Abstract
In this research, our aim was to assess the occurrence of Staphylococcus aureus in a Hungarian large-scale dairy farm during the S. aureus control program conducted in the course of our studies. Furthermore, the phenotypic and genotypic properties of the isolates (type of haemolysis, antibiotic susceptibility, staphylococcal enterotoxin (SE) gene carrying ability and spa type) were determined. S. aureus was detected in all bulk tank milk samples collected during this study. Two different spa types were identified among the 17 strains isolated in the farm. A total of 14 of the 17 studied strains (82%) showed β-haemolysis on blood agar, 2/17 strains (12%) expressed double zone and 1/17 strains (6%) showed weak β-haemolysis. All strains were susceptible to most antibiotics tested (cefoxitin, chloramphenicol, clindamycin, erythromycin, gentamicin, tetracycline and trimethoprim/sulphamethoxazole), but all strains were resistant to penicillin G. A total of 11 of the 17 strains (65%) were found to harbour seg, sei, selm, seln, selo genes; 4/17 strains (24%) harboured sei, selm, seln, selo genes and 2/17 strains (11%) harboured sei gene. Since the new SEs/SEls can also cause foodborne outbreaks potentially and all strains were found to be resistant to penicillin G, it is essential to decrease and keep the prevalence of S. aureus low in the dairy farm and the implementation of the S. aureus control program is also highly justified. The results showed that the S. aureus count decreased by the end of our studies, so the control program was proved to be effective.
1. Introduction
Currently, 53 validly published species and 24 subspecies can be distinguished within the genus Staphylococcus [1]. Amongst them, Staphylococcus aureus is one of the most common pathogens associated with contamination of raw milk and dairy products [2]. Staphylococcus spp. are spherical, non-spore-forming Gram-positive bacteria that are facultative anaerobes [3]. Contamination of raw milk with S. aureus can occur in the dairy farm, for example, from the skin, mucous membranes of dairy animals, milking equipment, milkers’ hands, or the milking parlour environment [4].
S. aureus strains that produce staphylococcal enterotoxins (SEs) can cause food poisoning in human [5]. The number of SEs seems to be growing year by year as more and more types are discovered. Only 14 different SEs were identified in 2003 [6]; according to Benkerroum [7], currently, more than 23 SEs and Staphylococcus enterotoxin-like proteins (SEl) can be distinguished. The current nomenclature of SEs uses the name “SE” followed by alphabetic characters in the order of discovery of enterotoxins. The first five SEs (SEA-SEE) are called classical enterotoxins. SEls show significant structural, biological and functional similarities to classical enterotoxins, with the difference that they either do not have the emetic activity characteristic of SEs or further testing is required to determine this [7,8]. SEs and SEls are globular, unique polypeptides with molecular weights ranging from 22,614 to 28,565 Da [3,9]. Genes encoding SEs are carried by mobile genetic elements such as phages, plasmids, and pathogenicity islands. Staphylococcus enterotoxin gene clusters (egc) are pathogenicity island-derived structures that also carry SE genes [10]. These toxins are heat resistant and also resistant to some enzymes [6,11,12,13].
Food poisoning is most commonly caused by the classical enterotoxins SEA and SEB, and among the new SEs/SEls by SEH [14,15]. Umeda et al. [16] suggest that new types of SE/SEls may be potential causes of foodborne outbreaks. An analysis of the causes of an outbreak in 2016 revealed, that S. aureus isolates that carried the seg, sei, sem, sen, seo, and selu genes without carrying the classical se genes may have caused food poisoning. It is, therefore, worth examining not only the genes of classical SEs but also the genes of the new types of SE/SEls. In 2018, a total of 114 outbreaks caused by SEs were reported in the European Union, of which 37 were confirmed outbreaks. None of the total 1124 human cases resulted in death [17].
The spa typing is a single locus DNA-sequencing method of the repeat region of the gene (spa), encoding protein A, in Staphylococci. This test method is reliable and accurate [18]. A grouping algorithm which is “based upon repeat patterns” (BURP) is recently described and is used for evaluating “the ability of spa typing to determine the clonal relatedness of a natural population of S. aureus strains” [19].
The α-, β-, γ-, and δ-haemolysins produced by Staphylococcus spp. are cytotoxic proteins that damage cell membranes [20] and have the effect of dissolving red blood cells of animal origin. α-haemolysin dissolves red blood cells in sheep and rabbit, and can damage tissue, and the β-haemolysin dissolves red blood cells in cattle and sheep [21].
Antibiotics are used to kill pathogenic microorganisms in the human body. It is important to ensure that the infected organism receives the appropriate amount of the antibiotic in an appropriate duration to prevent the development of resistance [22]. Foods (e.g., milk) that contain antibiotic-resistant microorganisms can be ideal tools for introducing antibiotic-resistant strains into the body [23]. Antibiotic resistance in microorganisms is an important health problem worldwide [24,25]. S. aureus is able to adapt to a wide range of environmental conditions and is able to rapidly become resistant to antibiotics [26], for example to β-lactam antibiotics [27].
In this study, our aims were to assess the occurrence of S. aureus in a Hungarian large-scale dairy farm and to monitor the phenotypic and genotypic properties of the bacterium (type of haemolysis, antibiotic susceptibility, enterotoxin gene carrying ability and spa type) during the S. aureus control program conducted in the course of our studies. In terms of the effective control against it, not only the determination of the quantitative data, but also the determination of the characteristics of the bacterium is of great importance.
2. Materials and Methods
2.1. Place and Date of Samplings
One dairy farm was involved in this study, which was located in the eastern part of Hungary. In the farm, Holstein Friesian cows were milked in milking parlour, and pre- and post-milking disinfection were also used. Total mixed ration (TMR) feeding was applied in the farm, and deep litter was used as keeping method. S. aureus control program was applied in the farm, during which, bulk milk samples were collected for our studies between February 2019 and December 2019. In the control program, the stables and the milking parlour were cleaned and disinfected more frequently, the infected cows were isolated and milked separately and their milk was destroyed.
For the microbiological examinations, a total of 35 bulk milk samples were collected in sterile plastic sample tubes from the same milk storage tank containing the milk of all the cows milked in the farm. There were a total of six sampling occasions and 42 to 84 days elapsed between sampling occasions. After sampling, the samples were delivered for analysis to the Microbiological Laboratory of the Institute of Food Science at the University of Debrecen.
2.2. Enumeration and Isolation of Staphylococcus aureus
The raw milk sample preparations and the microbiological examinations were performed as described earlier [28]. The bulk tank milk samples, the initial suspension and the tenfold serial dilutions were prepared in accordance with the MSZ EN ISO 6887-1:2017 [29] standard.
The amount of S. aureus was determined according to the MSZ EN ISO 6888-1:2008 [30] standard. Baird-Parker agar (BPA) (Biolab Ltd., Budapest, Hungary) was used for the tests, which were supplemented with egg yolk tellurit emulsion (LAB-KA Ltd., Karcag, Hungary). S. aureus was differentiated from other Staphylococcus spp. with latex agglutination test kit (Prolex™ Staph Xtra Latex Kit, Biolab Ltd., Budapest, Hungary).
Genetic level identification was also performed for each strain; the prevalence of species-specific thermonuclease gene (nuc) was tested by PCR method as described below. The primers (Table 1) used for the tests have been previously verified to be specific for S. aureus nuc gene [31].

Table 1.
Primers for amplification of genes encoding staphylococcal enterotoxins and thermonuclease.
2.3. Identification of Staphylococcus aureus with MALDI-TOF MS
As a further confirmation, the strains collected from the samples were identified with MALDI-TOF MS in the microbiological laboratory of the Wessling Hungary Kft in Budapest, Hungary. The S. aureus isolates were grown on Columbia Blood agar (Biolab Ltd., Budapest, Hungary) for the tests. A single colony was chosen from the agar and was applied directly to the target plate. One µl of formic acid (70%) was added dropwise to the samples on the target plate and was allowed to dry. Finally, 1 µL of α-HCCA (10 mg/mL) matrix solution was added dropwise to the samples. For the identification of isolates MALDI BioTyper 3.1. Bruker software was used. In the case of the S. aureus strain SA57A, SA57B and SA57C the indentification with the MALD-TOF MS was performed at the Department of Bacteriology, Mycology and Parasitology of the National Public Health Center.
2.4. Spa Typing
The spa typing tests were performed at the Department of Bacteriology, Mycology and Parasitology of the National Public Health Center. The method used by the laboratory is based on the 1.1 version of the description (DNA Sequencing of the spa Gene) of the European Network of Laboratories for Sequence Based Typing of Microbial Pathogens (SeqNet) [32]. BURP clustering for the spa types were performed with default parameters with Ridom Staphtype software (Ridom GmbH, Münster, Germany) as described by Mellmann et al. [19].
2.5. Haemolysis Testing
Haemolysis tests of collected S. aureus strains were performed according to Pereira et al. [33] on Columbia Blood agar.
2.6. Antibiotic Susceptibility Testing
The antibiotic susceptibility test of isolated S. aureus strains was performed in accordance with the Clinical and Laboratory Standards Institute guidelines [34]. The antibiotic disks applied for the tests were the following: cefoxitin (30 μg/disk), chloramphenicol (30 μg/disk), clindamycin (2 μg/disk), erythromycin (15 μg/disk), gentamicin (10 μg/disk), penicillin G (10U), tetracycline (30 μg/disk) and trimethoprim/sulphamethoxazole (1.25 + 23.75 μg/disk) (Biolab Ltd., Budapest, Hungary). Reference strain ATCC 25923 was used as control.
2.7. PCR Amplification of Staphylococcal Enterotoxin-Encoding Genes and Thermonuclease Gene
The PCR tests were performed according to Bianchi et al. [35], with some modifications in the protocol.
Reference strains ATCC 29213 (sea; seg; sei), ATCC 14458 (seb), ATCC 19095 (sec; seg; seh; sei), ATCC 23235 (sed; seg; sei; sej), ATCC 27664 (see) were used as positive control. A strain harbouring selm, seln, selo genes isolated from bulk milk was used as reference strain for the test of these genes.
Extraction of genomic DNA from S. aureus strains was performed with PrepMan™ Ultra Sample Preparation Reagent (Biocenter Ltd., Szeged, Hungary), according to the instructions of the producer.
For PCR tests, seven primer sets were prepared: Set 1. was designed to amplify sea, seb, sec, sed, see; Set 2. was designed to amplify seg, sei; Set 4. was designed to amplify selm, selo; Set 6. was designed to amplify seh and ser. Simplex PCR was used to detect sej (Set 3.) and seln (Set 5.) enterotoxins and the nuc gene (Set 7.). The primer sequences and amplicon sizes are shown in Table 1.
DNA amplification was carried out in a T100™ Thermal Cycler (Bio-Rad Hungary Ltd., Budapest, Hungary). The following amplification cycles were used: the initial denaturation was at 95 °C for five min, followed by 35 amplification cycles (denaturation at 95 °C for 30 s, annealing at 52 °C (56 °C for nuc) for 30 sec, extension at 72 °C for one min). The final extension was at 72 °C for 10 min. The amplified samples were analysed by electrophoresis at 120 V for 40 min with PowerPac Basic power supply (Bio-Rad Hungary Ltd., Hungary). One percent agarose gel (Lab Mark Ltd., Praha, Czech Republic) was used, and 5 µL/1000 mL Midori Green Advance dye (Nucleotest Bio Ltd., Budapest, Hungary) was added to the gel and 1×TBE buffer was used for the gel electrophoresis. A 100 bp ladder (GeneRuler™ 100 bp Plus DNA Ladder, Biocenter Ltd., Szeged, Hungary) was used as molecular weight marker. The bands were visualised with FluorChem M system (Bio-Science Ltd., Budapest, Hungary).
2.8. Statistical Analysis
The averages and standard deviations (SD) were calculated with SPSS v.22.0 [40] software which was also used for the logarithmic transformation (log10) of the amounts and variance analysis.
3. Results
3.1. Enumeration and Isolation of Staphylococcus aureus
Based on the results, S. aureus occurred in all bulk tank milk samples collected during this study. The mean values ranged from 2.0 to 3.5 log10 cfu/mL (Figure 1). In the study of Peles et al. [41] the mean S. aureus count was <2.7 log10 cfu/mL in the same (LF5) farm. In this study the S. aureus count in the samples collected during the sixth sampling (in December; 2.0 log10 cfu/mL) was lower (p < 0.05) than in the samples collected during all the other sampling occasions, including the first sampling (3.1 log10 cfu/mL), which was also performed in winter (February).
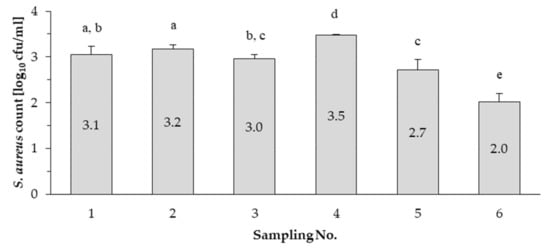
Figure 1.
The mean S. aureus count in bulk milk during the control program. The means marked with the same letters (a, b, c, d or e) do not differ significantly from each other (p > 0.05).
On the other hand, the results also show that the S. aureus count decreased by the end of the year, the amounts did not exceed the refusal limit (M = 2.7 log10 cfu/mL) set in the regulation of the Hungarian Ministry of Health 4/1998 (XI.11) in the case of the last two samplings.
Seventeen S. aureus strains were chosen (two to four strains from each sampling occasion) from bulk tank milk samples for further investigations. Fifteen strains (88%) were dark grey, and two strains (12%) were black on BPA and there were clear zones around the colonies. These results are consistent with the results of Peles et al. [41], because the six S. aureus isolates collected from the same farm (LF5) by the authors had similar properties.
In the case of all the 17 S. aureus strains isolated from bulk milk, the latex agglutination test was positive and the presence of nuc gene was revealed.
3.2. Identification of Staphylococcus aureus with MALDI-TOF MS
Table 2 shows the results of the identification obtained with the MALDI-TOF MS and the results of the spa typing. Based on the results (best score and best-matching isolate) of the tests with the MALDI-TOF MS device, all strains were confirmed as S. aureus and the isolates, except for the SA57A, SA57B and SA57C, matched the spectra of three S. aureus reference strains (1: S. aureus subsp. aureus DSM 20231T; 2: S. aureus subsp. aureus DSM 799; 3: S. aureus ATCC 33862 THL) in the Bruker MALDI Biotyper database. All isolates (except for SA57B) gave MS best scores ≥ 2.300, so the species were reliably identified. In the case of SA57B, the MS best score was 2.270, so the genus identification was secure, but the species identification was only probable.
3.3. Spa Typing
Two spa types (t164 and t1987) were identified among the 17 isolates originated from the bulk milk samples (Table 2). The t164 spa type had eight repeats, and the t1987 spa type had five repeats. The spa types detected in this study were singletons according to the BURP algorithm of the Ridom Staphtype software.

Table 2.
Identification of isolates by MALDI-TOF MS and the results of the spa typing.
Table 2.
Identification of isolates by MALDI-TOF MS and the results of the spa typing.
| Strain ID | Sampling No. | Best Score | Organism (Best Match) * | spa Type | spa Repeats |
|---|---|---|---|---|---|
| SA33 | 1. | 2.361 | 1 | t164 | r07r06r17r21r34r34r22r34 |
| SA34 | 1. | 2.374 | 1 | t164 | r07r06r17r21r34r34r22r34 |
| SA35A | 1. | 2.369 | 1 | t164 | r07r06r17r21r34r34r22r34 |
| SA35B | 1. | 2.374 | 2 | t164 | r07r06r17r21r34r34r22r34 |
| SA39A | 2. | 2.342 | 1 | t164 | r07r06r17r21r34r34r22r34 |
| SA39B | 2. | 2.312 | 3 | t164 | r07r06r17r21r34r34r22r34 |
| SA44 | 3. | 2.419 | 2 | t164 | r07r06r17r21r34r34r22r34 |
| SA45 | 3. | 2.359 | 1 | t164 | r07r06r17r21r34r34r22r34 |
| SA53A | 4. | 2.318 | 1 | t164 | r07r06r17r21r34r34r22r34 |
| SA53B | 4. | 2.410 | 1 | t164 | r07r06r17r21r34r34r22r34 |
| SA53D | 4. | 2.305 | 2 | t164 | r07r06r17r21r34r34r22r34 |
| SA54A | 5. | 2.302 | 2 | t164 | r07r06r17r21r34r34r22r34 |
| SA54B | 5. | 2.371 | 2 | t164 | r07r06r17r21r34r34r22r34 |
| SA54C | 5. | 2.500 | 1 | t1987 | r07r06r17r21r34 |
| SA57A | 6. | 2.333 | - | t164 | r07r06r17r21r34r34r22r34 |
| SA57B | 6. | 2.270 | - | t164 | r07r06r17r21r34r34r22r34 |
| SA57C | 6. | 2.417 | - | t164 | r07r06r17r21r34r34r22r34 |
* 1: S. aureus subsp. aureus DSM 20231T; 2: S. aureus subsp. aureus DSM 799; 3: S. aureus ATCC 33862 THL.
3.4. Haemolysis Testing
A total of 14 of the 17 studied strains (82%) showed β-haemolysis on blood agar, 2 of the 17 strains (12%) had incomplete haemolytic phenotype (expressed double zone) and 1 strain (6%) showed weak β-haemolysis. Table 3 shows the characteristics (tellurite reduction, lecithinase activity, haemolysis type, antibiotic susceptibility, enterotoxin gene harbouring ability) of S. aureus strains isolated from bulk milk.
3.5. Antibiotic Susceptibility Testing
Based on the results, all strains were susceptible to the antibiotic agents tested, except for the penicillin G (Table 3).
3.6. Staphylococcal Enterotoxin-Encoding Genes
All 17 strains were positive for one or more enterotoxin-encoding genes. Among the new types of SEs and SEls, five different enterotoxin-encoding genes were identified. A total of 11 out of 17 isolates (65%) were found to harbour seg, sei, selm, seln, selo genes; 4 of the 17 strains (24%) harboured sei, selm, seln, selo genes and 2 strains (11%) harboured the sei gene alone (Table 3). During the first and the last two samplings, it was found that different enterotoxin-producing strains could be isolated from the collected milk samples originated from the same milk tank in each sampling occasions.

Table 3.
Characteristics of S. aureus strains isolated from bulk milk.
Table 3.
Characteristics of S. aureus strains isolated from bulk milk.
| Strain ID | Sampling No. | Characteristics | ||||
|---|---|---|---|---|---|---|
| Tellurite Reduction | Lecithinase Activity | Hemolysis | Antibiotic Resistance * | Detected Enterotoxin Genes | ||
| SA33 | 1. | dark grey | + | β | R (P) | sei |
| SA34 | 1. | dark grey | + | β | R (P) | seg, sei, selm, seln, selo |
| SA35A | 1. | dark grey | + | β | R (P) | seg, sei, selm, seln, selo |
| SA35B | 1. | dark grey | + | β | R (P) | seg, sei, selm, seln, selo |
| SA39A | 2. | dark grey | + | β | R (P) | sei, selm, seln, selo |
| SA39B | 2. | dark grey | + | β | R (P) | sei, selm, seln, selo |
| SA44 | 3. | dark grey | + | β | R (P) | seg, sei, selm, seln, selo |
| SA45 | 3. | dark grey | + | β | R (P) | seg, sei, selm, seln, selo |
| SA53A | 4. | dark grey | + | β | R (P) | seg, sei, selm, seln, selo |
| SA53B | 4. | dark grey | + | β | R (P) | seg, sei, selm, seln, selo |
| SA53D | 4. | dark grey | + | β | R (P) | seg, sei, selm, seln, selo |
| SA54A | 5. | dark grey | + | α + β | R (P) | seg, sei, selm, seln, selo |
| SA54B | 5. | dark grey | + | α + β | R (P) | sei, selm, seln, selo |
| SA54C | 5. | dark grey | + | weak β | R (P) | sei |
| SA57A | 6. | dark grey | + | β | R (P) | seg, sei, selm, seln, selo |
| SA57B | 6. | black | + | β | R (P) | sei, selm, seln, selo |
| SA57C | 6. | black | + | β | R (P) | seg, sei, selm, seln, selo |
* R: resistant; P: penicillin G.
4. Discussion
According to Zeinhom et al. [42] the somatic cell count and the amount of pathogen microorganisms may increase in summer milk samples and this can be related to the heat stress of cows. Due to heat stress caused by unfavourable weather conditions the incidence of udder infections, such as S. aureus infections can be increasing. In our research, the fourth sampling occasion was in summer, and this may explain the increased S. aureus count (3.5 log10 cfu/mL) in bulk milk. It can also be stated that the S. aureus count decreased by the end of our research, and this is a possible outcome of the S. aureus control program.
The most common spa type was t164; 16/17 strains (94%) belonged to this type. The spa type of the SA54C strain was t1987, which differed from the t164 only in three spa repeats (r34r22r34). Therefore,, the t164 spa type occurred most frequently, the strain with t1987 spa type only occurred while testing the raw milk samples collected during the fifth sampling occasion. In their research, Hwang et al. [43] assorted the similar spa types (t164 and t1987) into one cluster and stated that evolutionary relationship can be displayed in each cluster between the isolates. However, unlike their study, we found that more than one spa type (t164 and t1987) was detected in a single dairy farm.
In the study of Peles et al. [41] three of the six strains (50%) isolated from bulk tank milk from LF5 farm showed weak haemolysis, two of the six strains (33%) isolated from udder quarter milk showed weak haemolysis and one strain (17%) originated from the same origin showed α-haemolysis on blood agar. These results are different from the results of our study. In the study of Morandi et al. [44], 50 of the 81 S. aureus strains (62%) isolated from dairy products made from cow milk showed β-haemolysis, while 29 strains (36%) showed double (α + β) haemolysis and α-haemolysis was occurred in the case of only two (2%) cow isolates. From the 148 S. aureus strains isolated from various food products in the study of Pereira et al. [33] 81% showed β-haemolysis, 11% showed γ-haemolysis and eight percent showed α-haemolysis on blood agar.
Peles et al. [41] also experienced in their study, that all S. aureus isolates collected from the LF5 dairy farm were resistant to penicillin G but were susceptible to the other antibiotics tested (methicillin; cefoxitin; lincomycin; tetracycline; erythromycin; trimethoprim/sulfamethoxazole). Similar results were published previously by other authors: isolates investigated by Abo-Shama [45] were susceptible to cefoxitin, Visciano et al. [46] published strains which were susceptible to gentamicin. In the study of Morandi et al. [44] none of the tested 81 strains isolated from cow dairy products showed resistance to methicillin. Pereira et al. [33] obtained that the isolates collected from bovine mastitis and raw cow’s milk were demonstrated to be the most susceptible to the tested antibiotics (erythromycin, gentamicin, tetracycline, chloramphenicol, ciprofloxacin, rifampicin, ampicillin, penicillin, oxacillin, vancomycin, nitrofurantoin). Among the S. aureus isolated from raw milk by André et al. [47] 71% were resistant to penicillin and 33% were resistant to tetracycline.
Food poisoning is most commonly caused by the classical enterotoxins SEA and SEB, and by the newly described enterotoxin SEH [14,15]. None of the strains examined in this study harboured enterotoxin genes encoding these enterotoxins. According to Umeda et al. [16] new SE/SEls (e.g., seg, sei, selm, seln, selo and selu) can also be potential causes of foodborne outbreaks. In this study, all the strains were found to harbour at least one of these genes; therefore, it is essential in the dairy farm to decrease the occurrence and keep the prevalence of S. aureus low, and the implementation of the S. aureus control program is highly justified, as well. However, it is worth noting that the S. aureus concentration of at least 105 cfu/mL in milk could produce enough enterotoxins to cause food poisoning [48] but this number of bacteria was not present in our samples. In the study of Peles et al. [41] two of the three S. aureus strains isolated from bulk tank milk harboured seb gene. One of the three strains isolated from udder quarter milk harboured seg and sei genes. The third strain isolated from bulk milk and the two other strains isolated from udder quarter milk did not harbour any of the tested enterotoxin genes (sea, seb, sec, sed, see, seg, seh, sei, sej, tsst). Morandi et al. [44] stated that 58 of the 81 S. aureus strains (72%) from dairy products made from cow milk were positive for se. Sea, sed, and sej were found more densely. Only two strains (2%) (isolated from milk and from cheese) were found to have the sec gene and two strains (2%) harboured seg and sei genes. In the study of Pereira et al. [33] seven of the 20 S. aureus strains (35%) isolated from raw cow’s milk harboured se. Three strains (43%) harboured seg and sei, two strains (29%) harboured sec (bovine), and two strains (28%) harboured sec (bovine) and seg genes. Karahan et al. [49] investigated SE genes (classical se-s and seg, seh, sej, sei) by multiplex PCR method in S. aureus strains isolated from bovine mastitis and found that 27 of the strains (29%) harboured one or more se genes, and also sei was the most common. In the study of Korpysa-Dzirba and Osek [50] five out of 66 isolates (7.6%) of S. aureus isolated from raw cow’s milk carried genes encoding classical SE-s. Of the 20 virulence genes investigated by Dai et al. [51] in the case of S. aureus strains isolated from pasteurised milk, seg, sei, and sem were detected the most frequently showing 41.7% of prevalence. sea, seb, sed, see, seu, seq, sej, ser, sek, and pvl were not detected, but sen, sec, sel, seo, sep, seh, and tsst were detected in the isolates.
Hwang et al. [43] found in their study that most of the isolates (seven of eight) with t164 spa type and three of the seven isolates with t1987 spa type harboured seg, sei, selm, seln and selo genes. Similarly, in our study, 11 of the 17 isolates with t164 spa type harboured these genes without harbouring classical enterotoxin genes. The isolate with t1987 spa type harboured only sei gene.
5. Conclusions
The results show that the mean S. aureus count decreased by the end of the year, and the amounts did not exceed the refusal limit in the case of the last two samplings. However, in winter lower amounts of pathogenic bacteria (e.g., coliform bacteria) can be detected in milk [52,53], the first sampling occasion was in winter similarly to the last sampling occasion and yet a significantly higher mean S. aureus count could be detected in milk than that obtained during the test of the samples collected in the last sampling occasion. Thus, the decreasing S. aureus count can be attributed to the effectiveness of the control program. Furthermore, based on our results, S. aureus strains with different characteristics could be detected from raw milk of a single dairy farm. Two different spa types (t164 and t1987) were identified among the 17 strains isolated in the dairy farm. The t1987 spa type isolate was detected during the tests of the milk samples collected during the fifth sampling occasion, but it could not be detected in the sixth sampling occasion. Differences between strains were also reflected in the diversity of haemolysis types, although there was no difference between strains in terms of antibiotic susceptibility. All strains were susceptible to most of the antibiotics tested but were resistant to penicillin G. None of the isolates harboured the classical enterotoxin genes, but all the strains were found to harbour at least one of the seg, sei, selm, seln and selo genes. Since the new SEs/SEls can also be potential cause of foodborne outbreaks, it is essential in the dairy farm to decrease the occurrence and keep the prevalence of S. aureus low.
Author Contributions
Conceptualization, F.M.P., B.B. and F.P.; data curation, F.M.P.; investigation, F.M.P., Á.P., K.D.S, B.H. and E.U.; methodology, K.P., G.K., E.A. and F.P.; supervision, B.B. and F.P.; writing—original draft, F.M.P.; writing—review and editing, F.M.P., K.P., G.K., E.A., B.H., E.U., B.B. and F.P. All authors have read and agreed to the published version of the manuscript.
Funding
The work/publication is supported by the EFOP-3.6.3-VEKOP-16-2017-00008 project. The project is co-financed by the European Union and the European Social Fund.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available within the article.
Acknowledgments
The authors would like to thank the managers and employees of the dairy farm for their helpful contribution, and to the employees of the Wessling Hungary Kft. for their professional assistance during the tests with the MALDI-TOF MS device.
Conflicts of Interest
The authors declare no conflict of interest.
References
- DSMZ (German Collection of Microorganisms and Cell Cultures) Genus Staphylococcus. Available online: https://www.bacterio.net/genus/staphylococcus (accessed on 12 May 2020).
- Akindolire, M.A.; Babalola, O.O.; Ateba, C.N. Detection of Antibiotic Resistant Staphylococcus aureus from Milk: A Public Health Implication. Int. J. Environ. Res. Public Health 2015, 12, 10254–10275. [Google Scholar] [CrossRef] [PubMed]
- Hennekinne, J.; Ostyn, A.; Guillier, F.; Herbin, S.; Prufer, A.; Dragacci, S. How should Staphylococcal food poisoning outbreaks be characterized? Toxins 2010, 2, 2106–2116. [Google Scholar] [CrossRef] [PubMed]
- Bergonier, D.; De Crémoux, R.; Rupp, R.; Lagriffoul, G.; Berthelot, X. Mastitis of dairy small ruminants. Vet. Res. 2003, 34, 689–716. [Google Scholar] [CrossRef] [PubMed]
- Hennekinne, J.; De Buyser, M.; Dragacci, S. Staphylococcus aureus and its food poisoning toxins: Characterization and outbreak investigation. FEMS Microbiol. Rev. 2011, 36, 815–836. [Google Scholar] [CrossRef] [PubMed]
- Loir, Y.L.; Baron, F.; Gautier, M. Staphylococcus aureus and food poisoning. Genet. Mol. Res. 2003, 2, 63–76. [Google Scholar] [PubMed]
- Benkerroum, N. Staphylococcal enterotoxins and enterotoxin-like toxins with special reference to dairy products: An overview. Crit. Rev. Food Sci. Nutr. 2018, 58, 1943–1970. [Google Scholar] [CrossRef]
- Lina, G.; Bohach, G.A.; Nair, S.P.; Hiramatsu, K.; Jouvin-Marche, E.; Mariuzza, R. Standard nomenclature for the superantigens expressed by Staphylococcus. J. Infect. Dis. 2004, 189, 2334–2336. [Google Scholar] [CrossRef]
- Schelin, J.; Wallin-Carlquist, N.; Cohn, M.T.; Lindqvist, R.; Barker, G.C. The formation of Staphylococcus aureus enterotoxin in food environments and advances in risk assessment. Virulence 2011, 2, 580–592. [Google Scholar] [CrossRef]
- Viçosa, G.N.; Moraes, P.M.; Yamazi, A.K.; Nero, L.A. Enumeration of coagulase and thermonuclease-positive Staphylococcus spp. in raw milk and fresh soft cheese: An evaluation of Baird-Parker agar, Rabbit Plasma Fibrinogen agar and the PetrifilmTM Staph Express count system. Food Microbiol. 2010, 27, 447–452. [Google Scholar] [CrossRef]
- Denny, C.B.; Tan, P.L.; Bohrer, C.W. Heat inactivation of staphylococcus enterotoxin A. J. Food Sci. 1966, 31, 762–767. [Google Scholar] [CrossRef]
- Genigeorgis, C.A. Present state of knowledge on staphylococcal intoxication. Int. J. Food Microbiol. 1989, 9, 327–360. [Google Scholar] [CrossRef]
- Gunvig, A.; Andresen, M.S.; Jacobsen, T.; Borggaard, C. Staphtox predictor—A dynamic mathematical model to predict formation of Staphylococcus enterotoxin during heating and fermentation of meat products. Int. J. Food Microbiol. 2018, 285, 81–97. [Google Scholar] [CrossRef] [PubMed]
- Argudín, M.Á.; Mendoza, M.C.; Rodicio, M.R. Food Poisoning and Staphylococcus aureus Enterotoxins. Toxins 2010, 2, 1751–1773. [Google Scholar] [CrossRef] [PubMed]
- Hnasko, R.; Lin, A.V.; McGarvey, J.A. Rapid Detection of Staphylococcal Enterotoxin-B by Lateral Flow Assay. Monoclon. Antib. Immunodiagn. Immunother. 2019, 38, 209–212. [Google Scholar] [CrossRef] [PubMed]
- Umeda, K.; Nakamura, H.; Yamamoto, K.; Nishina, N.; Yasufuku, K.; Hirai, Y.; Hirayama, T.; Goto, K.; Hase, A.; Ogasawara, J. Molecular and epidemiological characterization of staphylococcal foodborne outbreak of Staphylococcus aureus harboring seg, sei, sem, sen, seo, and selu genes without production of classical enterotoxins. Int. J. Food. Microbiol. 2017, 256, 30–35. [Google Scholar] [CrossRef] [PubMed]
- EFSA. The European Union summary report on trends and sources of zoonoses, zoonotic agents and foodborne outbreaks in 2018. EFSA J. 2019, 17, 5926. [Google Scholar] [CrossRef]
- Harmsen, D.; Claus, H.; Witte, W.; Rothgänger, J.; Claus, H.; Turnwald, D.; Vogel, U. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting using a novel software for spa-repeat determination and database management. J. Clin. Microbiol. 2003, 41, 5442–5448. [Google Scholar] [CrossRef]
- Mellmann, A.; Weniger, T.; Berssenbrügge, C.; Keckevoet, U.; Friedrich, A.W.; Harmsen, D.; Grundmann, H. Characterization of clonal relatedness among the natural population of Staphylococcus aureus strains by using spa sequence typing and the BURP (based upon repeat patterns) algorithm. J. Clin. Microbiol. 2008, 46, 2805–2808. [Google Scholar] [CrossRef]
- Vann, J.M.; Proctor, R.A. Cytotoxic effects of ingested Staphylococcus aureus on bovine endothelial cells: Role of S. aureus α-hemolysin. Microb. Pathog. 1988, 4, 443–453. [Google Scholar] [CrossRef]
- Medveczky, I.; Rusvai, M.; Varga, J.; Tuboly, S. Állatorvosi Járványtan I.—Állatorvosi Mikrobiológia, Bakteriológia, Virológia, Immunológia; Mezőgazda Kiadó: Budapest, Hungary, 1999; p. 612. [Google Scholar]
- Barcs, I. Az antibiotikum-érzékenység és-rezisztencia. In A Semmelweis Egyetem Egészségtudományi Kar Népegészségtani Intézetének kiadványa; Barcs, I., Ed.; Semmelweis Egyetem, Egészségtudományi Kar, Népegészségtani Intézet: Budapest, Hungary, 2009; p. 40. [Google Scholar]
- Angulo, F.J.; Nargund, V.N.; Chiller, T.C. Evidence of an association between use of anti-microbial agents in food animals and anti-microbial resistance among bacteria isolated from humans and the human health consequences of such resistance. J. Vet. Med. B 2004, 51, 374–379. [Google Scholar] [CrossRef]
- Carmeli, Y.; Troillet, N.; Karchmer, A.W.; Samore, M.H. Health and economic outcomes of antibiotic resistance in Pseudomonas aeruginosa. Arch. Intern. Med. 1999, 159, 1127–1132. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, S.E. The relationship between antimicrobial resistance and patient outcomes: Mortality, length of hospital stay, and health care costs. Clin. Infect. Dis. 2006, 42, S82–S89. [Google Scholar] [CrossRef] [PubMed]
- McCallum, N.; Berger-Bachi, B.; Senn, M.M. Regulation of antibiotic resistance in Staphylococcus aureus. Int. J. Med. Microbiol. 2010, 300, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Chambers, H.F.; DeLeo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Petróczki, F.M.; Tonamo, T.A.; Béri, B.; Peles, F. The effect of breed and stage of lactation on the microbiological status of raw milk. Acta Agrar. Debr. 2019, 1, 37–45. [Google Scholar] [CrossRef]
- MSZ EN ISO 6887-1. Élelmiszerek és Takarmányok Mikrobiológiája. A Vizsgálati Minták, az Alapszuszpenzió és a Decimális Hígítások Elkészítése Mikrobiológiai Vizsgálathoz. 1. Rész: Az Alapszuszpenzió és a Decimális Hígítások Elkészítésének Általános Szabályai; ISO: Geneva, Switzerland, 2017. [Google Scholar]
- MSZ EN ISO 6888-1. Élelmiszerek és Takarmányok Mikrobiológiája. Horizontális Módszer a Koagulázpozitív Sztafilokokkuszok (Staphylococcus Aureus és más Fajok) Számának Meghatározása. 1. Rész: Baird-Parker-agar Táptalajos Eljárás; ISO: Geneva, Switzerland, 2008. [Google Scholar]
- Brakstad, O.G.; Aasbakk, K.; Maeland, J.A. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J. Clin. Microbiol. 1992, 30, 1654–1660. [Google Scholar] [CrossRef]
- SeqNet (European Network of Laboratories for Sequence Based Typing of Microbial Pathogens) DNA Sequencing of the Spa Gene. 2004. Available online: https://www.seqnet.org/downloads.html (accessed on 1 October 2020).
- Pereira, V.; Lopes, C.; Castro, A.; Silva, J.; Gibbs, P.; Teixeira, P. Characterization for enterotoxin production, virulence factors, and antibiotic susceptibility of Staphylococcus aureus isolates from various foods in Portugal. Food Microbiol. 2009, 26, 278–282. [Google Scholar] [CrossRef]
- Clinical Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. In M100, 27th ed.; Replaces M100-S26; CLSI: Wayne, NJ, USA, 2017; 282p. [Google Scholar]
- Bianchi, D.M.; Gallina, S.; Bellio, A.; Chiesa, F.; Civera, T.; Decastelli, L. Enterotoxin gene profiles of Staphylococcus aureus isolated from milk and dairy products in Italy. Lett. Appl. Microbiol. 2014, 58, 190–196. [Google Scholar] [CrossRef]
- Mehrotra, M.; Wang, G.; Johnson, W.M. Multiplex PCR for Detection of Genes for Staphylococcus aureus Enterotoxins, Exfoliative Toxins, Toxic Shock Syndrome Toxin 1, and Methicillin Resistance. J. Clin. Microbiol. 2000, 38, 1032–1035. [Google Scholar] [CrossRef]
- Sharma, N.K.; Rees, C.E.D.; Dodd, C.E.R. Development of a single-reaction multiplex PCR toxin typing assay for Staphylococcus aureus strains. Appl. Environ. Microbiol. 2000, 66, 1347–1353. [Google Scholar] [CrossRef]
- Bania, J.; Dabrowska, A.; Bystron, J.; Korzekwa, K.; Chrzanowska, J.; Molenda, J. Distribution of newly described enterotoxin-like genes in Staphylococcus aureus from food. Int. J. Food Microbiol. 2006, 108, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.-C.; Liao, W.-W.; Fan, C.-M.; Pai, W.-Y.; Chiou, C.-S.; Tsen, H.-Y. PCR detection of Staphylococcal enterotoxins (SEs) N, O, P, Q, R, U, and survey of SE types in Staphylococcus aureus isolates from food-poisoning cases in Taiwan. Int. J. Food Microbiol. 2008, 121, 66–73. [Google Scholar] [CrossRef] [PubMed]
- SPSS 220 for Windows; SPSS Inc.: Chicago, IL, USA, 2013.
- Peles, F.; Wagner, M.; Varga, L.; Hein, I.; Rieck, P.; Gutser, K.; Keresztúri, P.; Kardos, G.; Turcsányi, I.; Béri, B.; et al. Characterization of Staphylococcus aureus strains isolated from bovine milk in Hungary. Int. J. Food Microbiol. 2007, 118, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Zeinhom, M.M.A.; Aziz, R.A.L.; Mohammed, A.N.; Bernabucci, U. Impact of seasonal conditions on quality and pathogens content of milk in Friesian cows. Asian Australas. J. Anim. Sci. 2016, 29, 1207–1213. [Google Scholar] [CrossRef]
- Hwang, S.Y.; Park, Y.K.; Koo, H.C.; Park, Y.H. spa typing and enterotoxin gene profile of Staphylococcus aureus isolated from bovine raw milk in Korea. J. Vet. Sci. 2010, 11, 125–131. [Google Scholar] [CrossRef][Green Version]
- Morandi, S.; Brasca, M.; Andrighetto, C.; Lombardi, A.; Lodi, R. Phenotypic and genotypic characterization of Staphylococcus aureus strains from Italian dairy products. Int. J. Microbiol. 2009, 2009, 1–7. [Google Scholar] [CrossRef]
- Abo-Shama, U.H. Prevalence and antimicrobial susceptibility of Staphylococcus aureus isolated from cattle, buffalo, sheep and goats raws milk in Sohag governorate, Egypt. Assiut. Vet. Med. J. 2014, 60, 63–72. [Google Scholar]
- Visciano, P.; Pomilio, F.; Tofalo, R.; Sacchini, L.; Saletti, M.A.; Tieri, E.; Schirone, M.; Suzzi, G. Detection of methicillin-resistant Staphylococcus aureus in dairy cow farms. Food Control 2014, 46, 532–538. [Google Scholar] [CrossRef]
- André, M.C.D.P.B.; Campos, M.R.H.; Borges, L.J.; Kipnis, A.; Pimenta, F.C.; Serafini, Á.B. Comparison of Staphylococcus aureus isolates from food handlers, raw bovine milk and Minas Frescal cheese by antibiogram and pulsed-field gel electrophoresis following SmaI digestion. Food Control 2008, 19, 200–207. [Google Scholar] [CrossRef]
- Karahan, M.; Açik, M.N.; Cetinkaya, B. Investigation of toxin genes by polymerase chain reaction in Staphylococcus aureus strains isolated from bovine mastitis in Turkey. Foodborne Pathog. Dis. 2009, 6, 1029–1035. [Google Scholar] [CrossRef]
- Korpysa-Dzirba, W.; Osek, J. Identification of genes encoding classical staphylococcal enterotoxins in Staphylococcus aureus isolated from raw milk. Bull. Vet. Inst. Pulawy 2011, 55, 55–58. [Google Scholar]
- Dai, J.; Wu, S.; Huang, J.; Wu, Q.; Zhang, F.; Zhang, J.; Wang, J.; Ding, Y.; Zhang, S.; Yang, X.; et al. Prevalence and characterization of Staphylococcus aureus isolated from pasteurized milk in China. Front. Microbiol. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Cziszter, L.T.; Acatincăi, S.; Neciu, F.C.; Neamţ, R.I.; Ilie, D.E.; Costin, L.I.; Gavojdian, D.; Tripon, I. The influence of season on the cow milk quantity, quality and hygiene. Sci. Pap. Anim. Sci. Biotechnol. 2012, 45, 305–312. [Google Scholar]
- Petróczki, F.M.; Béri, B.; Peles, F. The effect of season on the microbiological status of raw milk. Acta Agrar. Debr. 2020, 1, 95–99. [Google Scholar] [CrossRef]
- Hill, B.; Smythe, B.; Lindsay, D.; Shepherd, J. Microbiology of raw milk in New Zealand. Int. J. Food Microbiol. 2012, 157, 305–308. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).