Zealand Ministry of Health: Wellington, New Zealand, 2018. Available online: https://www.health.govt.nz/publication/drinking-water-standards-new-zealand-2005-revised-2018 (accessed on 12 January 2021).
- Sandery, M.; Stinear, T.; Kaucner, C. Detection of pathogenic Yersinia enterocolitica in environmental waters by PCR. J. Appl. MicroBiol. 1996, 80, 327–332. [Google Scholar] [CrossRef]
- Falcão, J.P.; Brocchi, M.; Proença-Módena, J.L.; Acrani, G.O.; Corrêa, E.F.; Falcão, D.P. Virulence characteristics and epidemiology of Yersinia enterocolitica and Yersiniae other than Y. pseudotuberculosis and Y. pestis isolated from water and sewage. J. Appl. MicroBiol. 2004, 96, 1230–1236. [Google Scholar] [CrossRef] [PubMed]
- Moriki, S.; Nobata, A.; Shibata, H.; Nagai, A.; Minami, N.; Taketani, T.; Fukushima, H. Familial outbreak of Yersinia enterocolitica serotype O9 biotype 2. J. Infect. Chemther. 2010, 16, 56–58. [Google Scholar] [CrossRef]
- Morse, D.L.; Shayegani, M.; Gallo, R.J. Epidemiologic investigation of a Yersinia camp outbreak linked to a food handler. Am. J. Public Health 1984, 74, 589–592. [Google Scholar] [CrossRef]
- Ratnam, S.; Mercer, E.; Picco, B.; Parsons, S.; Butler, R. A nosocomial outbreak of diarrheal disease due to Yersinia enterocolitica serotype O:5, biotype 1. J. Infect. Dis. 1982, 145, 242–247. [Google Scholar] [CrossRef]
- Theakston, E.P.; Baker, B.W.; Morris, A.J.; Woodfield, D.G.; Streat, S.J. Transfusion transmitted Yersinia enterocolitica infections in New Zealand. Aust. N. Z. Med. J. 1997, 27, 62–67. [Google Scholar] [CrossRef]
- Morley, S. Chief Medical Officer, NZBlood, Email Communication with J. Wright, December 2020
- New Zealand Blood. Annual Report 2010; New Zealand Blood: Wellington, New Zealand, 2010; Available online: https://www.nzblood.co.nz/assets/Haemovigilance/Haemovigilance-Annual-Report-2010.pdf (accessed on 30 November 2020).
- Watts, J. Senior Adviser Animal Health Surveillance and Incursion Investigation Biosecurity New Zealand Ministry for Primary Industries, Email Communication with J. Wright, December 2020
- Mair, N.S.; Fox, E.; Thal, E. Biochemical, pathogenicity and toxicity studies of type III strains of Yersinia pseudotuberculosis isolated from the cecal contents of pigs. Contrib. MicroBiol. Immunol. 1979, 5, 359–365. [Google Scholar]
- Tsubokura, M.; Otsuki, K.; Kawaoka, Y.; Maruyama, T. Characterization and pathogenicity of Yersinia pseudotuberculosis isolated from swine and other animals. J. Clin. MicroBiol. 1984, 19, 754–756. [Google Scholar] [CrossRef]
- Martins, C.H.; Bauab, T.M.; Falcão, D.P. Characteristics of Yersinia pseudotuberculosis isolated from animals in Brazil. J. Appl. MicroBiol. 1998, 85, 703–707. [Google Scholar] [CrossRef] [PubMed]
- Buhles, W.C., Jr.; Vanderlip, J.E.; Russell, S.W.; Alexander, N.L. Yersinia pseudotuberculosis infection: Study of an epizootic in squirrel monkeys. J. Clin. MicroBiol. 1981, 13, 519–525. [Google Scholar] [CrossRef]
- Warth, J.F.; Biesdorf, S.M.; de Souza, C. Yersinia pseudotuberculosis O III causes diarrhea in Brazilian cattle. Adv. Exp. Med. Biol. 2012, 954, 107–110. [Google Scholar] [CrossRef] [PubMed]
- Aleksić, S.; Bockemühl, J.; Wuthe, H.H. Epidemiology of Y. pseudotuberculosis in Germany, 1983–1993. Contrib. MicroBiol. Immunol. 1995, 13, 55–58. [Google Scholar]
- EnteroBase. Yersinia. Available online: https://enterobase.warwick.ac.uk/species/index/yersinia (accessed on 20 November 2020).
- Gupta, V.; Gulati, P.; Bhagat, N.; Dhar, M.S.; Virdi, J.S. Detection of Yersinia enterocolitica in food: An overview. Eur. J. Clin. MicroBiol. Infect. Dis. 2015, 34, 641–650. [Google Scholar] [CrossRef]
- Fukushima, H.; Shimizu, S.; Inatsu, Y. Yersinia enterocolitica and Yersinia pseudotuberculosis detection in foods. J. Pathog. 2011, 2011, 735308. [Google Scholar] [CrossRef] [PubMed]
- International Organization for Standardization (ISO). ISO/FDIS. 10273—Microbiology of the Food Chain—Horizontal Method for the Detection of Pathogenic Yersinia enterocolitica. 2017. Available online: https://infostore.saiglobal.com/en-us/Standards/ISO-10273-2017-588078_SAIG_ISO_ISO_1347019/ (accessed on 14 January 2021).
- European Centre for Disease Prevention and Control (EFSA). Technical specifications for harmonised national surveys of Yersinia enterocolitica in slaughtered pigs. EFSA J. 2009, 7, 1341. [Google Scholar]
- Van Damme, I.; Habib, I.; De Zutter, L. Yersinia enterocolitica in slaughter pig tonsils: Enumeration and detection by enrichment versus direct plating culture. Food MicroBiol. 2010, 27, 158–161. [Google Scholar] [CrossRef]
- Hallanvuo, S.; Herranen, M.; Jaakkonen, A.; Nummela, M.; Ranta, J.; Botteldoorn, N.; De Zutter, L.; Fredriksson-Ahomaa, M.; Hertwig, S.; Johannessen, G.S.; et al. Validation of ISO method 10273—Detection of pathogenic Yersinia enterocolitica in foods. Int. J. Food MicroBiol. 2018. [Google Scholar] [CrossRef]
- Wauters, G.; Goossens, V.; Janssens, M.; Vandepitte, J. New enrichment medium for isolation of pathogenic Yersinia enterocolitica serogroup O:3 in pork. Appl. Environ. MicroBiol. 1988, 54, 851–854. [Google Scholar] [CrossRef]
- Weagant, S.D.; Feng, P. Yersinia enterocolitica. Available online: https://www.fda.gov/food/foodscienceresearch/laboratorymethods/ucm072633.htm (accessed on 14 January 2021).
- Fukushima, H.; Gomyoda, M. Growth of Yersinia pseudotuberculosis and Yersinia enterocolitica biotype 3B serotype O3 inhibited on cefsulodin-Irgasan-novobiocin agar. J. Clin. MicroBiol. 1986, 24, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Jourdan, A.D.; Johnson, S.C.; Wesley, I.V. Development of a fluorogenic 5′ nuclease PCR assay for detection of the ail gene of pathogenic Yersinia enterocolitica. Appl. Environ. MicroBiol. 2000, 66, 3750–3755. [Google Scholar] [CrossRef]
- Boyapalle, S.; Wesley, I.V.; Hurd, H.S.; Reddy, P.G. Comparison of culture, multiplex, and 5′ nuclease polymerase chain reaction assays for the rapid detection of Yersinia enterocolitica in swine and pork products. J. Food Prot. 2001, 64, 1352–1361. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wu, V.C.H.; Fung, D.Y.C.; Oberst, R.B. Evaluation of a 5′-nuclease (TaqMan) assay with the thin agar layer oxyrase method for detection of Yersinia enterocolitica in ground pork samples. J. Food Prot. 2004, 67, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Mäde, D.; Reiting, R.; Strauch, E.; Ketteritzsch, K.; Wicke, A. A real-time PCR for detection of pathogenic Yersinia enterocolitica in food combined with an universal internal amplification control system. J. Verbr. Lebensm. 2008, 3, 141–151. [Google Scholar] [CrossRef]
- Kapperud, G.; Vardund, T.; Skjerve, E.; Hornes, E.; Michaelsen, T.E. Detection of pathogenic Yersinia enterocolitica in foods and water by immunomagnetic separation, nested polymerase chain reactions, and colorimetric detection of amplified DNA. Appl. Environ. MicroBiol. 1993, 59, 2938–2944. [Google Scholar] [CrossRef]
- Lambertz, S.T.; Nilsson, C.; Hallanvuo, S.; Lindblad, M. Real-time PCR method for detection of Yersinia enterocolitica in food. Appl. Environ. MicroBiol. 2008, 74, 6060–6067. [Google Scholar] [CrossRef]
- Nakajima, H.; Inoue, M.; Mori, T.; Itoh, K.-I.; Arakawa, E.; Watanabe, H. Detection and identification of Yersinia pseudotuberculosis and pathogenic Yersinia enterocolitica by an imporved polymerase chain reaction method. J. Clin. MicroBiol. 1992, 30, 2482–2486. [Google Scholar] [CrossRef]
- Kaneko, S.; Ishizaki, N.; Kokubo, Y. Detection of pathogenic Yersinia enterocolitica and Yersinia pseudotuberculosis from pork using the polymerase chain reaction. Contrib. MicroBiol. Immunol. 1995, 13, 153–155. [Google Scholar] [PubMed]
- Petsios, S.; Fredriksson-Ahomaa, M.; Sakkas, H.; Papadopoulou, C. Conventional and molecular methods used in the detection and subtyping of Yersinia enterocolitica in food. Int. J. Food MicroBiol. 2016, 237, 55–72. [Google Scholar] [CrossRef]
- International Organization for Standardization (ISO). ISO/TS 18867—Microbiology of the Food Chain-Polymerase Chain Reaction (PCR) for the Detection of Food-Borne Pathogens- Detection of Pathogenic Yersinia enterocolitica and Yersinia pseudotubculosi; ISO: Geneva, Switzerland, 2015. [Google Scholar]
- Bonardi, S.; Paris, A.; Bassi, L.; Salmi, F.; Bacci, C.; Riboldi, E.; Boni, E.; D’Incau, M.; Tagliabue, S.; Brindani, F. Detection, semiquantitative enumeration, and antimicrobial susceptibility of Yersinia enterocolitica in pork and chicken meats in Italy. J. Food Prot. 2010, 73, 1785–1792. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-z.; Duan, R.; Liang, J.-r.; Huang, Y.; Xiao, Y.-c.; Qiu, H.-y.; Wang, X.; Jing, H.-q. Real-time TaqMan PCR for Yersinia enterocolitica detection based on the ail and foxA genes. J. Clin. MicroBiol. 2014, 52, 4443–4444. [Google Scholar] [CrossRef]
- Seoane, A.; García Lobo, J.M. Cloning of chromosomal beta-lactamase genes from Yersinia enterocolitica. J. Gen. MicroBiol. 1991, 137, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Bonke, R.; Wacheck, S.; Stüber, E.; Meyer, C.; Märtlbauer, E.; Fredriksson-Ahomaa, M. Antimicrobial susceptibility and distribution of β-lactamase A (blaA) and β-lactamase B (blaB) genes in enteropathogenic Yersinia species. Microb. Drug Resist. 2011, 17, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Saraka, D.; Savin, C.; Kouassi, S.; Cisse, B.; Koffi, E.; Cabanel, N.; Bremont, S.; Faye-Kette, H.; Dosso, M.; Carniel, E. Yersinia enterocolitica, a neglected cause of human enteric infections in Cote d’Ivoire. PLoS Negl. Trop. Dis. 2017, 11, e0005216. [Google Scholar] [CrossRef]
- Bhaduri, S.; Wesley, I.; Richards, H.; Draughon, A.; Wallace, M. Clonality and antibiotic susceptibility of Yersinia enterocolitica isolated from U.S. market weight hogs. Foodborne Pathog. Dis. 2009, 6, 351–356. [Google Scholar] [CrossRef]
- Lai, C.-H.; Lin, J.-N.; Chen, Y.-H.; Chang, L.-L.; Huang, W.-Y.; Ku, H.-P.; Lin, H.-H. The first imported human case of Yersinia pseudotuberculosis serotype O1 septicemia presents with acute appendicitis-like syndrome in Taiwan. J. Formos. Med. Assoc. 2014, 113, 656–659. [Google Scholar] [CrossRef] [PubMed]
- Magistrali, C.F.; Cucco, L.; Pezzotti, G.; Farneti, S.; Cambiotti, V.; Catania, S.; Prati, P.; Fabbi, M.; Lollai, S.; Mangili, P.; et al. Characterisation of Yersinia pseudotuberculosis isolated from animals with yersiniosis during 1996–2013 indicates the presence of pathogenic and Far Eastern strains in Italy. Vet. MicroBiol. 2015, 180, 161–166. [Google Scholar] [CrossRef]
- Frazão, M.R.; Andrade, L.N.; Darini, A.L.C.; Falcão, J.P. Antimicrobial resistance and plasmid replicons in Yersinia enterocolitica strains isolated in Brazil in 30 years. Braz. J. Infect. Dis. 2017, 21, 477–480. [Google Scholar] [CrossRef] [PubMed]
- Lucero-Estrada, C.S.; Soria, J.M.; Favier, G.I.; Escudero, M.E. Evaluation of the pathogenic potential, antimicrobial susceptibility, and genomic relations of Yersinia enterocolitica strains from food and human origin. Can. J. MicroBiol. 2015, 61, 851–860. [Google Scholar] [CrossRef]
- Stock, I.; Wiedemann, B. An in-vitro study of the antimicrobial susceptibilities of Yersinia enterocolitica and the definition of a database. J. Antimicrob. Chemother. 1999, 43, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Health and Research Council New Zealand. Unravelling the Mysteries of Yersinosis. Available online: https://hrc.govt.nz/resources/research-repository/unravelling-mysteries-yersiniosis (accessed on 14 January 2021).

| Country | Year | Number of Cases | Biotype/Serotype a | Source | Reference |
|---|---|---|---|---|---|
| New Zealand | 2016 | 24 (21 confirmed, 3 probable) | YE biotype 2 | Not confirmed, suspected sushi | [34] |
| Sweden and Denmark | 2019 | 57 | YE 4, O:3 | Spinach | [80] |
| Japan | 2015 | 4 | YE O:8 | Not stated | [81] |
| Norway | 2014 | 133 | YE O:9 | Mixed salad | [82] |
| Norway | 2011 | 21 | YE O:9 | Mixed salad | [83] |
| United States of America | 2011 | 16 | YE | Pasteurized milk | [84] |
| Australia | 2009 | 3 | YE | Roast pork, Barbequed pork | [85] |
| Norway | 2006 | 11 | YE O:9 | Processed pork | [86] |
| Japan | 2004 | 16 | YE O:8 | Salad (containing apple, cucumbers, ham, potato, carrots and mayonnaise) | [87] |
| United States of America | 2002–2003 | 9 | YE O:3 | Chitterlings (pig intestine) | [88] |
| New Zealand | 2014 | 220 | YP | Not confirmed, suspected produce | [5] |
| Finland | 2014 | 55 | YP O:1 | Raw milk | [89] |
| Finland | 2006 | 104 | YP O:1 | Raw carrots | [90] |
| Finland | 2004 | 53 | YP O:1 | Raw carrots | [91] |
| Finland | 2003 | 111 | YP O:1 | Raw carrots | [92] |
| Country | Year | Species (Outbreak) a | Risk Factors | OR/aOR/mOR (95% CI)/p Value b | Reference |
|---|---|---|---|---|---|
| New Zealand | 1995–1996 | YE | Consumption of pork | OR 1.34 (1.03–1.75) | [71] |
| Eating food from a sandwich bar | OR 1.18 (1.09–1.27) | ||||
| Sweden | 2004 | YE | Eating food prepared from raw pork products | OR 3.0 (1.8–5.1) | [93] |
| Eating treated sausage | OR 1.9 (1.1–3.3) | ||||
| Use of a baby’s dummy | OR 1.9 (1.1–3.2) | ||||
| Contact with domestic animals | OR 2.0 (1.2–3.4) | ||||
| Sweden | 2019 | YE (Outbreak) | Eaten spinach | aOR 1.4 (0.5–3.7) | [80] |
| Denmark | Eaten spinach | aOR 113 (3.7–3400) | |||
| Norway | 2014 | YE O:9 | Eaten salad | OR: 10.26 (0.85–123.57) | [82] |
| Finland | 2006 | YE (Bioserogroups 3–4/O:3, 2/O:9) | Eating or tasting raw or medium done pork | OR 6.6 (1.7–24.9) | [99] |
| Eating in a canteen | OR 3.5 (1.6–7.9) | ||||
| Eating in a restaurant | OR 6.1 (1.4–27.2) | ||||
| YE Biotype 1A | Eating game meat | OR 0.5 (0.2–0.9) | |||
| Consumption of milk and milk products | OR 0.4 (0.1–1.0) | ||||
| Consumption of imported fruits and berries | OR 3.5 (1.2–10.5) | ||||
| Consumption of lettuce and cabbage | OR 0.3 (0.1–0.8) | ||||
| Germany | 2009–2010 | YE | Consumption of raw minced pork | aOR: 4.7 (3.5–6.3) | [100] |
| Preparation of minced pork in the household | aOR: 1.4 (1.1–1.9) | ||||
| Playing in a sandbox | aOR 1.7 (1.3–2.4) | ||||
| Contact with birds | aOR 1.7 (1.1–2.6) | ||||
| Finland | 1998 | YP (Outbreak) | Consumption of iceberg lettuce | mOR: 3.8 (1.3–9.4) | [101] |
| Finland | 2001 | YP | Consumption of iceberg lettuce | mOR: 5.7 (1.6–47.7) | [102] |
| Finland | 2014 | YP (Outbreak) | Consumption of raw milk from a producer | mOR: 22.2 (3.6–∞) | [89] |
| Raw milk in general | mOR: 16.9 (2.6–∞) | ||||
| Norway | 1998–1990 | YE | Consumption of pork items | p = 0.02 | |
| Consumption of sausage | p = 0.03 |
| Country | Sample Tested | YE/YP | Prevalence (%) | Biotype (BT), Serotype | Reference |
|---|---|---|---|---|---|
| New Zealand | Tonsils | YE | 57/200 (28.5% | BT4, O:3; 2, O:5, 27; BT1A | [103] |
| YP | 62/200 (31%) | ||||
| Belgium | Tonsils | YE | 199/360 (55.3%) | Not reported | [111] |
| Feces at slaughter | 92/360 (25.6%) | ||||
| Carcass | 143/360 (39.7%) | ||||
| Tonsils | YP | 5/360 (1.4% | |||
| Feces at slaughter | 2/360 (0.6%) | ||||
| Carcass | 1/360 (0.3%) | ||||
| Brazil | Carcass | YE | 1/400 (0.3%) | BT4, O:3 | [114] |
| Tonsils | 5/100 (5%) | ||||
| Lymph nodes | 2/90 (2%) | ||||
| China | Tonsils | YE | 878/4495 (19.53%) | BT2, O:9; BT4, O:3; BT2, O:3; BT1A | [115] |
| Intestinal contents | 93/1239 (7.5%) | ||||
| Feces | 161/3039 (5.3%) | ||||
| Tonsils | YP | 4/4495 (0.08%) | |||
| Intestinal contents | 0/1239 | ||||
| Feces | 0/3039 | ||||
| Finland | Tonsils | YE | 234/388 (60.3%) PCR only | - | [116] |
| Intestinal samples | 94/356 (26.4%) By culture | BT4, O:3 | |||
| France | Tonsils | YE | 414/3120 (13.7%) | BT3, 4, 5 | [117] |
| Germany | Fecal during rearing period (final) fattening unit) | YE | 96/491 (19.6%) | BT4, O:3 | [118] |
| Feces at slaughter | 2/379 (0.5%) | BT4, O:3 | |||
| Tonsils | 143/372 (38.4%) | BT4, O:3; 2, O:9 | |||
| Lymph nodes | 13/346 (3.8%) | BT4, O:3 | |||
| Carcass (before chilling) | 1/393 (0.3%) | BT4, O:3 | |||
| Carcass (after chilling) | 0/383 | BT4, O:3 | |||
| Italy | Fecal (cecal contents) | YE | 77/451 (17.1%) | BT2, O:9; BT4, O:3; BT1A | [119] |
| Tonsils | 27/250 (10.8%) | BT4, O:3; BT1A | |||
| Carcass | 11/451 (2.4%) | BT2, O:9; BT4, O:3; BT1A | |||
| Scalding water | 4/34 (11.8%) | BT4, O:3; BT1A | |||
| Italy | Tonsils | YE | 55/201 (27.4%) | BT4, O:3 | [120]) |
| YP | 4/201 (2.0%) | Serotypes O:3, O:1 | |||
| Italy | Carcase swabs: finishing pigs | YE | 0/126 (0%) | - | [121] |
| Carcase swabs: piglets | 0/35 (0%) | ||||
| Colon contents: finishing pigs | 15/126 (11.9%) | BT1A; BT 2, O:5; BT4, O:3 | |||
| Colon contents: piglets | 3/35 (8.6%) | ||||
| Tonsils: finishing pigs | 4/126 (3.2%) | BT2, O:5; BT4, O:3 | |||
| Tonsils: piglets | 0/35 (0%) | ||||
| Lymph nodes: finishing pigs | 3/126 (2.8% | BT1A; BT4, O:3 | |||
| Lymph nodes: piglets | 1/35 (2.8%) | ||||
| Ireland | Rectal and environmental swabs | YE | 3/576 (0.52%) | BT2, O:9; BT1A | [122] |
| Rectal swab at abattoir | 1/20 (5%) | BT2, O:9 | |||
| Carcass | 0/20 | - | |||
| Norway | Carcass (before chilling) | YE | 6/60 (10%) | BT4, O:3; BT2, O:9 | [112] |
| Carcass (after chilling) | 5/60 (8.3%) | ||||
| South Korea | Carcass (at slaughter) | YE | 0/100 (0%) | Not reported | [123] |
| Pork samples | YE | 0/300 (0%) | Not reported | ||
| Sweden | Fecal (at farm) | YE | 32/105 (30.5%) | BT4, O:3; BT2, O:9 | [124] |
| Animal | YE Bioserotype a | YP a | Total | |||||
|---|---|---|---|---|---|---|---|---|
| BT1A | BT2/3 O:9 | BT2/3 O:5, 27 | BT2/3 O:1, 2, 3 | BT4 O:3 | BT5 O:2, 3 | |||
| Cattle | 7 | 25 | 21 | 34 | 17 | 104 | ||
| Sheep | 4 | 3 | 1 | 2 | 77 | 1 | 88 | |
| Goats | 3 | 55 | 58 | |||||
| Deer | 12 | 9 | 9 | 5 | 35 | |||
| Alpaca | 3 | 1 | 4 | 3 | 11 | |||
| Dogs | 5 | 4 | 9 | 6 | 2 | 26 | ||
| Cats | 1 | 2 | 1 | 4 | ||||
| Pigs | 3 | 1 | 4 | |||||
| Horses | 3 | 1 | 1 | 5 | ||||
| Birds | 5 | 7 | 12 | |||||
| Total | 42 | 44 | 51 | 2 | 6 | 174 | 28 | 347 |
| Animal | YE BT2/3 O:9 ST12 a | YE BT5 O:3 b ST13 a | YP ST19 a | YP ST43 a | Total |
|---|---|---|---|---|---|
| Cattle | 1 | 2 | 87 | 90 | |
| Unspecified | 2 | 2 | |||
| Budgerigar | 1 | 1 | |||
| Total | 1 | 2 | 89 | 1 | 93 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rivas, L.; Strydom, H.; Paine, S.; Wang, J.; Wright, J. Yersiniosis in New Zealand. Pathogens 2021, 10, 191. https://doi.org/10.3390/pathogens10020191
Rivas L, Strydom H, Paine S, Wang J, Wright J. Yersiniosis in New Zealand. Pathogens. 2021; 10(2):191. https://doi.org/10.3390/pathogens10020191
Chicago/Turabian StyleRivas, Lucia, Hugo Strydom, Shevaun Paine, Jing Wang, and Jackie Wright. 2021. "Yersiniosis in New Zealand" Pathogens 10, no. 2: 191. https://doi.org/10.3390/pathogens10020191
APA StyleRivas, L., Strydom, H., Paine, S., Wang, J., & Wright, J. (2021). Yersiniosis in New Zealand. Pathogens, 10(2), 191. https://doi.org/10.3390/pathogens10020191





