Tracking the Transcription Kinetic of SARS-CoV-2 in Human Cells by Reverse Transcription-Droplet Digital PCR
Abstract
:1. Introduction
2. Materials and Methods
2.1. SARS-CoV-2 Sequences and Alignment
2.2. Primers and Dual-Labeled Hydrolysis Probes
2.3. Viruses and Cells
2.4. Virus Infection and Collection of Cell Lysate
2.5. Preparation of cDNA Templates
2.6. Endogenous Control and Absolute Quantification by ddPCR
2.7. Validation of the Primers and Probes on Clinical Specimens by RT-QPCR
2.8. Data and Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- WHO Website. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 26 August 2021).
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. China Medical Treatment Expert Group for Covid-19. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef] [PubMed]
- Mariano, G.; Farthing, R.J.; Lale-Farjat, S.L.M.; Bergeron, J.R.C. Structural Characterization of SARS-CoV-2: Where We Are, and Where We Need to Be. Front. Mol. Biosci. 2020, 7, 605236. [Google Scholar] [CrossRef] [PubMed]
- Hui, K.P.Y.; Cheung, M.C.; Perera, R.A.P.M.; Ng, K.C.; Bui, C.H.T.; Ho, J.C.W.; Ng, M.M.T.; Kuok, D.I.T.; Shih, K.C.; Tsao, S.W.; et al. Tropism, replication competence, and innate i mMune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: An analysis in ex-vivo and in-vitro cultures. Lancet. Respir. Med. 2020, 8, 687–695. [Google Scholar] [CrossRef]
- V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Wong, N.A.; Saier, M.H., Jr. The SARS-Coronavirus Infection Cycle: A Survey of Viral Membrane Proteins, Their Functional Interactions and Pathogenesis. Int. J. Mol. Sci. 2021, 22, 1308. [Google Scholar] [CrossRef] [PubMed]
- Knoops, K.; Kikkert, M.; Worm, S.H.; Zevenhoven-Dobbe, J.C.; van der Meer, Y.; Koster, A.J.; Mo mMaas, A.M.; Snijder, E.J. Sars-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008, 6, e226. [Google Scholar] [CrossRef] [PubMed]
- Snijder, E.J.; Limpens, R.; de Wilde, A.H.; de Jong, A.W.M.; Zevenhoven-Dobbe, J.C.; Maier, H.J.; Faas, F.; Koster, A.J.; Barcena, M. A unifying structural and functional model of the coronavirus replication organelle: Tracking down rna synthesis. PLoS Biol. 2020, 18, e3000715. [Google Scholar] [CrossRef] [PubMed]
- Telwatte, S.; Martin, H.A.; Marczak, R.; Fozouni, P.; Vallejo-Gracia, A.; Kumar, G.R.; Murray, V.; Lee, S.; Ott, M.; Wong, J.K.; et al. Novel RT-ddPCR assays for measuring the levels of subgenomic and genomic SARS-CoV-2 transcripts. Methods. 2021, 18, S1046-2023(21)00103-1. [Google Scholar] [CrossRef]
- Telwatte, S.; Kumar, N.; Vallejo-Gracia, A.; Kumar, G.R.; Lu, C.M.; Ott, M.; Wong, J.K.; Yukl, S.A. Novel RT-ddPCR Assays for determining the transcriptional profile of SARS-CoV-2. J. Virol. Methods. 2021, 114115. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Lee, J.Y.; Yang, J.S.; Kim, J.W.; Kim, V.N.; Chang, H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2021, 181, 914–921.e10. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.C.; Laperriere, G.; Germain, H. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: From variable nonsense to publication quality data. Sci. Rep. 2017, 7, 2409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.B.; Choi, H.; Lee, G.D.; Lee, J.; Lee, S.; Kim, Y.; Cho, S.Y.; Lee, D.G.; Kim, M. Analytical and Clinical Performance of Droplet Digital PCR in the Detection and Quantification of SARS-CoV-2. Mol. Diagn. Ther. 2021. In press. [Google Scholar]
- Zella, D.; Giovanetti, M.; Benedetti, F.; Unali, F.; Spoto, S.; Guarino, M.; Angeletti, S.; Ciccozzi, M. The variants question: What is the problem? J. Med. Virol. 2021. In press. [Google Scholar] [CrossRef] [PubMed]
- Nyaruaba, R.; Li, C.; Mwaliko, C.; Mwau, M.; Odiwuor, N.; Muturi, E.; Muema, C.; Xiong, J.; Li, J.; Yu, J.; et al. Developing multiplex ddPCR assays for SARS-CoV-2 detection based on probe mix and amplitude based multiplexing. Expert Rev. Mol. Diagn. 2021, 21, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, M.A.; Sethna, P.B.; Brian, D.A. Bovine coronavirus mRNA replication continues throughout persistent infection in cell culture. J. Virol. 1990, 64, 4108–4114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
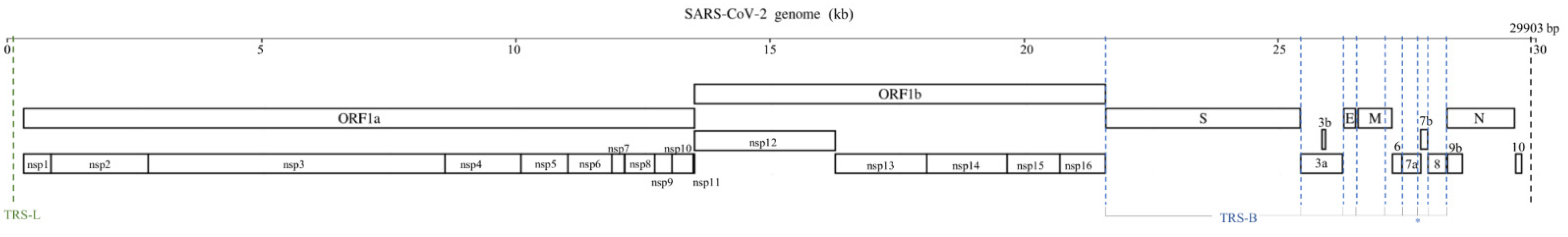
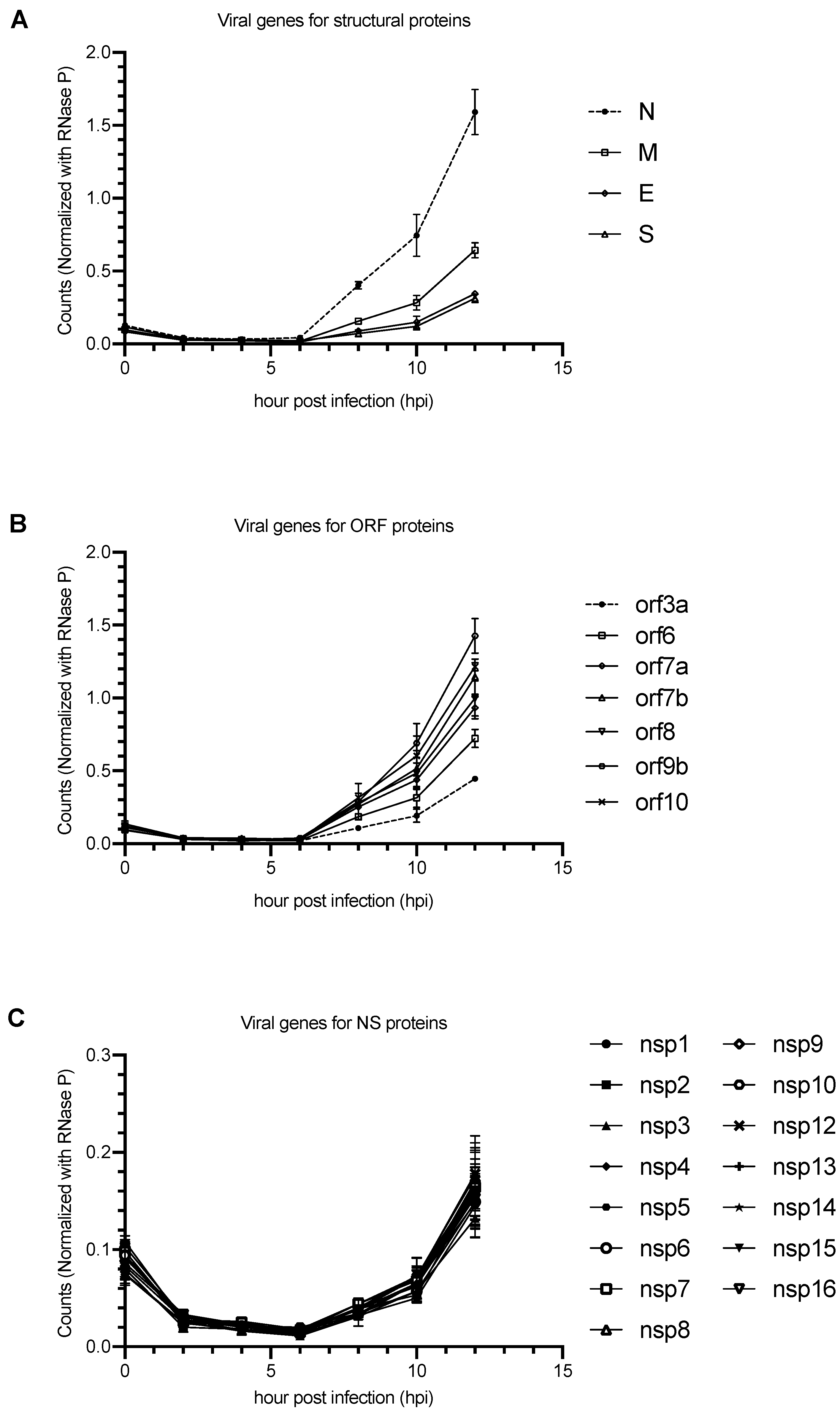
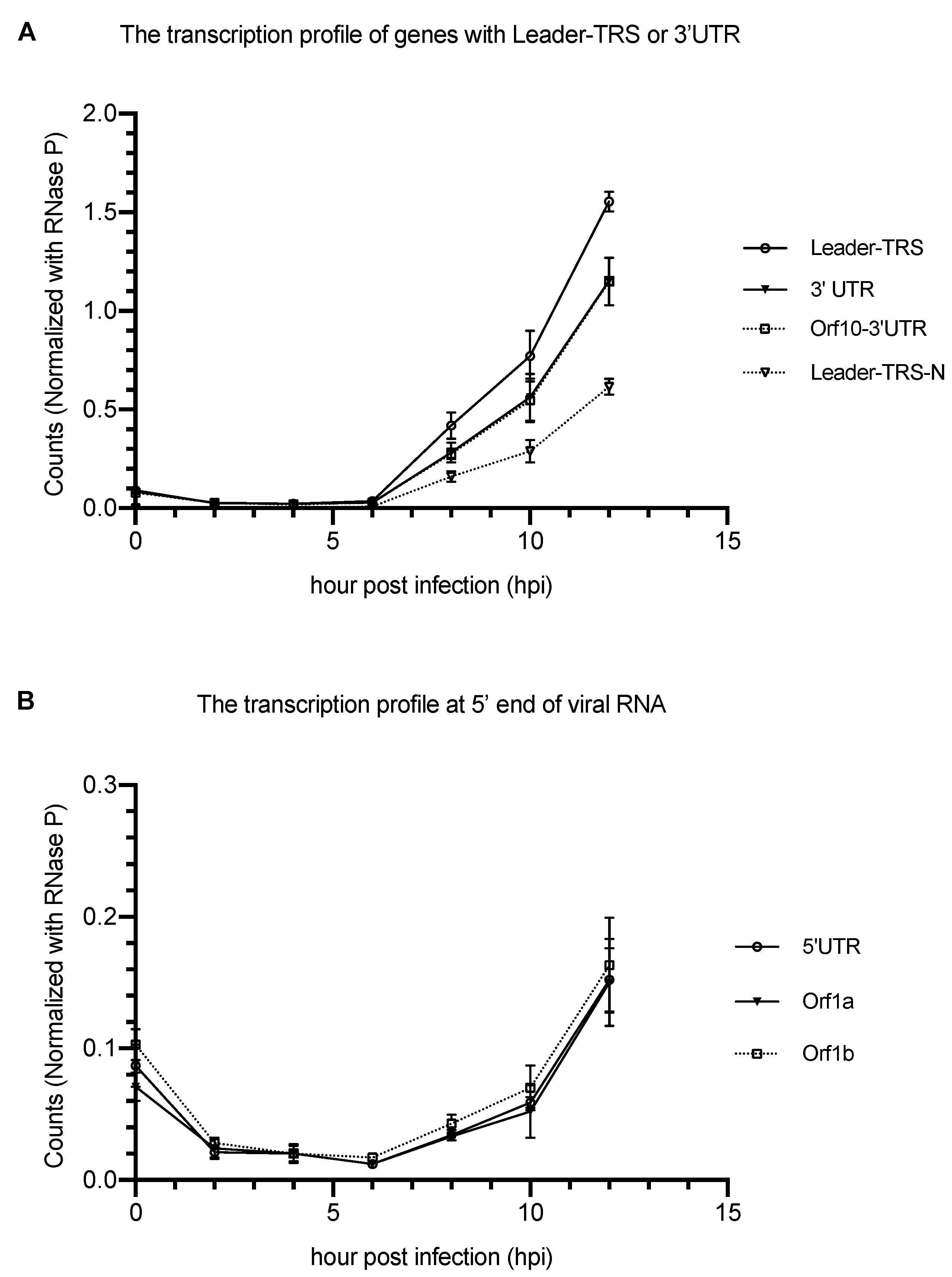
| Gene Categories | Target Regions | Primer/Probe | Sequence (5′ to 3′) | Position | Amplicon Size |
|---|---|---|---|---|---|
| Structural Proteins | S (A) | Forward | GTGACATCTCTGGCATTAATGC | 25062–25173 | 112 |
| Reverse | CCAAGTTCTTGGAGATCGATGAG | ||||
| Probe | TGGCAACCTCATTGAGGCGGTC | ||||
| E | Forward | GGTACGTTAATAGTTAATAGCGTAC | 26272–26395 | 124 | |
| Reverse | GACTCACGTTAACAATATTGCAG | ||||
| Probe | TCCTTACTGCGCTTCGATTGTGTG | ||||
| M | Forward | GTGGACATCTTCGTATTGCTG | 26959–27081 | 123 | |
| Reverse | CACGCTGCGAAGCTCCCAA | ||||
| Probe | CAACAGTGATTTCTTTAGGCAGGTCC | ||||
| N (A) | Forward | GAAGTCACACCTTCGGGAAC | 29240–29323 | 84 | |
| Reverse | GACTTGATCTTTGAAATTTGGATCT | ||||
| Probe | TGGTTGACCTACACAGGTGCCATC | ||||
| Nonstructural Proteins (NSP) | orf1a (nsp2–3) | Forward | CCCTTGCACCTAATATGATGG | 2667–2762 | 96 |
| Reverse | CTTCTATCACAGTGTCATCACC | ||||
| Probe | CTCAAAGGCGGTGCACCAACAAAG | ||||
| orf1b (nsp12–13) | Forward | CACTTCAAGGTATTGGGAACC | 16173–16304 | 132 | |
| Reverse | GGTCTACGTATGCAAGCACC | ||||
| Probe | CAGTCTTACAGGCTGTTGGGGCTT | ||||
| nsp1 | Forward | TTCAACGAGAAAACACACGTCC | 287–407 | 121 | |
| Reverse | CTTTAAGATGTTGACGTGCCTC | ||||
| Probe | CTTTGGAGACTCCGTGGAGGAGG | ||||
| nsp2 | Forward | GTATTAACGGGCTTATGTTGCTC | 2616–2706 | 91 | |
| Reverse | GTGAAGGTATTGTTTGTTACCATC | ||||
| Probe | CAGAAAAGTACTGTGCCCTTGCACC | ||||
| nsp3 (A) | Forward | GACATAGAAGTTACTGGCGATAG | 8249–8367 | 119 | |
| Reverse | GCATTAATATGACGCGCACTAC | ||||
| Probe | CATGACACCCCGTGACCTTGG | ||||
| nsp4 | Forward | GCTACAGAGAAGCTGCTTGTT | 9939–10051 | 113 | |
| Reverse | CAAAACAGCTGAGGTGATAGAG | ||||
| Probe | CATCAGAACCTGAGTTACTGAAGTC | ||||
| nsp5 | Forward | GGAGTTCATGCTGGCACAGA | 10562–10685 | 124 | |
| Reverse | CAGCGTACAACCAAGCTAAAAC | ||||
| Probe | ACAAGCAGCTGGTACGGACACAAC | ||||
| nsp6 | Forward | GTGTTATGTATGCATCAGCTGTAG | 11310–11404 | 95 | |
| Reverse | ATTCATAAGTGTCCACACTCTCC | ||||
| Probe | CACCATCATCATACACAGTTCTTGC | ||||
| nsp7 | Forward | GTCAGATGTAAAGTGCACATCAG | 11851–11945 | 95 | |
| Reverse | ACTGGACACATTGAGCCCACA | ||||
| Probe | CTCAGTTTTGCAACAACTCAGAGTAG | ||||
| nsp8 | Forward | GGCTAAATCTGAATTTGACCGTG | 12223–12296 | 74 | |
| Reverse | GGGTCATAGCTTGATCAGCC | ||||
| Probe | CCAACTTACGTTGCATGGCTGCA | ||||
| nsp9 | Forward | CTAAGAGTGATGGAACTGGTAC | 12855–12938 | 84 | |
| Reverse | CTTTAGGACCTTTAGGTGTGTCT | ||||
| Probe | CCTACAAGGTGGTTCCAGTTCTG | ||||
| nsp10 | Forward | TGCTGTAGATGCTGCTAAAGCT | 13025–13169 | 88 | |
| Reverse | TGTGTGTACACAACATCTTAACAC | ||||
| Probe | TGGTTGTCCCCCACTAGCTAGA | ||||
| nsp12 | Forward | GTCATGTGTGGCGGTTCACT | 15439–15510 | 72 | |
| Reverse | AGCATAAGCAGTTGTGGCATC | ||||
| Probe | CCTGATGAGGTTCCACCTGGTTTAAC | ||||
| nsp13 | Forward | CTATAGGTCCAGACATGTTCCTC | 17528–17602 | 75 | |
| Reverse | CCAAAGCACTCACAGTGTCAAC | ||||
| Probe | CAGCAGGACAACGCCGACAAGTTC | ||||
| nsp14 | Forward | GTATAACACGTTGCAATTTAGGTG | 19457–19604 | 148 | |
| Reverse | GTGTTCCAGAGGTTATAAGTATC | ||||
| Probe | TCAGCTGGCTTTAGCTTGTGGGTT | ||||
| nsp15 | Forward | GCATTTGAGCTTTGGGCTAAGC | 19780–19861 | 82 | |
| Reverse | CAGCAATGTCCACACCCAAAT | ||||
| Probe | CAACATTAAACCAGTACCAGAGGTG | ||||
| nsp16 | Forward | CAGGTACAGCTGTTTTAAGACAG | 20897–20977 | 81 | |
| Reverse | CATCAGAGACAAAGTCATTAAGATC | ||||
| Probe | CAGCGTACCCGTAGGCAACC | ||||
| Accessory Proteins | orf3a | Forward | CAAGGTGAAATCAAGGATGCTAC | 25441–25517 | 77 |
| Reverse | GGGAGTGAGGCTTGTATCGG | ||||
| Probe | CTTCAGATTTTGTTCGCGCTACTGC | ||||
| orf6 | Forward | GTTTCATCTCGTTGACTTTCAGG | 27204–27289 | 86 | |
| Reverse | CAAGATTCCAAATGGAAACTTTAAAAG | ||||
| Probe | CCTCATAATAATTAGTAATATCTCTGC | ||||
| orf7a | Forward | GCTTTAGCACTCAATTTGCTTTTGC | 27566–27640 | 75 | |
| Reverse | AAACTGATCTGGCACGTAACTG | ||||
| Probe | TGTCCTGACGGCGTAAAACACGTC | ||||
| orf7b | Forward | GCTTTTTAGCCTTTCTGCTATTCC | 27790–27884 | 95 | |
| Reverse | GGCGTGACAAGTTTCATTATGATC | ||||
| Probe | CTTTTGGTTCTCACTTGAACTGC | ||||
| orf8 | Forward | CAGCACCTTTAATTGAATTGTGC | 28054–28193 | 140 | |
| Reverse | CACTACAAGACTACCCAATTTAGG | ||||
| Probe | CCCATTCAGTACATCGATATCGG | ||||
| orf9b | Forward | CCCAATAATACTGCGTCTTGG | 28409–28492 | 84 | |
| Reverse | TGGAACGCCTTGTCCTCGAG | ||||
| Probe | CACCGCTCTCACTCAACATGGC | ||||
| orf10 | Forward | TGGGCTATATAAACGTTTTCGCT | 29559–29642 | 84 | |
| Reverse | GTGCTATGTAGTTACGAGAATTC | ||||
| Probe | CCGTTTACGATATATAGTCTACTC | ||||
| Others | Leader–TRS | Forward | TTAAAGGTTTATACCTTCCCAGG | 2–75 | 74 |
| Reverse | GTTCGTTTAGAGAACAGATCTAC | ||||
| Probe | AACAAACCAACCAACTTTCGATCTCT | ||||
| 5′ UTR | Forward | GACAGGACACGAGTAACTCG | 155–229 | 75 | |
| Reverse | TGCTGATGATCGGCTGCAAC | ||||
| Probe | CTGCAGGCTGCTTACGGTTTCG | ||||
| 3′ UTR | Forward | CACCACATTTTCACCGAGGC | 29719–29795 | 77 | |
| Reverse | CCATATAGGCAGCTCTCCC | ||||
| Probe | CTGTACACTCGATCGTACTCCGC | ||||
| Leader–TRS–N | Forward | CCCAGGTAACAAACCAACCAAC | 19–28332 | N/A | |
| Reverse | GGTCCACCAAACGTAATGCG | ||||
| Probe | CCCCAAAATCAGCGAAATGCACC | ||||
| orf10-3′ UTR | Forward | GAATTCTCGTAACTACATAGCAC | 29620–29743 | 124 | |
| Reverse | GCGTGGCCTCGGTGAAAATG | ||||
| Probe | CATTAGGGAGGACTTGAAAGAGCC | ||||
| S (B) | Forward | GTTCTTGTGGATCCTGCTGC | 25305–25378 | 74 | |
| Reverse | GTAATGTAATTTGACTCCTTTGAGC | ||||
| Probe | TGATGAAGACGACTCTGAGCCAG | ||||
| N (B) | Forward | CTCATCACGTAGTCGCAACAG | 28831–28940 | 110 | |
| Reverse | GCAGCAAAGCAAGAGCAGCA | ||||
| Probe | CCTGCTAGAATGGCTGGCAATGGC | ||||
| nsp3 (B) | Forward | CGTTAAAGATTTCATGTCATTGTCTG | 8407–8513 | 107 | |
| Reverse | CTTGTCTAGTAGTTGCACATGTC | ||||
| Probe | CTACGAAAACAAATACGTAGTGCTGCT | ||||
| Endogenous Controls | RNase P | Forward | AGATTTGGACCTGCGAGCG | 28–114 | 87 |
| Reverse | GCAACAACTGAATAGCCAAGG | ||||
| Probe | TTCTGACCTGAAGGCTCTGCGCG |
| Sample (CT Value) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| S | 14.6 | 16.9 | 15.9 | 25.1 | 24.5 | 25.8 | 26.6 | 26.4 | 19.5 | 16.6 | 15.7 | 13.7 |
| E | 17.5 | 19.9 | 18.4 | 28.1 | 27.7 | 28.8 | 29.9 | 29.4 | 22.3 | 19.3 | 18.0 | 16.5 |
| M | 17.7 | 19.6 | 19.1 | 27.8 | 26.1 | 29.0 | 29.8 | 29.5 | 22.3 | 19.5 | 18.8 | 16.9 |
| N | 12.8 | 15.4 | 15.5 | 22.8 | 23.6 | 24.6 | 25.1 | 25.4 | 17.1 | 13.9 | 13.3 | 11.9 |
| orf1a | 14.4 | 16.5 | 15.6 | 24.3 | 24.5 | 24.9 | 26.5 | 26.3 | 19.7 | 16.7 | 15.8 | 13.4 |
| orf1b | 13.4 | 16.2 | 15.7 | 24.0 | 23.7 | 25.1 | 25.6 | 25.6 | 19.3 | 16.4 | 15.5 | 13.4 |
| nsp1 | 14.0 | 16.6 | 15.6 | 24.4 | 24.2 | 25.6 | 26.0 | 26.4 | 19.8 | 16.9 | 16.0 | 13.7 |
| nsp2 | 14.9 | 17.4 | 16.6 | 25.2 | 24.4 | 26.0 | 26.3 | 26.8 | 20.3 | 17.5 | 16.8 | 14.2 |
| nsp3 | 14.8 | 17.2 | 16.7 | 24.9 | 23.7 | 26.0 | 26.0 | 26.6 | 19.9 | 17.3 | 16.4 | 13.9 |
| nsp4 | 15.8 | 18.0 | 18.0 | 26.3 | 24.4 | 26.9 | 27.6 | 27.5 | 21.1 | 18.3 | 17.5 | 15.1 |
| nsp5 | 15.4 | 18.0 | 18.1 | 25.6 | 24.4 | 26.8 | 27.6 | 27.8 | 21.1 | 18.4 | 17.6 | 15.2 |
| nsp6 | 18.5 | 19.5 | 19.6 | 28.0 | 28.3 | 28.7 | 30.5 | 30.5 | 23.7 | 20.5 | 20.9 | 17.7 |
| nsp7 | 15.5 | 17.6 | 16.8 | 25.4 | 24.5 | 25.9 | 27.2 | 26.9 | 20.9 | 17.9 | 17.3 | 15.0 |
| nsp8 | 14.3 | 17.1 | 16.3 | 24.9 | 24.4 | 25.5 | 26.5 | 26.3 | 20.2 | 17.4 | 16.8 | 14.2 |
| nsp9 | 15.0 | 17.4 | 16.6 | 25.4 | 25.2 | 26.3 | 26.8 | 27.3 | 20.6 | 17.9 | 17.3 | 14.7 |
| nsp10 | 14.9 | 17.6 | 17.3 | 25.4 | 25.4 | 26.5 | 27.2 | 27.5 | 20.8 | 18.0 | 17.2 | 14.7 |
| nsp12 | 14.9 | 17.7 | 17.4 | 25.6 | 25.0 | 26.2 | 27.1 | 26.5 | 20.5 | 18.1 | 17.5 | 14.8 |
| nsp13 | 14.0 | 16.6 | 16.3 | 24.4 | 24.0 | 24.9 | 26.2 | 27.2 | 19.7 | 17.0 | 16.2 | 13.9 |
| nsp14 | 14.1 | 16.8 | 16.6 | 24.5 | 24.2 | 25.4 | 26.2 | 26.2 | 19.8 | 17.0 | 16.2 | 13.9 |
| nsp15 | 15.6 | 18.0 | 17.5 | 25.9 | 25.8 | 26.6 | 27.4 | 27.2 | 21.1 | 18.4 | 17.5 | 15.3 |
| nsp16 | 13.6 | 16.4 | 15.9 | 24.2 | 23.5 | 24.6 | 25.8 | 25.4 | 19.4 | 16.4 | 15.6 | 13.6 |
| orf3a | 15.3 | 17.3 | 15.6 | 25.1 | 25.6 | 25.8 | 27.4 | 27.4 | 19.5 | 16.5 | 15.8 | 13.9 |
| orf6 | 16.8 | 18.9 | 18.0 | 27.2 | 28.1 | 28.9 | 29.9 | 29.6 | 21.0 | 18.1 | 17.7 | 15.6 |
| orf7a | 13.3 | 16.2 | 15.4 | 24.0 | 24.5 | 25.2 | 26.2 | 25.8 | 18.3 | 15.0 | 14.4 | 12.7 |
| orf7b | 13.8 | 15.9 | 14.8 | 23.9 | 24.3 | 25.1 | 25.9 | 25.6 | 17.9 | 14.8 | 14.6 | 12.3 |
| orf8 | 14.2 | 16.5 | 15.2 | 24.1 | 24.6 | 25.3 | 26.5 | 25.9 | 20.2 | 17.2 | 16.5 | 14.9 |
| orf9b | 12.8 | 14.7 | 13.9 | 22.6 | 23.5 | 23.7 | 25.3 | 24.8 | 15.7 | 12.9 | 13.0 | 10.6 |
| orf10 | 16.0 | 17.9 | 16.9 | 25.7 | 27.3 | 27.2 | 29.2 | 28.5 | 19.8 | 16.4 | 16.0 | 14.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Au, K.-K.; Chen, C.; Chan, Y.-M.; Wong, W.W.S.; Lv, H.; Mok, C.K.P.; Chow, C.-K. Tracking the Transcription Kinetic of SARS-CoV-2 in Human Cells by Reverse Transcription-Droplet Digital PCR. Pathogens 2021, 10, 1274. https://doi.org/10.3390/pathogens10101274
Au K-K, Chen C, Chan Y-M, Wong WWS, Lv H, Mok CKP, Chow C-K. Tracking the Transcription Kinetic of SARS-CoV-2 in Human Cells by Reverse Transcription-Droplet Digital PCR. Pathogens. 2021; 10(10):1274. https://doi.org/10.3390/pathogens10101274
Chicago/Turabian StyleAu, Ka-Ki, Chunke Chen, Yee-Man Chan, Winsome Wing Sum Wong, Huibin Lv, Chris Ka Pun Mok, and Chun-Kin Chow. 2021. "Tracking the Transcription Kinetic of SARS-CoV-2 in Human Cells by Reverse Transcription-Droplet Digital PCR" Pathogens 10, no. 10: 1274. https://doi.org/10.3390/pathogens10101274
APA StyleAu, K.-K., Chen, C., Chan, Y.-M., Wong, W. W. S., Lv, H., Mok, C. K. P., & Chow, C.-K. (2021). Tracking the Transcription Kinetic of SARS-CoV-2 in Human Cells by Reverse Transcription-Droplet Digital PCR. Pathogens, 10(10), 1274. https://doi.org/10.3390/pathogens10101274






