Evaluation of Antibody Response in Symptomatic and Asymptomatic COVID-19 Patients and Diagnostic Assessment of New IgM/IgG ELISA Kits
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design, Ethical Compliance, and Sample Collection
2.2. Serum Samples
2.3. SARS-CoV-2 Antibodies Detection Using Enzyme-Linked Immunosorbent Assay (ELISA)
2.4. Statistical Analysis
3. Results
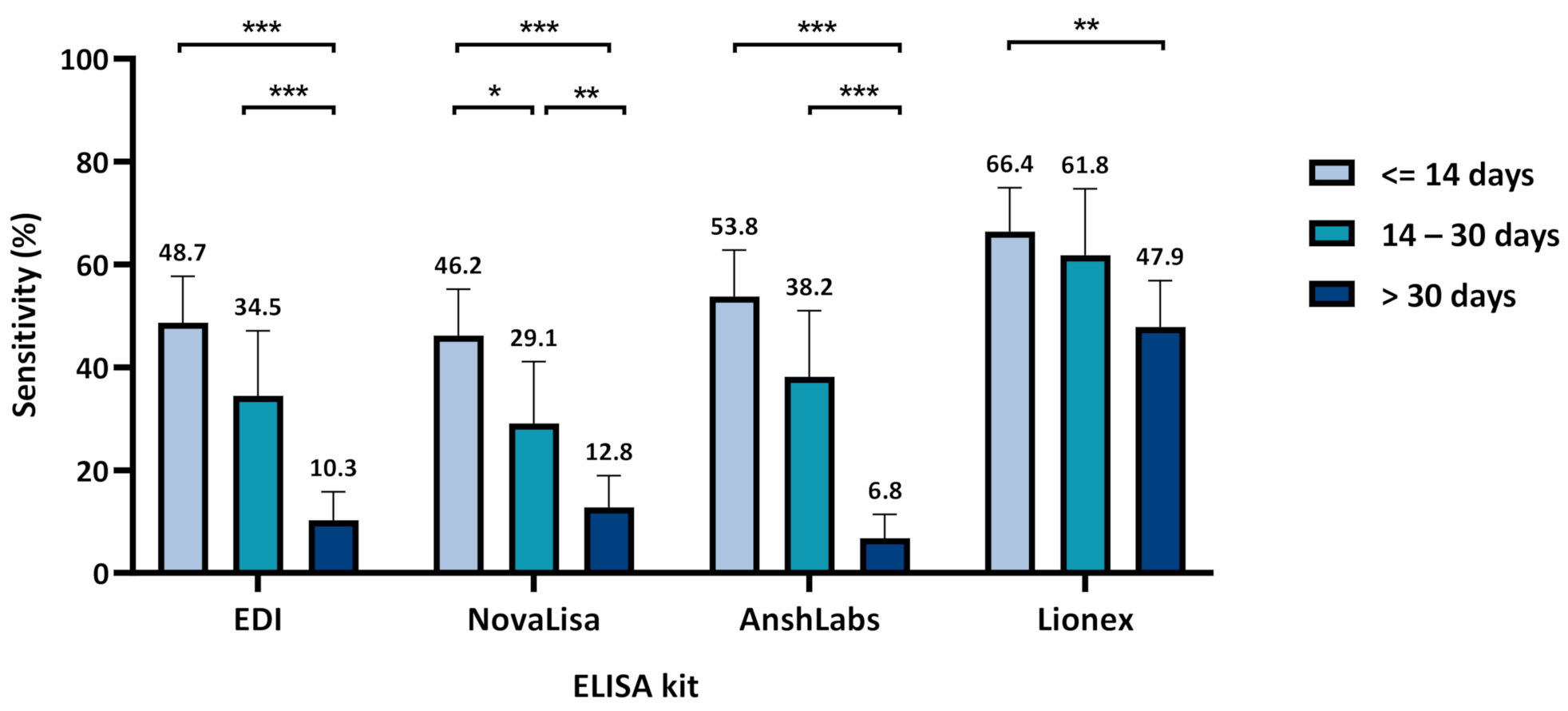
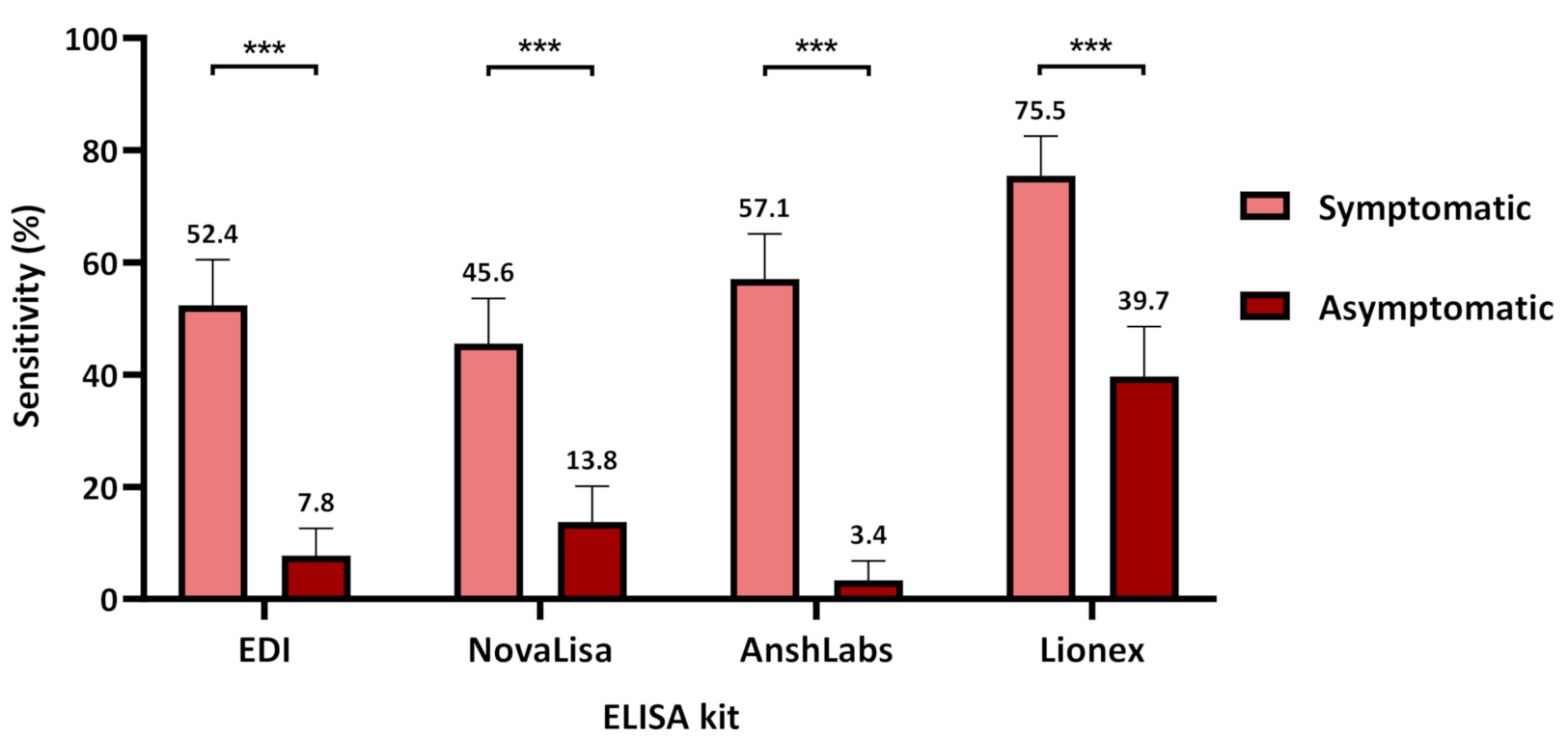
3.1. Diagnostic Assessment of IgM ELISA Kits
3.1.1. Assays Assessment at Different Time Intervals from Sample Collection
3.1.2. Assays Assessment in Symptomatic and Asymptomatic COVID-19 Patients
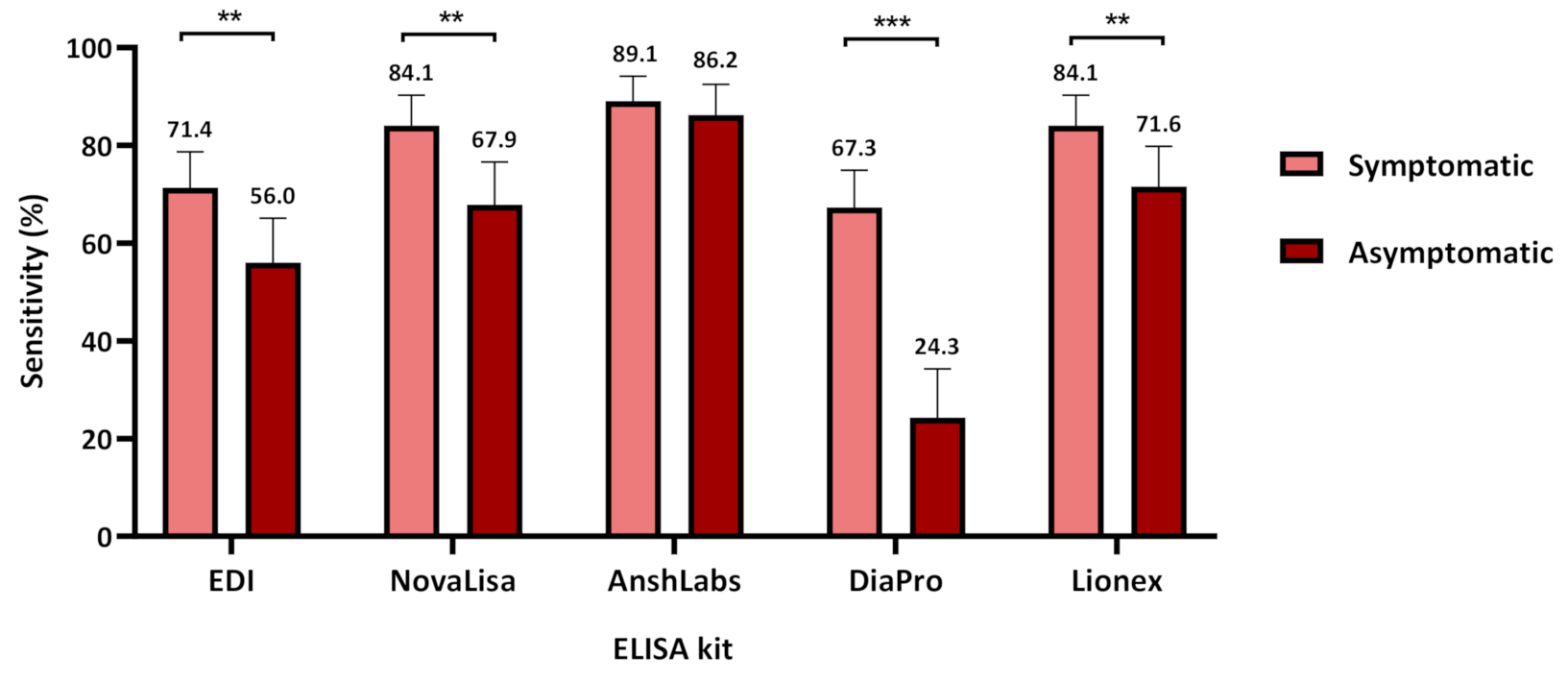
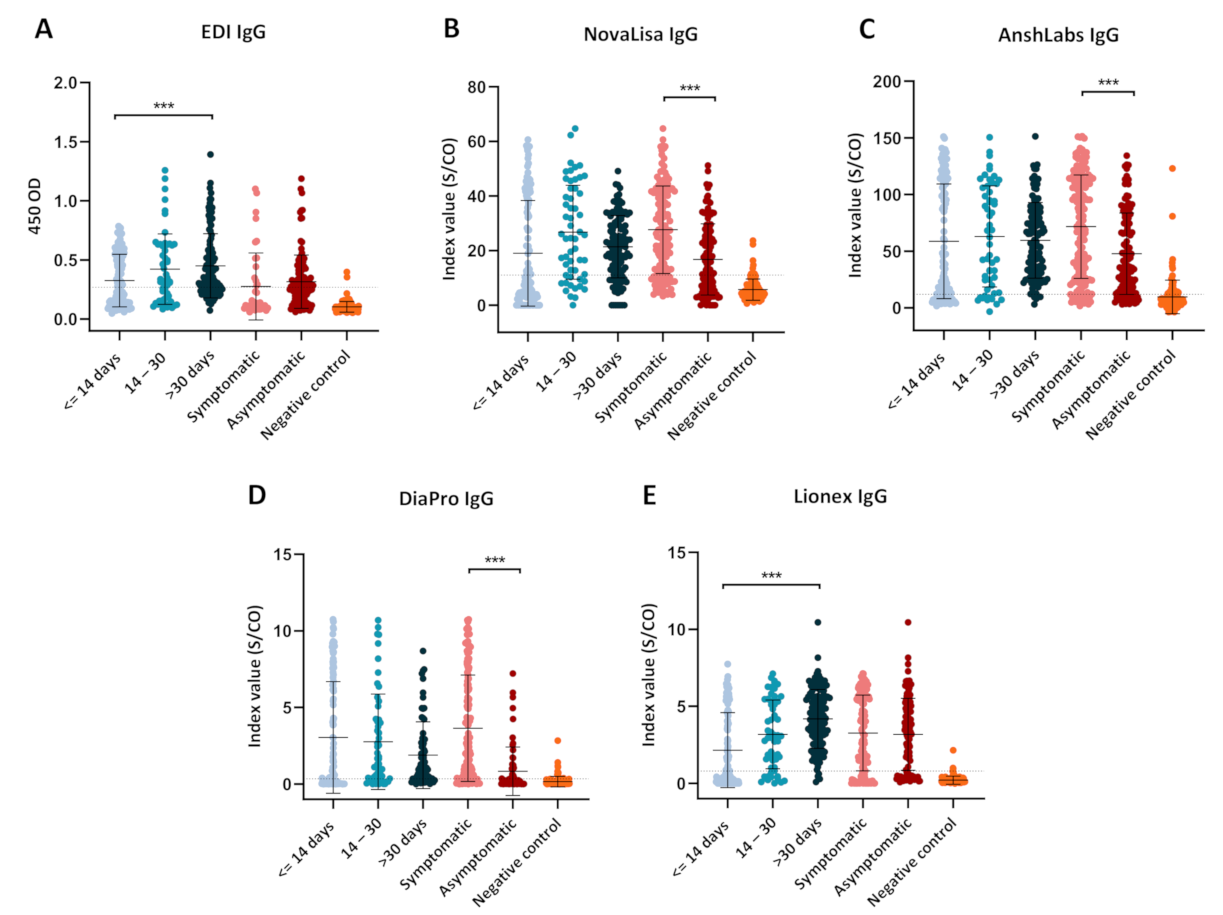
3.2. Diagnostic Assessment of IgG ELISA Kits
3.2.1. Assays Assessment at Different Time Intervals of Sample Collection
3.2.2. Assays Assessment in Symptomatic and Asymptomatic COVID-19 Patients
3.3. Specificity According to the Negative Control Subgroups for IgM and IgG ELISA
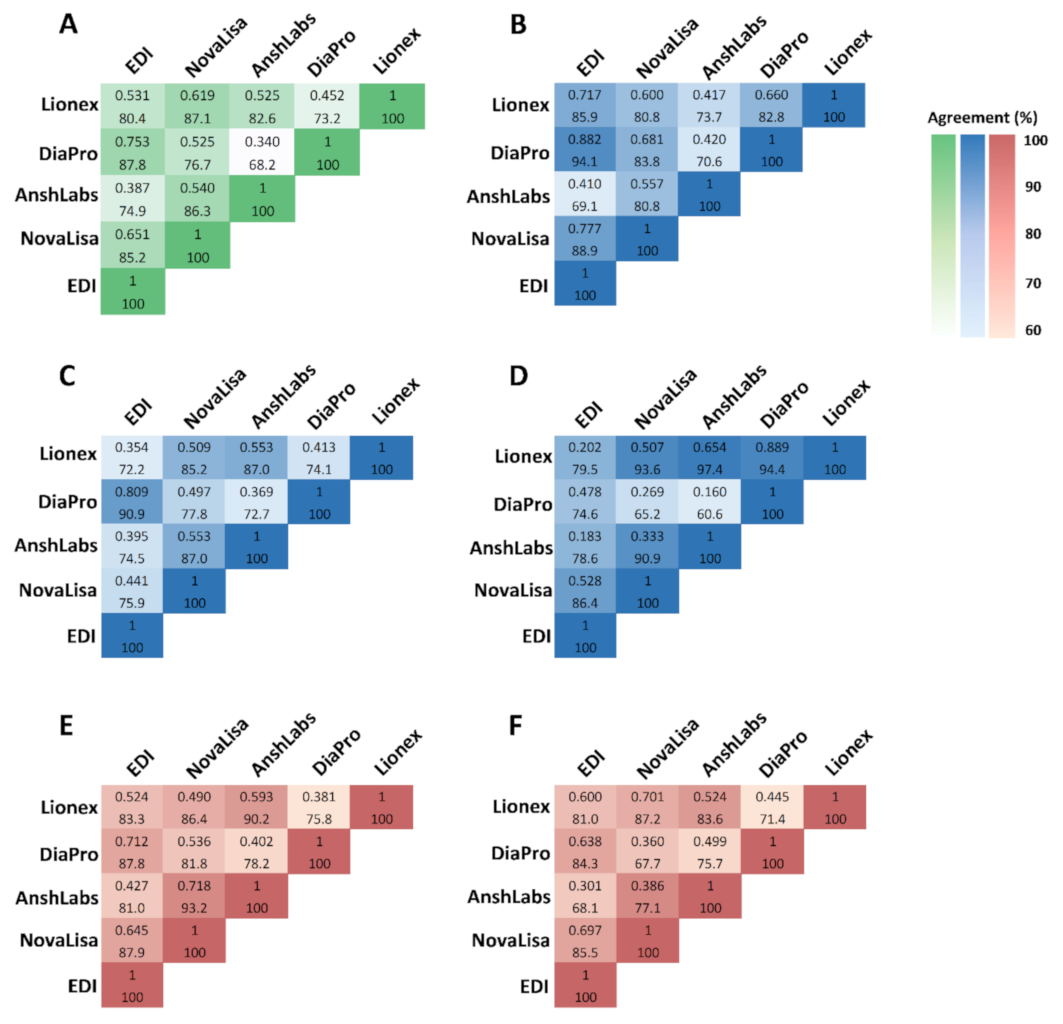
3.4. Agreement of IgM and IgG ELISA Kit
3.5. Negative Samples by All IgG ELISA Kits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dobaño, C.; Vidal, M.; Santano, R.; Jiménez, A.; Chi, J.; Barrios, D.; Ruiz-Olalla, G.; Melero, N.R.; Carolis, C.; Parras, D.; et al. Highly sensitive and specific multiplex antibody assays to quantify immunoglobulins M, A and G against SARS-CoV-2 antigens. bioRxiv 2020. [Google Scholar] [CrossRef] [PubMed]
- Adams, E.R.; Anand, R.; Andersson, M.I.; Auckland, K.; Baillie, J.K.; Barnes, E.; Bell, J.; Berry, T.; Bibi, S.; Carroll, M.; et al. Evaluation of antibody testing for SARS-Cov-2 using ELISA and lateral flow immunoassays. medRxiv 2020. [Google Scholar] [CrossRef]
- Karpiński, T.M.; Ożarowski, M.; Seremak-Mrozikiewicz, A.; Wolski, H.; Wlodkowic, D. The 2020 race towards SARS-CoV-2 specific vaccines. Theranostics 2021, 11, 1690–1702. [Google Scholar] [CrossRef] [PubMed]
- Sewell, H.F.; Agius, R.M.; Kendrick, D.; Stewart, M. Covid-19 vaccines: Delivering protective immunity. BMJ 2020, 371, m4838. [Google Scholar] [CrossRef]
- Winter, A.K.; Hegde, S.T. The important role of serology for COVID-19 control. Lancet Infect. Dis. 2020, 20, 758–759. [Google Scholar] [CrossRef]
- Younes, N.; Al-Sadeq, D.W.; Al-Jighefee, H.; Younes, S.; Al-Jamal, O.; Daas, H.I.; Yassine, H.M.; Nasrallah, G.K. Challenges in Laboratory Diagnosis of the Novel Coronavirus SARS-CoV-2. Viruses 2020, 12, 582. [Google Scholar] [CrossRef]
- Wang, Q.; Du, Q.; Guo, B.; Mu, D.; Lu, X.; Ma, Q.; Guo, Y.; Fang, L.; Zhang, B.; Zhang, G.; et al. A Method To Prevent SARS-CoV-2 IgM False Positives in Gold Immunochromatography and Enzyme-Linked Immunosorbent Assays. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef]
- Long, Q.-X.; Liu, B.-Z.; Deng, H.-J.; Wu, G.-C.; Deng, K.; Chen, Y.-K.; Liao, P.; Qiu, J.-F.; Lin, Y.; Cai, X.-F.; et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020, 26, 845–848. [Google Scholar] [CrossRef]
- Jin, Y.; Wang, M.; Zuo, Z.; Fan, C.; Ye, F.; Cai, Z.; Wang, Y.; Cui, H.; Pan, K.; Xu, A. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int J. Infect. Dis 2020, 94, 49–52. [Google Scholar] [CrossRef]
- Chan, J.F.-W.; Yuan, S.; Kok, K.-H.; To, K.K.-W.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.-Y.; Poon, R.W.-S.; et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef]
- Rothe, C.; Schunk, M.; Sothmann, P.; Bretzel, G.; Froeschl, G.; Wallrauch, C.; Zimmer, T.; Thiel, V.; Janke, C.; Guggemos, W.; et al. Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. N. Engl. J. Med. 2020, 382, 970–971. [Google Scholar] [CrossRef]
- Hu, Z.; Song, C.; Xu, C.; Jin, G.; Chen, Y.; Xu, X.; Ma, H.; Chen, W.; Lin, Y.; Zheng, Y.; et al. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci. China Life Sci. 2020, 63, 706–711. [Google Scholar] [CrossRef] [PubMed]
- Al-Sadeq, D.W.; Nasrallah, G.K. The incidence of the novel coronavirus SARS-CoV-2 among asymptomatic patients: A systematic review. Int. J. Infect. Dis. 2020, 98, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Planning and Statistics Authority—State of Qatar. Qatar Monthly Statistics. 2020. Available online: https://www.psa.gov.qa/en/pages/default.aspx (accessed on 26 May 2020).
- Planning and Statistics Authority—State of Qatar. The Simplified Census of Population, Housing & Establishments. 2019. Available online: https://www.psa.gov.qa/en/statistics/Statistical%20Releases/Population/Population/2018/Population_social_1_2018_AE.pdf (accessed on 2 April 2020).
- World Population Review. Qatar Population (Live). 2020. Available online: https://worldpopulationreview.com/countries/qatar-population/ (accessed on 2 April 2020).
- Priya Dsouza Communications. Population of Qatar by Nationality—2019 Report. Available online: https://priyadsouza.com/population-of-qatar-by-nationality-in-2017/ (accessed on 17 January 2021).
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. Bull. Eur. Sur Les Mal. Transm. Eur. Commun. Dis. Bull. 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed]
- Nasrallah, G.K.; Al Absi, E.S.; Ghandour, R.; Ali, N.H.; Taleb, S.; Hedaya, L.; Ali, F.; Huwaidy, M.; Husseini, A. Seroprevalence of hepatitis E virus among blood donors in Qatar (2013–2016). Transfusion 2017, 57, 1801–1807. [Google Scholar] [CrossRef]
- Smatti, M.K.; Yassine, H.M.; AbuOdeh, R.; AlMarawani, A.; Taleb, S.A.; Althani, A.A.; Nasrallah, G.K. Prevalence and molecular profiling of Epstein Barr virus (EBV) among healthy blood donors from different nationalities in Qatar. PLoS ONE 2017, 12, e0189033. [Google Scholar] [CrossRef]
- Humphrey, J.M.; Al-Absi, E.S.; Hamdan, M.M.; Okasha, S.S.; Al-Trmanini, D.M.; El-Dous, H.G.; Dargham, S.R.; Schieffelin, J.; Abu-Raddad, L.J.; Nasrallah, G.K. Dengue and chikungunya seroprevalence among Qatari nationals and immigrants residing in Qatar. PLoS ONE 2019, 14, e0211574. [Google Scholar] [CrossRef]
- Al-Qahtani, A.A.; Alabsi, E.S.; AbuOdeh, R.; Thalib, L.; El Zowalaty, M.E.; Nasrallah, G.K. Prevalence of anelloviruses (TTV, TTMDV, and TTMV) in healthy blood donors and in patients infected with HBV or HCV in Qatar. Virol. J. 2016, 13, 208. [Google Scholar] [CrossRef]
- Dargham, S.R.; Nasrallah, G.K.; Al-Absi, E.S.; Mohammed, L.I.; Al-Disi, R.S.; Nofal, M.Y.; Abu-Raddad, L.J. Herpes Simplex Virus Type 2 Seroprevalence Among Different National Populations of Middle East and North African Men. Sex. Transm. Dis. 2018, 45, 482–487. [Google Scholar] [CrossRef]
- Nasrallah, G.K.; Dargham, S.R.; Mohammed, L.I.; Abu-Raddad, L.J. Estimating seroprevalence of herpes simplex virus type 1 among different Middle East and North African male populations residing in Qatar. J. Med. Virol 2018, 90, 184–190. [Google Scholar] [CrossRef]
- Smatti, M.K.; Nasrallah, G.K.; Al Thani, A.A.; Yassine, H.M. Measuring influenza hemagglutinin (HA) stem-specific antibody-dependent cellular cytotoxicity (ADCC) in human sera using novel stabilized stem nanoparticle probes. Vaccine 2020, 38, 815–821. [Google Scholar] [CrossRef]
- Al Kahlout, R.A.; Nasrallah, G.K.; Farag, E.A.; Wang, L.; Lattwein, E.; Muller, M.A.; El Zowalaty, M.E.; Al Romaihi, H.E.; Graham, B.S.; Al Thani, A.A.; et al. Comparative Serological Study for the Prevalence of Anti-MERS Coronavirus Antibodies in High- and Low-Risk Groups in Qatar. J. Immunol. Res. 2019, 2019, 1386740. [Google Scholar] [CrossRef] [PubMed]
- Van Elslande, J.; Houben, E.; Depypere, M.; Brackenier, A.; Desmet, S.; André, E.; Van Ranst, M.; Lagrou, K.; Vermeersch, P. Diagnostic performance of seven rapid IgG/IgM antibody tests and the Euroimmun IgA/IgG ELISA in COVID-19 patients. Clin. Microbiol. Infect. 2020, 26, 1082–1087. [Google Scholar] [CrossRef] [PubMed]
- Meyer, B.; Torriani, G.; Yerly, S.; Mazza, L.; Calame, A.; Arm-Vernez, I.; Zimmer, G.; Agoritsas, T.; Stirnemann, J.; Spechbach, H.; et al. Validation of a commercially available SARS-CoV-2 serological Immunoassay. medRxiv 2020. [Google Scholar] [CrossRef] [PubMed]
- Fleiss, J.L.; Levin, B.; Paik, M.C. The measurement of interrater agreement. Stat. Methods Rates Proportions 1981, 2, 22–23. [Google Scholar]
- Serre-Miranda, C.; Nobrega, C.; Roque, S.; Canto-Gomes, J.; Silva, C.S.; Vieira, N.; Barreira-Silva, P.; Alves-Peixoto, P.; Cotter, J.; Reis, A.; et al. Performance assessment of 11 commercial serological tests for SARS-CoV-2 on hospitalized COVID-19 patients. medRxiv 2020. [Google Scholar] [CrossRef]
- Kweon, O.J.; Lim, Y.K.; Kim, H.R.; Kim, M.-C.; Choi, S.-H.; Chung, J.-W.; Lee, M.-K. Antibody kinetics and serologic profiles of SARS-CoV-2 infection using two serologic assays. PLoS ONE 2020, 15, e0240395. [Google Scholar] [CrossRef]
- Ainsworth, M.; Andersson, M.; Auckland, K.; Baillie, J.K.; Barnes, E.; Beer, S.; Beveridge, A.; Bibi, S.; Blackwell, L.; Borak, M.; et al. Performance characteristics of five immunoassays for SARS-CoV-2: A head-to-head benchmark comparison. Lancet Infect. Dis. 2020, 20, 1390–1400. [Google Scholar] [CrossRef]
- Chaudhuri, S.; Thiruvengadam, R.; Chattopadhyay, S.; Mehdi, F.; Kshetrapal, P.; Shrivastava, T.; Desiraju, B.K.; Batra, G.; Kang, G.; Bhatnagar, S. Comparative evaluation of SARS-CoV-2 IgG assays in India. J. Clin. Virol. 2020, 131, 104609. [Google Scholar] [CrossRef]
- Cramer, A.; Goodman, N.; Cross, T.; Gant, V.; Dziadzio, M. Analytical evaluation and critical appraisal of early commercial SARS-CoV-2 immunoassays for routine use in a diagnostic laboratory. Br. J. Biomed. Sci 2020. [Google Scholar] [CrossRef]
- Michel, M.; Bouam, A.; Edouard, S.; Fenollar, F.; Di Pinto, F.; Mège, J.-L.; Drancourt, M.; Vitte, J. Evaluating ELISA, Immunofluorescence, and Lateral Flow Assay for SARS-CoV-2 Serologic Assays. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Whitman, J.D.; Hiatt, J.; Mowery, C.T.; Shy, B.R.; Yu, R.; Yamamoto, T.N.; Rathore, U.; Goldgof, G.M.; Whitty, C.; Woo, J.M.; et al. Evaluation of SARS-CoV-2 serology assays reveals a range of test performance. Nat. Biotechnol. 2020, 38, 1174–1183. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Wang, T.; Zhang, B.; Luo, Y.; Mao, L.; Wang, F.; Wu, S.; Sun, Z. Detection of IgM and IgG antibodies in patients with coronavirus disease 2019. Clin. Transl. Immunol. 2020, 9, e01136. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liang, B.; Chen, C.; Wang, H.; Fang, Y.; Shen, S.; Yang, X.; Wang, B.; Chen, L.; Chen, Q.; et al. SARS-CoV-2 infection induces sustained humoral immune responses in convalescent patients following symptomatic COVID-19. medRxiv 2020. [Google Scholar] [CrossRef]
- Zhao, J.; Yuan, Q.; Wang, H.; Liu, W.; Liao, X.; Su, Y.; Wang, X.; Yuan, J.; Li, T.; Li, J.; et al. Antibody Responses to SARS-CoV-2 in Patients With Novel Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 71, 2027–2034. [Google Scholar] [CrossRef] [PubMed]
- Huynh, A.; Arnold, D.M.; Kelton, J.G.; Smith, J.W.; Moore, J.C.; Chetty, V.T.; Stacey, H.D.; Ang, J.C.; Chagla, Z.; Harvey, B.J.; et al. Development of a serological assay to identify SARS-CoV-2 antibodies in COVID-19 patients. medRxiv 2020. [Google Scholar] [CrossRef]
- Coste, A.T.; Jaton, K.; Papadimitriou-Olivgeris, M.; Greub, G.; Croxatto, A. Comparison of SARS-CoV-2 serological tests with different antigen targets. medRxiv 2020. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.S.; Hock, K.G.; Logsdon, N.M.; Hayes, J.E.; Gronowski, A.M.; Anderson, N.W.; Farnsworth, C.W. Clinical Performance of Two SARS-CoV-2 Serologic Assays. Clin. Chem. 2020, 66, 1055–1062. [Google Scholar] [CrossRef] [PubMed]
- Tay, M.Z.; Poh, C.M.; Rénia, L.; MacAry, P.A.; Ng, L.F.P. The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 2020, 20, 363–374. [Google Scholar] [CrossRef]
- Liu, W.; Liu, L.; Kou, G.; Zheng, Y.; Ding, Y.; Ni, W.; Wang, Q.; Tan, L.; Wu, W.; Tang, S.; et al. Evaluation of Nucleocapsid and Spike Protein-Based Enzyme-Linked Immunosorbent Assays for Detecting Antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Ren, L.; Yang, S.; Xiao, M.; Chang, D.; Yang, F.; Dela Cruz, C.S.; Wang, Y.; Wu, C.; Xiao, Y.; et al. Profiling Early Humoral Response to Diagnose Novel Coronavirus Disease (COVID-19). Clin. Infect. Dis. 2020, 71, 778–785. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, M.; Shen, C.; Wang, F.; Yuan, J.; Li, J.; Zhang, M.; Wang, Z.; Xing, L.; Wei, J.; et al. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. medRxiv 2020. [Google Scholar] [CrossRef]
- Lee, C.Y.-P.; Lin, R.T.P.; Renia, L.; Ng, L.F.P. Serological Approaches for COVID-19: Epidemiologic Perspective on Surveillance and Control. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.W.; Faulkner, N.; Cornish, G.H.; Rosa, A.; Harvey, R.; Hussain, S.; Ulferts, R.; Earl, C.; Wrobel, A.G.; Benton, D.J.; et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science 2020, 370, 1339. [Google Scholar] [CrossRef] [PubMed]
- Tré-Hardy, M.; Wilmet, A.; Beukinga, I.; Favresse, J.; Dogné, J.-M.; Douxfils, J.; Blairon, L. Analytical and clinical validation of an ELISA for specific SARS-CoV-2 IgG, IgA, and IgM antibodies. J. Med. Virol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Kohler, H.; Nara, P. A Novel Hypothesis for Original Antigenic Sin in the Severe Disease of SARS-CoV-2 Infection. Monoclon. Antibodies Immunodiagn. Immunother. 2020, 39, 107–111. [Google Scholar] [CrossRef]
- Fierz, W.; Walz, B. Antibody Dependent Enhancement Due to Original Antigenic Sin and the Development of SARS. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef]
- Midgley, C.M.; Bajwa-Joseph, M.; Vasanawathana, S.; Limpitikul, W.; Wills, B.; Flanagan, A.; Waiyaiya, E.; Tran, H.B.; Cowper, A.E.; Chotiyarnwon, P.; et al. An In-Depth Analysis of Original Antigenic Sin in Dengue Virus Infection. J. Virol. 2011, 85, 410. [Google Scholar] [CrossRef]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef]
- Robertson, L.J.; Moore, J.S.; Blighe, K.; Ng, K.Y.; Quinn, N.; Jennings, F.; Warnock, G.; Sharpe, P.; Clarke, M.; Maguire, K.; et al. Laboratory evaluation of SARS-CoV-2 antibodies: Detectable IgG up to 20 weeks post infection. medRxiv 2020. [Google Scholar] [CrossRef]
- Oved, K.; Olmer, L.; Shemer-Avni, Y.; Wolf, T.; Supino-Rosin, L.; Prajgrod, G.; Shenhar, Y.; Payorsky, I.; Cohen, Y.; Kohn, Y.; et al. Multi-center nationwide comparison of seven serology assays reveals a SARS-CoV-2 non-responding seronegative subpopulation. EClinicalMedicine 2020, 29. [Google Scholar] [CrossRef]
- Yassine, H.M.; Al-Jighefee, H.; Al-Sadeq, D.W.; Dargham, S.R.; Younes, S.N.; Shurrab, F.; Marei, R.M.; Hssain, A.A.; Taleb, S.; Alhussain, H.; et al. Performance evaluation of five ELISA kits for detecting anti-SARS-COV-2 IgG antibodies. Int. J. Infect. Dis. 2021, 102, 181–187. [Google Scholar] [CrossRef] [PubMed]



| Negative Controls | COVID-19 Patients | |||||
|---|---|---|---|---|---|---|
| No (%) | Median (IQR 2) | Range | No (%) | Median (IQR 2) | Range | |
| Age (years) | ||||||
| All | 119 (100) | 36.0 (15.0) | 20.0–69.0 | 291 (100) | 43.0 (21.0) | 12.0–91.0 |
| 10–30 | 23 (19.3) | 52 (17.9) | ||||
| 31–60 | 82 (68.9) | 195 (67.0) | ||||
| 60+ | 2 (1.7) | 27 (9.3) | ||||
| Gender | ||||||
| Female | 57 (49.6) | 33 (11.3) | ||||
| Male | 59 (51.3) | 242 (83.2) | ||||
| Symptomatic | 147 (55.9) | |||||
| Asymptomatic | 116 (44.1) | |||||
| DPSO/DPD 1 | ||||||
| ≤14 days | 119 (40.9) | 8.0 (6.5) | 0–14 | |||
| 14–30 days | 55 (18.9) | 19.5 (7.5) | 14–30 | |||
| >30 days | 117 (40.2) | - | - | |||
| Assay | Manufacturer | Detected Antibody | Principle of Detection | Antigen/Antibody Coating the Plate | Reported Sensitivity | Reported Specificity |
|---|---|---|---|---|---|---|
| EDI™ Novel Coronavirus COVID-19 ELISA Kit | Epitope Diagnostics, Inc. | IgM | Capture ELISA | Anti-human IgM specific capture antibody | 45% (vs. RT-PCR 1) | 100% (vs. PCR) |
| IgG | Indirect ELISA | Recombinant full length nucleocapsid protein | 100% (vs. RT-PCR) | 100% (vs. PCR) | ||
| NovaLisa® SARS-CoV-2 ELISA | NovaLisa Immundiagnostica GmbH | IgM | Indirect ELISA | Recombinant nucleocapsid antigen | 0–30% (<11 days) 40% (≥12 days) (vs. RT-PCR) | 100% |
| IgG | Indirect ELISA | Recombinant nucleocapsid antigen | 8–40% (<11 days) 100% (≥12 days) (vs. RT-PCR) | 99.3% | ||
| AnshLabs SARS-CoV-2 ELISA | AnshLabs | IgM | Capture ELISA | Anti-human IgM specific capture antibody | 100% (vs. CLIA 2) 40% (vs. RT-PCR) | 98.5% (vs. CLIA) 100% (vs. PCR) |
| IgG | Indirect ELISA | Recombinant nucleocapsid and spike antigens | 95% (vs. CLIA) 83.6% (vs. RT-PCR) | 98.3% (vs. CLIA) 91.3% (vs. PCR) | ||
| DiaPro COVID-19 ELISA | Diagnostic Bioprobes | IgG | Indirect ELISA | Recombinant nucleocapsid and spike antigens | ≥98% (vs. RT-PCR) | ≥98% |
| Lionex COVID-19 ELISA | Lionex Diagnostics and Therapeutics | IgM | Indirect ELISA | Recombinant S1 antigen | 62.5% (vs. RT-PCR) | 97.9% |
| IgG | Indirect ELISA | Recombinant S1 antigen | >84% (vs. RT-PCR) | 99.35% |
| Control Subgroup | No. of Samples | IgG ELISA | IgM ELISA | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EDI | NovaLisa | AnshLabs | DiaPro | Lionex | EDI | NovaLisa | AnshLabs | Lionex | ||
| Other coronaviruses (SARS-CoV, MERS-CoV, HCoV-229E, NL63, OC43, and HKU1) | 20 | 19/20 (95.0%) | 17/20 (85.0%) | 17/20 (85.0%) | 19/20 (95.0%) | 20/20 (100%) | 20/20 (100%) | 20/20 (100%) | 17/20 (85.0%) | 15/20 (75.0%) |
| Non-CoV respiratory viruses (H1N1 influenza and RSV) | 28 | 28/28 (100%) | 19/28 (67.9%) | 14/28 (50.0%) | 26/28 (92.9%) | 27/28 (96.4%) | 28/28 (100%) | 25/28 (89.3%) | 27/28 (96.4%) | 26/28 (92.9%) |
| Non-respiratory viruses (HEV, HGV, HCV, HBV, DENV, WNV, CHIKV, B19, HSV-1, HSV-2, EBV, HHV-6, and HHV-8) | 65 | 64/65 (98.5%) | 58/65 (89.2%) | 54/65 (83.1%) | 64/65 (98.5%) | 63/65 (96.9%) | 64/65 (98.5%) | 62/65 (95.4%) | 62/65 (95.4%) | 59/65 (90.8%) |
| Antinuclear antibodies (ANAs) | 6 | 6/6 (100%) | 6/6 (100%) | 5/6 (83.3%) | 6/6 (100%) | 6/6 (100%) | 6/6 (100%) | 6/6 (100%) | 6/6 (100%) | 5/6 (83.3%) |
| Overall Specificity | 119 | 98.3% (117/119:96.0–100) | 96.6% (115/119:93.4–99.9) | 84.0% (100/119:77.5–90.6) | 75.6% (90/119:67.9–83.3) | 97.5% (116/119:94.7–100) | 99.2% (118/119:97.5–100) | 89.1% (106/119:83.5–94.7) | 95.0% (113/119:91.0–98.9) | 88.2% (105/119:82.4–92.0) |
| Sample No. | Days between Sample Collection and Symptoms Onset/Diagnosis | Hospitalized/Non-Hospitalized | Disease Status | Disease Severity |
|---|---|---|---|---|
| 1 | ≤14 days | Hospitalized | Symptomatic | Mild |
| 2 | ≤14 days | Hospitalized | Symptomatic | Mild |
| 3 | ≤14 days | Hospitalized | Symptomatic | Mild |
| 4 | ≤14 days | Hospitalized | Symptomatic | Critical |
| 5 | ≤14 days | Hospitalized | Symptomatic | Mild |
| 6 | ≤14 days | Hospitalized | Symptomatic | Severe |
| 7 | ≤14 days | Hospitalized | Symptomatic | Critical |
| 8 | ≤14 days | Hospitalized | Symptomatic | Critical |
| 9 | ≤14 days | Non-hospitalized | Asymptomatic | - |
| 10 | ≤14 days | Non-hospitalized | Asymptomatic | - |
| 11 | ≤14 days | Non-hospitalized | Asymptomatic | - |
| 12 | ≤14 days | Non-hospitalized | Asymptomatic | - |
| 13 | ≤14 days | Non-hospitalized | Asymptomatic | - |
| 14 | ≤14 days | Non-hospitalized | Asymptomatic | - |
| 15 | ≤14 days | Non-hospitalized | Asymptomatic | - |
| 16 | ≤14 days | Non-hospitalized | Asymptomatic | - |
| 17 | ≤14 days | Non-hospitalized | Asymptomatic | - |
| 18 | 14–30 days | Hospitalized | Symptomatic | Critical |
| 19 | 14–30 days | Non-hospitalized | Symptomatic | Severe |
| 20 | 14–30 days | Non-hospitalized | Asymptomatic | - |
| 21 | >30 days | Non-hospitalized | Asymptomatic | Severe |
| 22 | >30 days | Non-hospitalized | Symptomatic | Mild |
| 23 | >30 days | Non-hospitalized | Asymptomatic | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Jighefee, H.T.; Yassine, H.M.; Al-Nesf, M.A.; Hssain, A.A.; Taleb, S.; Mohamed, A.S.; Maatoug, H.; Mohamedali, M.; Nasrallah, G.K. Evaluation of Antibody Response in Symptomatic and Asymptomatic COVID-19 Patients and Diagnostic Assessment of New IgM/IgG ELISA Kits. Pathogens 2021, 10, 161. https://doi.org/10.3390/pathogens10020161
Al-Jighefee HT, Yassine HM, Al-Nesf MA, Hssain AA, Taleb S, Mohamed AS, Maatoug H, Mohamedali M, Nasrallah GK. Evaluation of Antibody Response in Symptomatic and Asymptomatic COVID-19 Patients and Diagnostic Assessment of New IgM/IgG ELISA Kits. Pathogens. 2021; 10(2):161. https://doi.org/10.3390/pathogens10020161
Chicago/Turabian StyleAl-Jighefee, Hadeel T., Hadi M. Yassine, Maryam A. Al-Nesf, Ali A. Hssain, Sara Taleb, Ahmed S. Mohamed, Hassen Maatoug, Mohamed Mohamedali, and Gheyath K. Nasrallah. 2021. "Evaluation of Antibody Response in Symptomatic and Asymptomatic COVID-19 Patients and Diagnostic Assessment of New IgM/IgG ELISA Kits" Pathogens 10, no. 2: 161. https://doi.org/10.3390/pathogens10020161
APA StyleAl-Jighefee, H. T., Yassine, H. M., Al-Nesf, M. A., Hssain, A. A., Taleb, S., Mohamed, A. S., Maatoug, H., Mohamedali, M., & Nasrallah, G. K. (2021). Evaluation of Antibody Response in Symptomatic and Asymptomatic COVID-19 Patients and Diagnostic Assessment of New IgM/IgG ELISA Kits. Pathogens, 10(2), 161. https://doi.org/10.3390/pathogens10020161







