Role of m6A mRNA Methylation in Plant Defense
Abstract
1. Introduction
2. Insights into m6A mRNA Modifications in Plants
3. m6A RNA Modifications During Fungal Infection
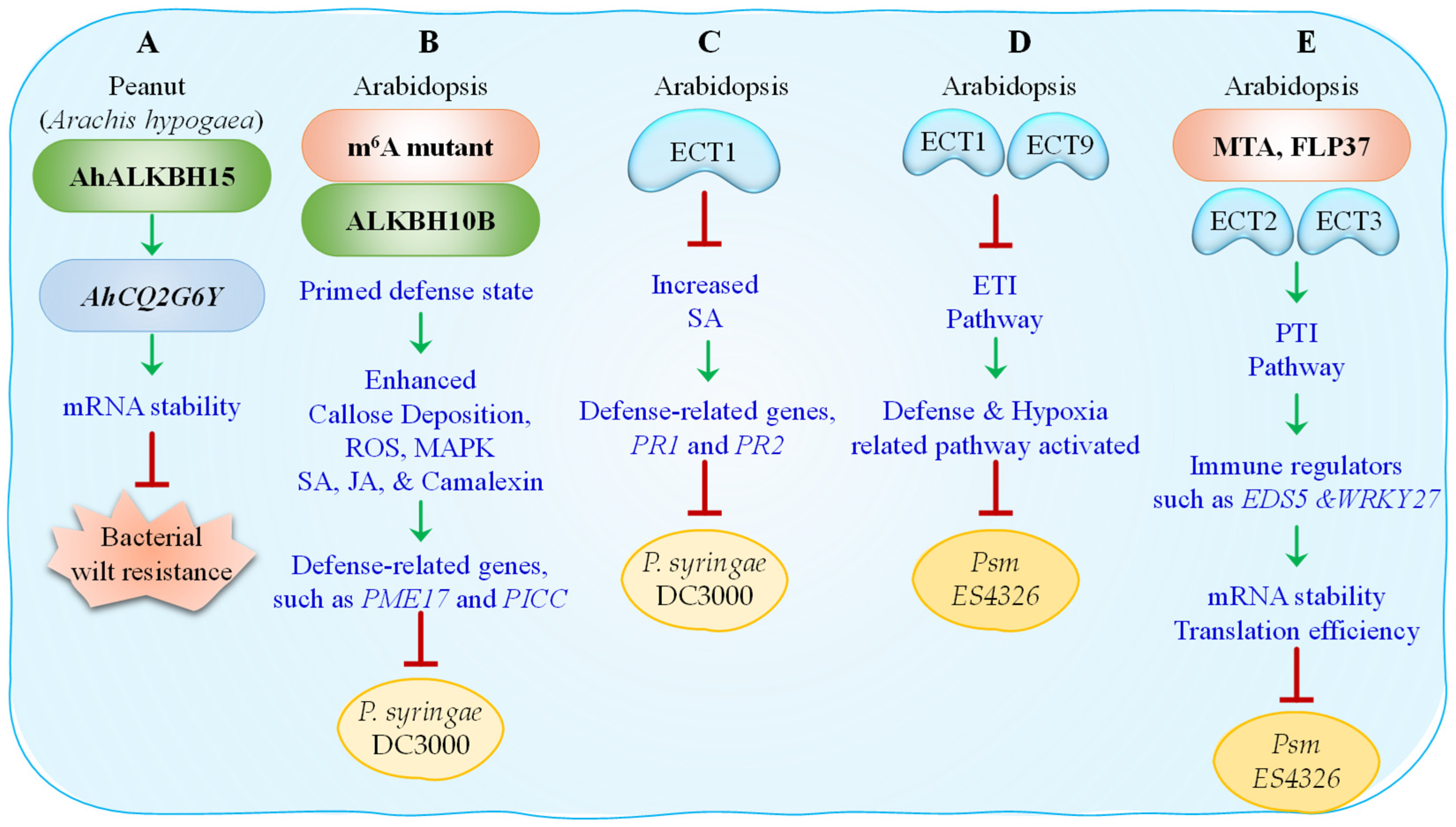
4. m6A RNA Modifications During Bacterial Infections
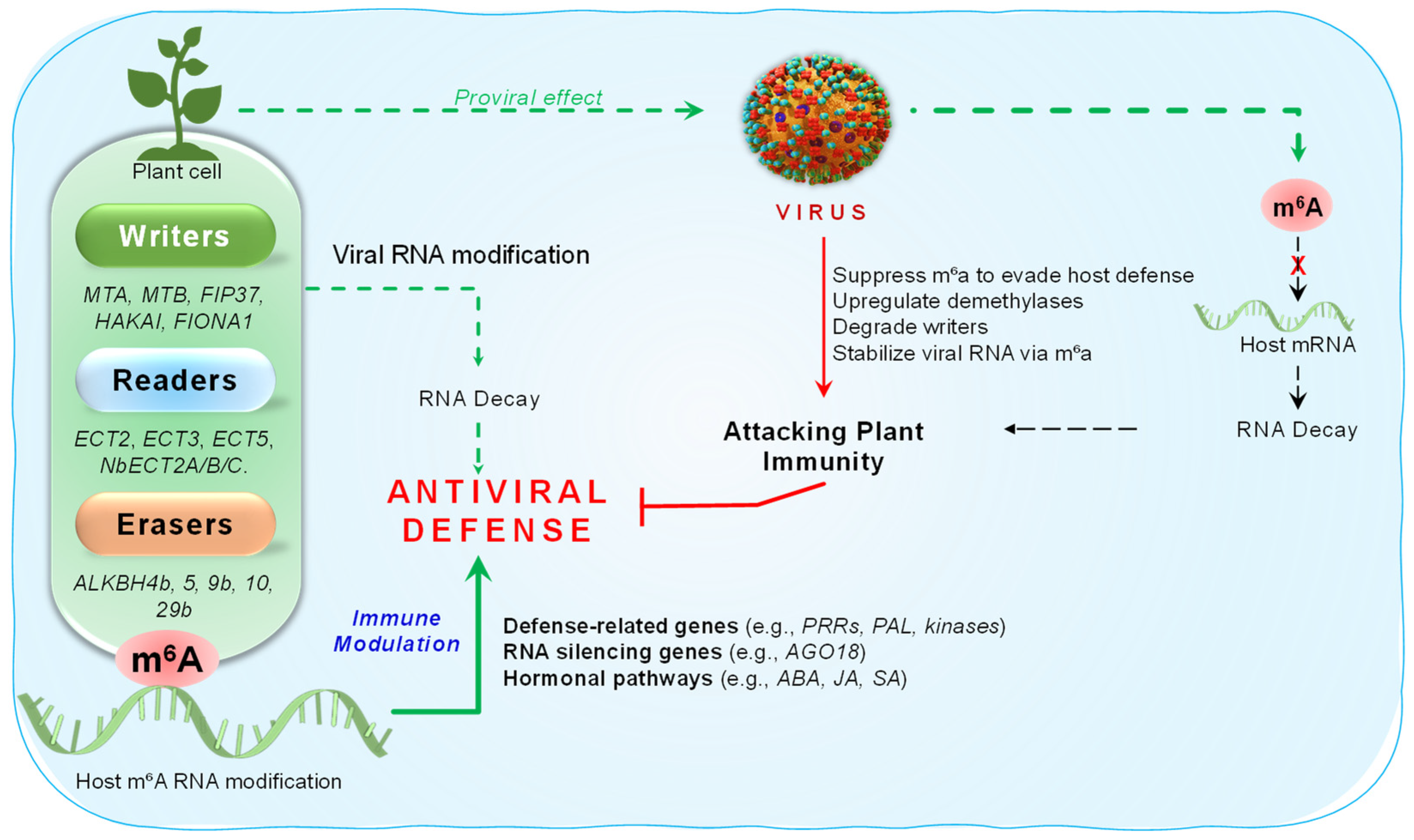
5. m6A RNA Modifications During Viral Infection
6. m6A RNA Modifications During Insect and Nematode Infestations
7. Limitations and Future Perspectives
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ramirez-Prado, J.S.; Abulfaraj, A.A.; Rayapuram, N.; Benhamed, M.; Hirt, H. Plant Immunity: From Signaling to Epigenetic Control of Defense. Trends Plant Sci. 2018, 23, 833–844. [Google Scholar] [CrossRef]
- Dangl, J.L.; Jones, J.D.G. Plant pathogens and integrated defence responses to infection. Nature 2001, 411, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J. Contrasting Mechanisms of Defense Against Biotrophic and Necrotrophic Pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef]
- Savary, S.; Ficke, A.; Aubertot, J.-N.; Hollier, C. Crop losses due to diseases and their implications for global food production losses and food security. Food Secur. 2012, 4, 519–537. [Google Scholar] [CrossRef]
- Ristaino, J.B.; Anderson, P.K.; Bebber, D.P.; Brauman, K.A.; Cunniffe, N.J.; Fedoroff, N.V.; Finegold, C.; Garrett, K.A.; Gilligan, C.A.; Jones, C.M.; et al. The persistent threat of emerging plant disease pandemics to global food security. Proc. Natl. Acad. Sci. USA 2021, 118, e2022239118. [Google Scholar] [CrossRef]
- Boller, T.; Felix, G. A Renaissance of Elicitors: Perception of Microbe-Associated Molecular Patterns and Danger Signals by Pattern-Recognition Receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef]
- Ding, B.; Wang, G.-L. Chromatin versus pathogens: The function of epigenetics in plant immunity. Front. Plant Sci. 2015, 6, 675. [Google Scholar] [CrossRef]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- Wang, Y.; Pruitt, R.N.; Nürnberger, T.; Wang, Y. Evasion of plant immunity by microbial pathogens. Nat. Rev. Microbiol. 2022, 20, 449–464. [Google Scholar] [CrossRef] [PubMed]
- Espinas, N.A.; Saze, H.; Saijo, Y. Epigenetic Control of Defense Signaling and Priming in Plants. Front. Plant Sci. 2016, 7, 1201. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, N.; Srivastava, R.; Verma, A.; Mohan Rai, K.; Singh, B.; Chandra Verma, P. Unravelling Cotton Nonexpressor of Pathogenesis-Related 1(NPR1)-Like Genes Family: Evolutionary Analysis and Putative Role in Fiber Development and Defense Pathway. Plants 2020, 9, 999. [Google Scholar] [CrossRef]
- Brooks, A.N.; Turkarslan, S.; Beer, K.D.; Yin Lo, F.; Baliga, N.S. Adaptation of cells to new environments. WIREs Syst. Biol. Med. 2011, 3, 544–561. [Google Scholar] [CrossRef]
- Periyasamy, S.; Gray, A.; Kille, P. The bottom–up approach to defining life: Deciphering the functional organization of biological cells via multi-objective representation of biological complexity from molecules to cells. Front. Physiol. 2013, 4, 369. [Google Scholar] [CrossRef]
- Srivastava, R.; Ahn, S.H. Modifications of RNA polymerase II CTD: Connections to the histone code and cellular function. Biotechnol. Adv. 2015, 33, 856–872. [Google Scholar] [CrossRef]
- Srivastava, R.; Rai, K.M.; Srivastava, R. Plant Biosynthetic Engineering Through Transcription Regulation: An Insight into Molecular Mechanisms During Environmental Stress; Springer Nature: Singapore, 2018; pp. 51–72. [Google Scholar]
- Chisolm, D.A.; Weinmann, A.S. Connections Between Metabolism and Epigenetics in Programming Cellular Differentiation. Annu. Rev. Immunol. 2018, 36, 221–246. [Google Scholar] [CrossRef]
- Srivastava, R.; Rai, K.M.; Srivastava, M.; Kumar, V.; Pandey, B.; Singh, S.P.; Bag, S.K.; Singh, B.D.; Tuli, R.; Sawant, S.V. Distinct Role of Core Promoter Architecture in Regulation of Light Mediated Responses in Plant Genes. Mol. Plant 2014, 7, 626–641. [Google Scholar] [CrossRef]
- Shoshani, O.; Zipori, D. Stress as a fundamental theme in cell plasticity. Biochim. Biophys. Acta (BBA)—Gene Regul. Mech. 2015, 1849, 371–377. [Google Scholar] [CrossRef]
- Srivastava, R.; Lodhi, N. DNA Methylation Malleability and Dysregulation in Cancer Progression: Understanding the Role of PARP1. Biomolecules 2022, 12, 417. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.; Singh, U.M.; Dubey, N.K. Histone Modifications by different histone modifiers: Insights into histone writers and erasers during chromatin modification. J. Biol. Sci. Med. 2016, 2, 45–54. [Google Scholar]
- Srivastava, R.; Srivastava, R.; Ahn, S.H. The Epigenetic Pathways to Ribosomal DNA Silencing. Microbiol. Mol. Biol. Rev. 2016, 80, 545–563. [Google Scholar] [CrossRef] [PubMed]
- Kiran, K.; Ansari, S.A.; Srivastava, R.; Lodhi, N.; Chaturvedi, C.P.; Sawant, S.V.; Tuli, R. The TATA-box sequence in the basal promoter contributes to determining light-dependent gene expression in plants. Plant Physiol. 2006, 142, 364–376. [Google Scholar] [CrossRef]
- Lodhi, N.; Ranjan, A.; Singh, M.; Srivastava, R.; Singh, S.P.; Chaturvedi, C.P.; Ansari, S.A.; Sawant, S.V.; Tuli, R. Interactions between upstream and core promoter sequences determine gene expression and nucleosome positioning in tobacco PR-1a promoter. Biochim. Biophys. Acta (BBA) 2008, 1779, 634–644. [Google Scholar] [CrossRef]
- Lodhi, N.; Singh, M.; Srivastava, R.; Sawant, S.V.; Tuli, R. Epigenetic malleability at core promoter initiates tobacco PR-1a expression post salicylic acid treatment. Mol. Biol. Rep. 2023, 50, 417–431. [Google Scholar] [CrossRef]
- Pandey, B.; Prakash, P.; Verma, P.C.; Srivastava, R. Regulated Gene Expression by Synthetic Modulation of the Promoter Architecture in Plants; Elsevier: Amsterdam, The Netherlands, 2019; pp. 235–255. [Google Scholar] [CrossRef]
- Srivastava, R.; Srivastava, R.; Singh, U.M. Understanding the patterns of gene expression during climate change. In Climate Change Effect on Crop Productivity; CRC Press, Taylor & Francis Group: Boca Raton, FL, USA, 2014; pp. 279–328. [Google Scholar]
- Furlan, M.; de Pretis, S.; Pelizzola, M. Dynamics of transcriptional and post-transcriptional regulation. Brief. Bioinform. 2021, 22, bbaa389. [Google Scholar] [CrossRef]
- Lancaster, C.L.; Moberg, K.H.; Corbett, A.H. Post-Transcriptional Regulation of Gene Expression and the Intricate Life of Eukaryotic mRNAs. Wiley Interdiscip. Rev. RNA 2025, 16, e70007. [Google Scholar] [CrossRef]
- Ahi, E.P.; Singh, P. Emerging Orchestrator of Ecological Adaptation: M(6)A Regulation of Post-Transcriptional Mechanisms. Mol. Ecol. 2024, 34, e17545. [Google Scholar] [CrossRef]
- Song, J.; Yi, C. Chemical Modifications to RNA: A New Layer of Gene Expression Regulation. ACS Chem. Biol. 2017, 12, 316–325. [Google Scholar] [CrossRef]
- Saletore, Y.; Chen-Kiang, S.; Mason, C.E. Novel RNA regulatory mechanisms revealed in the epitranscriptome. RNA Biol. 2013, 10, 342–346. [Google Scholar] [CrossRef]
- Li, C.; Chen, K.; Li, X.; Xiong, X. Epitranscriptome-epigenome interactions in development and disease mechanisms. Trends Genet. 2025, 41, 691–705. [Google Scholar] [CrossRef]
- Cappannini, A.; Ray, A.; Purta, E.; Mukherjee, S.; Boccaletto, P.; Moafinejad, S.N.; Lechner, A.; Barchet, C.; Klaholz, B.P.; Stefaniak, F.; et al. MODOMICS: A database of RNA modifications and related information. 2023 update. Nucleic Acids Res. 2023, 52, D239–D244. [Google Scholar] [CrossRef]
- Liu, W.-W.; Zheng, S.-Q.; Li, T.; Fei, Y.-F.; Wang, C.; Zhang, S.; Wang, F.; Jiang, G.-M.; Wang, H. RNA modifications in cellular metabolism: Implications for metabolism-targeted therapy and immunotherapy. Signal Transduct. Target. Ther. 2024, 9, 70. [Google Scholar] [CrossRef]
- Dominissini, D. Genomics and Proteomics. Roadmap to the epitranscriptome. Science 2014, 346, 1192. [Google Scholar] [CrossRef]
- Fray, R.G.; Simpson, G.G. The Arabidopsis epitranscriptome. Curr. Opin. Plant Biol. 2015, 27, 17–21. [Google Scholar] [CrossRef]
- Shen, L.; Ma, J.; Li, P.; Wu, Y.; Yu, H. Recent advances in the plant epitranscriptome. Genome Biol. 2023, 24, 43. [Google Scholar] [CrossRef]
- Song, P.; Cai, Z.; Jia, G. Principles, functions, and biological implications of m(6)A in plants. RNA 2024, 30, 491–499. [Google Scholar] [CrossRef]
- Zheng, H.; Li, S.; Zhang, X.; Sui, N. Functional Implications of Active N6-Methyladenosine in Plants. Front. Cell Dev. Biol. 2020, 8, 291. [Google Scholar] [CrossRef]
- Tang, J.; Chen, S.; Jia, G. Detection, regulation, and functions of RNA N6-methyladenosine modification in plants. Plant Commun. 2023, 4, 100546. [Google Scholar] [CrossRef]
- Brodersen, P.; Arribas-Hernández, L. The m6A-YTH regulatory system in plants: A status. Curr. Opin. Plant Biol. 2024, 82, 102650. [Google Scholar] [CrossRef]
- Ma, K.; Han, J.; Zhang, Z.; Li, H.; Zhao, Y.; Zhu, Q.; Xie, Y.; Liu, Y.-G.; Chen, L. OsEDM2L mediates m6A of EAT1 transcript for proper alternative splicing and polyadenylation regulating rice tapetal degradation. J. Integr. Plant Biol. 2021, 63, 1982–1994. [Google Scholar] [CrossRef]
- Gao, Z.; Yang, Q.; Shen, H.; Guo, P.; Xie, Q.; Chen, G.; Hu, Z. The knockout of SlMTC impacts tomato seed size and reduces resistance to salt stress in tomato. Plant Sci. 2024, 349, 112228. [Google Scholar] [CrossRef]
- Xu, C.; Wu, Z.; Duan, H.C.; Fang, X.; Jia, G.; Dean, C. R-loop resolution promotes co-transcriptional chromatin silencing. Nat. Commun. 2021, 12, 1790. [Google Scholar] [CrossRef]
- Wang, X.; Jiang, B.; Gu, L.; Chen, Y.; Mora, M.; Zhu, M.; Noory, E.; Wang, Q.; Lin, C. A photoregulatory mechanism of the circadian clock in Arabidopsis. Nat. Plants 2021, 7, 1397–1408. [Google Scholar] [CrossRef]
- Cheng, P.; Bao, S.; Li, C.; Tong, J.; Shen, L.; Yu, H. RNA N(6)-methyladenosine modification promotes auxin biosynthesis required for male meiosis in rice. Dev. Cell 2022, 57, 246–259.e244. [Google Scholar] [CrossRef]
- Shim, S.; Lee, H.G.; Lee, H.; Seo, P.J. H3K36me2 is highly correlated with m(6) A modifications in plants. J. Integr. Plant Biol. 2020, 62, 1455–1460. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3′ UTRs and near Stop Codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef]
- Shi, Y.; Yang, S.; Pei, T.; Xu, Y.; Zhao, Y.; Xue, H.; Ma, X.; Ren, H.; Liu, X. The m6A writers, readers, and erasers regulate plant development and respond to biotic/abiotic stresses. Epigenet. Insights 2025, 18, e008. [Google Scholar] [CrossRef]
- Luo, G.-Z.; MacQueen, A.; Zheng, G.; Duan, H.; Dore, L.C.; Lu, Z.; Liu, J.; Chen, K.; Jia, G.; Bergelson, J.; et al. Unique features of the m6A methylome in Arabidopsis thaliana. Nat. Commun. 2014, 5, 5630. [Google Scholar] [CrossRef]
- Wei, L.-H.; Song, P.; Wang, Y.; Lu, Z.; Tang, Q.; Yu, Q.; Xiao, Y.; Zhang, X.; Duan, H.-C.; Jia, G. The m6A Reader ECT2 Controls Trichome Morphology by Affecting mRNA Stability in Arabidopsis. Plant Cell 2018, 30, 968–985. [Google Scholar] [CrossRef]
- Zhang, F.; Zhang, Y.-C.; Liao, J.-Y.; Yu, Y.; Zhou, Y.-F.; Feng, Y.-Z.; Yang, Y.-W.; Lei, M.-Q.; Bai, M.; Wu, H.; et al. The subunit of RNA N6-methyladenosine methyltransferase OsFIP regulates early degeneration of microspores in rice. PLoS Genet. 2019, 15, e1008120. [Google Scholar] [CrossRef]
- Zheng, H.; Sun, X.; Li, J.; Song, Y.; Song, J.; Wang, F.; Liu, L.; Zhang, X.; Sui, N. Analysis of N6-methyladenosine reveals a new important mechanism regulating the salt tolerance of sweet sorghum. Plant Sci. 2021, 304, 110801. [Google Scholar] [CrossRef]
- Deng, H.; Cheema, J.; Zhang, H.; Woolfenden, H.; Norris, M.; Liu, Z.; Liu, Q.; Yang, X.; Yang, M.; Deng, X.; et al. Rice In Vivo RNA Structurome Reveals RNA Secondary Structure Conservation and Divergence in Plants. Mol. Plant 2018, 11, 607–622. [Google Scholar] [CrossRef]
- Cai, J.; Hu, J.; Amara, U.; Park, S.J.; Li, Y.; Jeong, D.; Lee, I.; Xu, T.; Kang, H. Arabidopsis N6-methyladenosine methyltransferase FIONA1 regulates floral transition by affecting the splicing of FLC and the stability of floral activators SPL3 and SEP3. J. Exp. Bot. 2023, 74, 864–877. [Google Scholar] [CrossRef]
- Xu, T.; Wu, X.; Wong, C.E.; Fan, S.; Zhang, Y.; Zhang, S.; Liang, Z.; Yu, H.; Shen, L. FIONA1-Mediated m6A Modification Regulates the Floral Transition in Arabidopsis. Adv. Sci. 2022, 9, 2103628. [Google Scholar] [CrossRef]
- Zhang, T.-y.; Wang, Z.-q.; Hu, H.-c.; Chen, Z.-q.; Liu, P.; Gao, S.-q.; Zhang, F.; He, L.; Jin, P.; Xu, M.-z.; et al. Transcriptome-Wide N6-Methyladenosine (m6A) Profiling of Susceptible and Resistant Wheat Varieties Reveals the Involvement of Variety-Specific m6A Modification Involved in Virus-Host Interaction Pathways. Front. Microbiol. 2021, 12, 656302. [Google Scholar] [CrossRef]
- Huang, T.; He, W.-J.; Li, C.; Zhang, J.-B.; Liao, Y.-C.; Song, B.; Yang, P. Transcriptome-wide analyses of RNA m6A methylation in hexaploid wheat reveal its roles in mRNA translation regulation. Front. Plant Sci. 2022, 13, 917335. [Google Scholar] [CrossRef]
- Hou, N.; Li, C.; He, J.; Liu, Y.; Yu, S.; Malnoy, M.; Mobeen Tahir, M.; Xu, L.; Ma, F.; Guan, Q. MdMTA-mediated m6A modification enhances drought tolerance by promoting mRNA stability and translation efficiency of genes involved in lignin deposition and oxidative stress. New Phytol. 2022, 234, 1294–1314. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.; Li, C.; Hu, S.; Yu, J.; Song, S. Transcriptome-wide N6-methyladenosine profiling of rice callus and leaf reveals the presence of tissue-specific competitors involved in selective mRNA modification. RNA Biol. 2014, 11, 1180–1188. [Google Scholar] [CrossRef]
- Arribas-Hernández, L.; Rennie, S.; Köster, T.; Porcelli, C.; Lewinski, M.; Staiger, D.; Andersson, R.; Brodersen, P. Principles of mRNA targeting via the Arabidopsis m(6)A-binding protein ECT2. eLife 2021, 10, e72375. [Google Scholar] [CrossRef]
- Li, D.; Zhang, H.; Hong, Y.; Huang, L.; Li, X.; Zhang, Y.; Ouyang, Z.; Song, F. Genome-Wide Identification, Biochemical Characterization, and Expression Analyses of the YTH Domain-Containing RNA-Binding Protein Family in Arabidopsis and Rice. Plant Mol. Biol. Rep. 2014, 32, 1169–1186. [Google Scholar] [CrossRef]
- Nguyen, T.K.H.; Kang, H. Reading m(6)A marks in mRNA: A potent mechanism of gene regulation in plants. J. Integr. Plant Biol. 2024, 66, 2586–2599. [Google Scholar] [CrossRef]
- Yin, S.; Ao, Q.; Tan, C.; Yang, Y. Genome-wide identification and characterization of YTH domain-containing genes, encoding the m6A readers, and their expression in tomato. Plant Cell Rep. 2021, 40, 1229–1245. [Google Scholar] [CrossRef]
- Hou, Y.; Sun, J.; Wu, B.; Gao, Y.; Nie, H.; Nie, Z.; Quan, S.; Wang, Y.; Cao, X.; Li, S. CPSF30-L-mediated recognition of mRNA m6A modification controls alternative polyadenylation of nitrate signaling-related gene transcripts in Arabidopsis. Mol. Plant 2021, 14, 688–699. [Google Scholar] [CrossRef]
- Amara, U.; Hu, J.; Cai, J.; Kang, H. FLK is an mRNA m6A reader that regulates floral transition by modulating the stability and splicing of FLC in Arabidopsis. Mol. Plant 2023, 16, 919–929. [Google Scholar] [CrossRef]
- Xiang, Y.; Zhang, D.; Li, L.; Xue, Y.-X.; Zhang, C.-Y.; Meng, Q.-F.; Wang, J.; Tan, X.-L.; Li, Y.-L. Detection, distribution, and functions of RNA N6-methyladenosine (m6A) in plant development and environmental signal responses. Front. Plant Sci. 2024, 15, 1429011. [Google Scholar] [CrossRef]
- Zhou, L.; Tian, S.; Qin, G. RNA methylomes reveal the m(6)A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening. Genome Biol. 2019, 20, 156. [Google Scholar] [CrossRef]
- Tang, J.; Lei, D.; Yang, J.; Chen, S.; Wang, X.; Huang, X.; Zhang, S.; Cai, Z.; Zhu, S.; Wan, J.; et al. OsALKBH9-mediated m(6)A demethylation regulates tapetal PCD and pollen exine accumulation in rice. Plant Biotechnol. J. 2024, 22, 2410–2423. [Google Scholar] [CrossRef]
- Cui, C.; Ma, Z.; Wan, H.; Gao, J.; Zhou, B. GhALKBH10 negatively regulates salt tolerance in cotton. Plant Physiol. Biochem. PPB 2022, 192, 87–100. [Google Scholar] [CrossRef]
- Liu, S.; Xiang, D. New understandings of the genetic regulatory relationship between non-coding RNAs and m6A modification. Front. Genet. 2023, 14, 1270983. [Google Scholar] [CrossRef]
- Pan, T. N6-methyl-adenosine modification in messenger and long non-coding RNA. Trends Biochem. Sci. 2013, 38, 204–209. [Google Scholar] [CrossRef]
- Lodhi, N.; Srivastava, R. Dynamics and Malleability of Plant DNA Methylation During Abiotic Stresses. Epigenomes 2025, 9, 31. [Google Scholar] [CrossRef]
- Liang, Z.; Shen, L.; Cui, X.; Bao, S.; Geng, Y.; Yu, G.; Liang, F.; Xie, S.; Lu, T.; Gu, X.; et al. DNA N6-Adenine Methylation in Arabidopsis thaliana. Dev. Cell 2018, 45, 406–416.e403. [Google Scholar] [CrossRef]
- Li, D.; Du, J.; Gao, M.; He, C. Identification of AtALKBH1A and AtALKBH1D as DNA N6-adenine demethylases in Arabidopsis thaliana. Plant Sci. 2024, 342, 112055. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Ma, A.; Lei, J.; He, C. METTL4-mediated N(6)-methyladenine DNA modification regulates thermotolerance in Arabidopsis thaliana. Plant Sci. 2024, 338, 111916. [Google Scholar] [CrossRef]
- Karanthamalai, J.; Chodon, A.; Chauhan, S.; Pandi, G. DNA N(6)-Methyladenine Modification in Plant Genomes-A Glimpse into Emerging Epigenetic Code. Plants 2020, 9, 247. [Google Scholar] [CrossRef]
- Hu, J.; Xu, T.; Kang, H. Crosstalk between RNA m6A modification and epigenetic factors in plant gene regulation. Plant Commun. 2024, 5, 101037. [Google Scholar] [CrossRef]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-Triggered Immunity: From Pathogen Perception to Robust Defense. Annu. Rev. Plant Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef]
- Prall, W.; Sheikh, A.H.; Bazin, J.; Bigeard, J.; Almeida-Trapp, M.; Crespi, M.; Hirt, H.; Gregory, B.D. Pathogen-induced m6A dynamics affect plant immunity. Plant Cell 2023, 35, 4155–4172. [Google Scholar] [CrossRef]
- Furci, L.; Berthelier, J.; Saze, H. RNA N6-adenine methylation dynamics impact Hyaloperonospora arabidopsidis resistance in Arabidopsis. Plant Physiol. 2024, 196, 745–753. [Google Scholar] [CrossRef]
- Song, Z.; Yang, Q.; Dong, B.; Wang, S.; Xue, J.; Liu, N.; Zhou, X.; Li, N.; Dandekar, A.M.; Cheng, L.; et al. Nanopore RNA direct sequencing identifies that m6A modification is essential for sorbitol-controlled resistance to Alternaria alternata in apple. Dev. Cell 2025, 60, 1439–1453.e1435. [Google Scholar] [CrossRef]
- Cerav, E.N.; Wu, N.; Akkaya, M.S. Transcriptome-Wide N6-Methyladenosine (m6A) Methylation Analyses in a Compatible Wheat–Puccinia striiformis f. sp. tritici Interaction. Plants 2024, 13, 982. [Google Scholar] [CrossRef]
- Han, C.; Zhang, F.; Qiao, X.; Zhao, Y.; Qiao, Q.; Huang, X.; Zhang, S. Multi-Omics Analysis Reveals the Dynamic Changes of RNA N6-Methyladenosine in Pear (Pyrus bretschneideri) Defense Responses to Erwinia amylovora Pathogen Infection. Front. Microbiol. 2022, 12, 803512. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Liu, C.; Meng, F.; Hu, L.; Fu, X.; Yang, Z.; Wang, N.; Jiang, Q.; Zhang, X.; Ma, F. The m6A reader MhYTP2 regulates MdMLO19 mRNA stability and antioxidant genes translation efficiency conferring powdery mildew resistance in apple. Plant Biotechnol. J. 2022, 20, 511–525. [Google Scholar] [CrossRef]
- Guo, T.; Bao, R.; Yang, Z.; Fu, X.; Hu, L.; Wang, N.; Liu, C.; Ma, F. The m(6) A reader MhYTP2 negatively modulates apple Glomerella leaf spot resistance by binding to and degrading MdRGA2L mRNA. Mol. Plant Pathol. 2023, 24, 1287–1299. [Google Scholar] [CrossRef]
- Ren, Z.; Tang, B.; Xing, J.; Liu, C.; Cai, X.; Hendy, A.; Kamran, M.; Liu, H.; Zheng, L.; Huang, J.; et al. MTA1-mediated RNA m6A modification regulates autophagy and is required for infection of the rice blast fungus. New Phytol. 2022, 235, 247–262. [Google Scholar] [CrossRef]
- Zhang, F.; Zhang, B.; Cui, T.; Chen, S.; Zhang, C.; Wang, Z.; Liu, X. The novel roles of RNA m6A modification in regulating the development, infection, and oxidative DNA damage repair of Phytophthora sojae. PLoS Pathog. 2024, 20, e1012553. [Google Scholar] [CrossRef]
- Zhao, L.; Wei, X.; Chen, F.; Yuan, L.; Chen, B.; Li, R. N6-methyladenosine RNA methyltransferase CpMTA1 mediates CpAphA mRNA stability through a YTHDF1-dependent m6A modification in the chestnut blight fungus. PLoS Pathog. 2024, 20, e1012476. [Google Scholar] [CrossRef]
- Zhao, L.; Wei, X.; Chen, F.; Chen, B.; Li, R. m(6)A demethylase CpALKBH regulates CpZap1 mRNA stability to modulate the development and virulence of chestnut blight fungus. mBio 2025, 16, e0184424. [Google Scholar] [CrossRef]
- Zhao, K.; Li, Z.; Ke, Y.; Ren, R.; Cao, Z.; Li, Z.; Wang, K.; Wang, X.; Wang, J.; Ma, Q.; et al. Dynamic N-methyladenosine RNA modification regulates peanut resistance to bacterial wilt. New Phytol. 2024, 242, 231–246. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.P.; Liu, K.; Kim, E.Y.; Medina-Puche, L.; Dong, H.; Di, M.; Singh, R.M.; Li, M.; Qi, S.; Meng, Z.; et al. The m6A reader ECT1 drives mRNA sequestration to dampen salicylic acid–dependent stress responses in Arabidopsis. Plant Cell 2023, 36, 746–763. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Niu, R.; Zhou, Y.; Tang, Z.; Xu, G.; Zhou, G. ECT9 condensates with ECT1 and regulates plant immunity. Front. Plant Sci. 2023, 14, 1140840. [Google Scholar] [CrossRef]
- Chen, T.; Greene, G.H.; Motley, J.; Mwimba, M.; Luo, G.-Z.; Xu, G.; Karapetyan, S.; Xiang, Y.; Liu, C.; He, C.; et al. m6A modification plays an integral role in mRNA stability and translation during pattern-triggered immunity. Proc. Natl. Acad. Sci. USA 2024, 121, e2411100121. [Google Scholar] [CrossRef]
- Mishra, J.; Srivastava, R.; Trivedi, P.K.; Verma, P.C. Effect of virus infection on the secondary metabolite production and phytohormone biosynthesis in plants. 3 Biotech 2020, 10, 547. [Google Scholar] [CrossRef]
- Zvereva, A.S.; Pooggin, M.M. Silencing and Innate Immunity in Plant Defense Against Viral and Non-Viral Pathogens. Viruses 2012, 4, 2578–2597. [Google Scholar] [CrossRef]
- Mandadi, K.K.; Scholthof, K.-B.G. Plant Immune Responses Against Viruses: How Does a Virus Cause Disease? Plant Cell 2013, 25, 1489–1505. [Google Scholar] [CrossRef] [PubMed]
- Baquero-Perez, B.; Geers, D.; Diez, J. From A to m(6)A: The Emerging Viral Epitranscriptome. Viruses 2021, 13, 1049. [Google Scholar] [CrossRef]
- He, Y.; Li, L.; Yao, Y.; Li, Y.; Zhang, H.; Fan, M. Transcriptome-wide N6-methyladenosine (m6A) methylation in watermelon under CGMMV infection. BMC Plant Biol. 2021, 21, 516. [Google Scholar] [CrossRef]
- Zhang, K.; Zhuang, X.; Dong, Z.; Xu, K.; Chen, X.; Liu, F.; He, Z. The dynamics of N(6)-methyladenine RNA modification in interactions between rice and plant viruses. Genome Biol. 2021, 22, 189. [Google Scholar] [CrossRef]
- Dong, Z.; Liu, N.; Zhang, Z.; Wang, H.; Li, S.; Zhan, B. N6-Methyladenosine Modification Regulates Prunus Necrotic Ringspot Virus Infection in Cucumis sativus. J. Agric. Food Chem. 2025, 73, 11586–11597. [Google Scholar] [CrossRef]
- Zhang, T.; Shi, C.; Hu, H.; Zhang, Z.; Wang, Z.; Chen, Z.; Feng, H.; Liu, P.; Guo, J.; Lu, Q.; et al. N6-methyladenosine RNA modification promotes viral genomic RNA stability and infection. Nat. Commun. 2022, 13, 6576. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Shi, J.; Yu, L.; Zhao, X.; Ran, L.; Hu, D.; Song, B. N6-methyl-adenosine level in Nicotiana tabacum is associated with tobacco mosaic virus. Virol. J. 2018, 15, 87. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Ge, L.; Li, Z.; Zhou, X.; Li, F. Pepino mosaic virus antagonizes plant m(6)A modification by promoting the autophagic degradation of the m(6)A writer HAKAI. Abiotech 2023, 4, 83–96. [Google Scholar] [CrossRef]
- Yue, J.; Lu, Y.; Sun, Z.; Guo, Y.; San León, D.; Pasin, F.; Zhao, M. Methyltransferase-like (METTL) homologues participate in Nicotiana benthamiana antiviral responses. Plant Signal. Behav. 2023, 18, 2214760. [Google Scholar] [CrossRef]
- Wu, W.; Wang, X.; Liang, X.; Huang, X.; Nawaz, M.A.; Jing, C.; Fan, Y.; Niu, J.; Wu, J.; Feng, X. Characterization of the m6A Regulatory Gene Family in Phaseolus vulgaris L. and Functional Analysis of PvMTA in Response to BCMV Infection. Int. J. Mol. Sci. 2025, 26, 2748. [Google Scholar] [CrossRef]
- Martínez-Pérez, M.; Aparicio, F.; López-Gresa, M.P.; Bellés, J.M.; Sánchez-Navarro, J.A.; Pallás, V. Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc. Natl. Acad. Sci. USA 2017, 114, 10755–10760. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Pérez, M.; Gómez-Mena, C.; Alvarado-Marchena, L.; Nadi, R.; Micol, J.L.; Pallas, V.; Aparicio, F. The m6A RNA Demethylase ALKBH9B Plays a Critical Role for Vascular Movement of Alfalfa Mosaic Virus in Arabidopsis. Front. Microbiol. 2021, 12, 745576. [Google Scholar] [CrossRef]
- Martínez-Pérez, M.; Aparicio, F.; Arribas-Hernández, L.; Tankmar, M.D.; Rennie, S.; von Bülow, S.; Lindorff-Larsen, K.; Brodersen, P.; Pallas, V. Plant YTHDF proteins are direct effectors of antiviral immunity against an N6-methyladenosine-containing RNA virus. EMBO J. 2023, 42, e113378. [Google Scholar] [CrossRef]
- Yue, J.; Wei, Y.; Sun, Z.; Chen, Y.; Wei, X.; Wang, H.; Pasin, F.; Zhao, M. AlkB RNA demethylase homologues and N-methyladenosine are involved in Potyvirus infection. Mol. Plant Pathol. 2022, 23, 1555–1564. [Google Scholar] [CrossRef]
- Sha, T.; Li, Z.; Xu, S.; Su, T.; Shopan, J.; Jin, X.; Deng, Y.; Lyu, X.; Hu, Z.; Zhang, M.; et al. eIF2Bβ confers resistance to Turnip mosaic virus by recruiting ALKBH9B to modify viral RNA methylation. Plant Biotechnol. J. 2024, 22, 3205–3217. [Google Scholar] [CrossRef]
- He, H.; Ge, L.; Chen, Y.; Zhao, S.; Li, Z.; Zhou, X.; Li, F. m(6)A modification of plant virus enables host recognition by NMD factors in plants. Sci. China Life Sci. 2024, 67, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Wu, N.; Zhang, L.; Wang, X. RNA N6-methyladenosine modification suppresses replication of rice black streaked dwarf virus and is associated with virus persistence in its insect vector. Mol. Plant Pathol. 2021, 22, 1070–1081. [Google Scholar] [CrossRef] [PubMed]
- Alvarado-Marchena, L.; Marquez-Molins, J.; Martinez-Perez, M.; Aparicio, F.; Pallás, V. Mapping of Functional Subdomains in the atALKBH9B m6A-Demethylase Required for Its Binding to the Viral RNA and to the Coat Protein of Alfalfa Mosaic Virus. Front. Plant Sci. 2021, 12, 701683. [Google Scholar] [CrossRef]
- Alvarado-Marchena, L.; Martínez-Pérez, M.; Úbeda, J.R.; Pallas, V.; Aparicio, F. Impact of the Potential m6A Modification Sites at the 3′UTR of Alfalfa Mosaic Virus RNA3 in the Viral Infection. Viruses 2022, 14, 1718. [Google Scholar] [CrossRef]
- Choudhary, A.; Srivastava, R.; Srivastava, R.; Verma, P.C. Transgenic Plant Technology: An Insight into Insect Resistance; Springer: Singapore, 2021; pp. 141–159. [Google Scholar]
- Mishra, D.K.; Srivastava, R.; Pandey, B.K.; Verma, P.C.; Sawant, S.V. Identification and validation of the wound and insect bite early-inducible promoter from Arabidopsis thaliana. 3 Biotech 2022, 12, 74. [Google Scholar] [CrossRef]
- Walling, L.L. Adaptive Defense Responses to Pathogens and Insects. In Advances in Botanical Research; Academic Press: Cambridge, MA, USA, 2009; Volume 51, pp. 551–612. [Google Scholar]
- Aerts, N.; Pereira Mendes, M.; Van Wees, S.C.M. Multiple levels of crosstalk in hormone networks regulating plant defense. Plant J. 2021, 105, 489–504. [Google Scholar] [CrossRef]
- de Bruxelles, G.L.; Roberts, M.R. Signals Regulating Multiple Responses to Wounding and Herbivores. Crit. Rev. Plant Sci. 2001, 20, 487–521. [Google Scholar] [CrossRef]
- Adedayo, A.A.; Richard, M.; Mari, A.; and Babalola, O.O. The biochemical and molecular mechanisms of plants: A review on insect herbivory. Plant Signal. Behav. 2025, 20, 2439248. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.; Liu, Z.; Shen, H.; Wu, D. Damage-Associated Molecular Pattern-Triggered Immunity in Plants. Front. Plant Sci. 2019, 10, 646. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Tan, X.; He, Z.; Jiang, L.; Li, Y.; Yang, L.; Hoffmann, A.A.; Zhao, C.; Fang, J.; Ji, R. Transcriptome-wide-methyladenosine profiling reveals growth–defense trade-offs in the response of rice to brown planthopper (Nilaparvata lugens) infestation. Pest Manag. Sci. 2024, 80, 5364–5376. [Google Scholar] [CrossRef]
- Li, S.; Tan, X.-Y.; He, Z.; Shen, C.; Li, Y.-L.; Qin, L.; Zhao, C.-Q.; Luo, G.-H.; Fang, J.-C.; Ji, R. The dynamics of N6-methyladenine RNA modification in resistant and susceptible rice varieties responding to rice stem borer damage. Insect Sci. 2025, 32, 530–550. [Google Scholar] [CrossRef]
- Yao, S.; Song, Y.; Cheng, X.; Wang, D.; Li, Q.; Zhang, J.; Chen, Q.; Yu, Q.; Ji, K. Transcriptome-Wide Identification of m6A Writers, Erasers and Readers and Their Expression Profiles under Various Biotic and Abiotic Stresses in Pinus massoniana Lamb. Int. J. Mol. Sci. 2024, 25, 7987. [Google Scholar] [CrossRef]
- Han, X.; Shi, Q.; He, Z.; Song, W.; Chen, Q.; Qi, Z. Transcriptome-wide N(6)-methyladenosine (m(6)A) methylation in soybean under Meloidogyne incognita infection. Abiotech 2022, 3, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Qin, R.; Huang, M.; Jiang, Y.; Jiang, D.; Chang, D.; Xie, Y.; Dou, Y.; Wu, L.; Wei, L.; Wang, M.; et al. N6-Methyladenosine (m6A) Sequencing Reveals Heterodera glycines-Induced Dynamic Methylation Promoting Soybean Defense. Phytopathology 2024, 114, 1612–1625. [Google Scholar] [CrossRef] [PubMed]
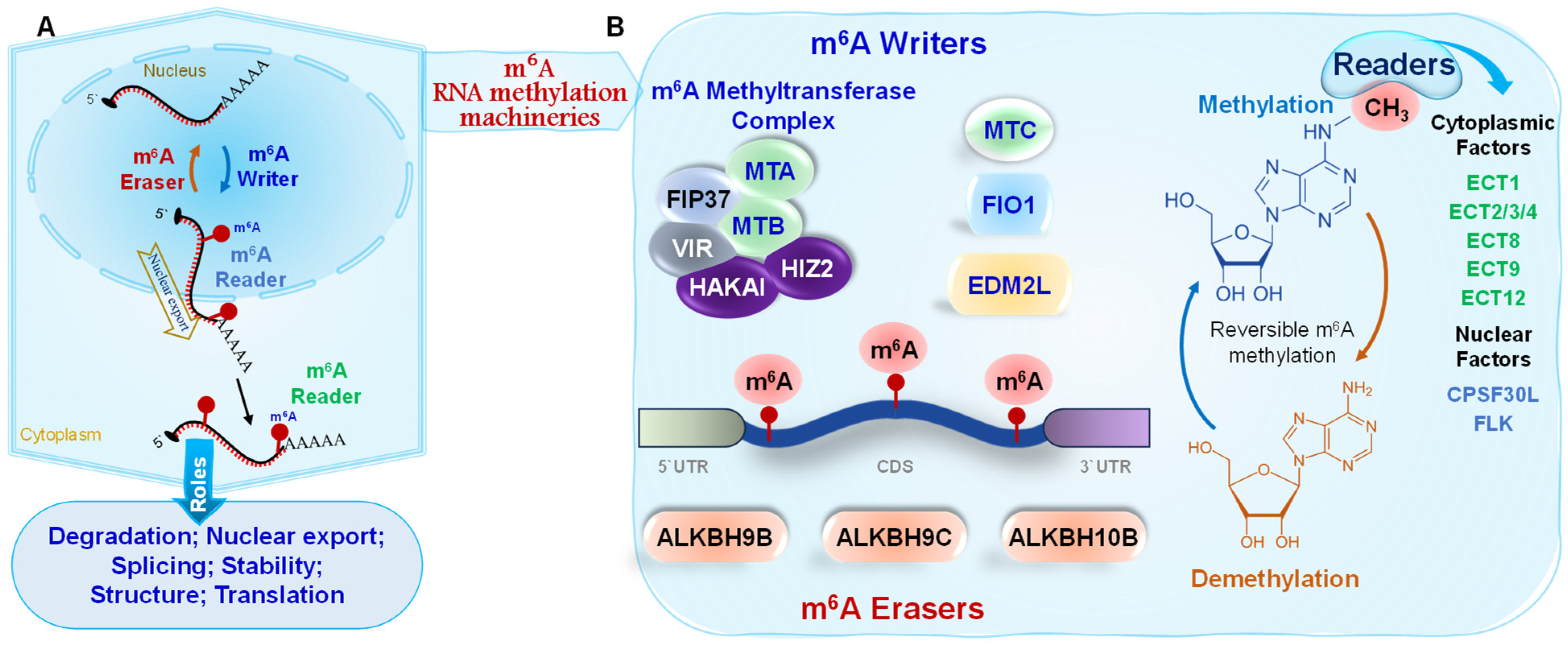

| Gene # | Organism | Type/Role | Function in Immunity/Virulence | Pathogen Context |
|---|---|---|---|---|
| FUNGUS INFECTION | ||||
| MTA | Arabidopsis | Writer | mta mutants show stronger resistance; m6A normally reduces defense signaling | Botrytis cinerea |
| HAKAI1 | Arabidopsis | Writer | hakai-1 mutants exhibit constitutive basal defense and enhanced resistance | Hyaloperonospora arabidopsidis |
| MdVIR1, MdVIR2 | Malus domestica (apple) | Writers | Enhance defense | Alternaria alternata |
| MdWRKY79 | M. domestica | Transcription factor (TF) | Stabilized by m6A, promoting immune response | A. alternata |
| MdNLR16 | M. domestica | NLR resistance | Stabilized by m6A, enhancing immunity | A. alternata |
| TaALKBH11B | Triticum aestivum (wheat) | Eraser | Upregulated during early infection | Puccinia striiformis f. sp. tritici (Pst) |
| TaALKBH4B | T. aestivum | Eraser | Peaks during colonization | Pst |
| TaECT25, TaECT31, TaECT21 | T. aestivum | Readers | Downregulated at early infection | Pst |
| TaVIR-D, TaVIR-A, TaHAKAI1-A | T. aestivum | Writers | Reduced activity during infection | Pst |
| TaFIP37-2D | T. aestivum | Writer | Continuously induced; central in defense regulation | Pst |
| MTA, ALKBH5B, ECT9 | Pyrus communis (pear) | Writer, Eraser, Reader | Core network regulating PTI and SAR immune genes | Erwinia amylovora |
| WRKYs, Ser/Thr kinases | P. communis | Defense genes | m6A-modified, positively correlated with higher expression | E. amylovora |
| MhYTP2 | Malus hupehensis | Reader (ECT2 homolog) | Enhances resistance to P. leucotricha; | Podosphaera leucotricha, Colletotrichum fructicola |
| MdMTA, MdMTB, MdFIP37 | M. domestica | Writers | Downregulated by MhYTP2 overexpression | P. leucotricha |
| MdALKBH6 | M. domestica | Eraser | Downregulated by MhYTP2 overexpression | P. leucotricha |
| MdMLO19, MdMLO19-X1 | M. domestica | Susceptibility genes | Downregulated via m6A-mediated degradation | P. leucotricha |
| MdGDH1L | M. domestica | Antioxidant enzyme | m6A enhances translation → higher antioxidant activity | P. leucotricha |
| MdRGA2L | M. domestica | NBS-LRR resistance | Promotes SA biosynthesis via ICS1; suppressed by MhYTP2 | C. fructicola |
| BACTERIAL INFECTION | ||||
| AhSAM1/2 | Arachis hypogaea | SAM synthases | Provide methyl donor for m6A; upregulated in resistant line H108 | Ralstonia solanacearum (bacterial wilt) |
| AhMTA1/2/4/5 | A. hypogaea | Writers | Core m6A methyltransferases; upregulated in resistant line | R. solanacearum |
| AhALKBH2/14/15/18 | A. hypogaea | Erasers | Dynamic regulation of defense transcripts | R. solanacearum |
| AhALKBH15 | A. hypogaea | Eraser | demethylates defense gene AhCQ2G6Y, enhancing stability and resistance | R. solanacearum |
| AhECT6–10, AhECT13 | A. hypogaea | Readers | Bind m6A-modified RNAs; regulate immune transcript fate | R. solanacearum |
| AhCQ2G6Y | A. hypogaea | Defense gene | Stabilized by AhALKBH15 demethylation; elevated expression suppresses pathogen growth | R. solanacearum |
| MTA | Arabidopsis | Writer | mta mutants show enhanced resistance; | Pseudomonas syringae pv. tomato DC3000, Psm ES4326 |
| FIP37-4 | Arabidopsis | Writer subunit | Mutants show enhanced resistance; impaired PTI; delayed transcript decay | P. syringae DC3000, elf18 treatment |
| VIR-2 | Arabidopsis | Writer subunit | Enhances resistance | P. syringae |
| ALKBH10B | Arabidopsis | Eraser | Overexpression enhances resistance; increases basal defenses (ROS, callose, SA, JA, camalexin) | P. syringae DC3000 |
| PME17, PICC | Arabidopsis | Defense genes | regulatory protein PICC role in callose deposition; Show higher expression in absence of m6A, | P. syringae |
| CPL3, SCREW3, VAD1 | Arabidopsis | Defense-related genes | Destabilized post-elicitation in m6A-deficient plants | P. syringae |
| ECT1 | Arabidopsis | Reader | Negative regulator of SA-mediated defense; forms condensates to degrade SA-induced PR1/PR2 transcripts | P. syringae DC3000 |
| PR1, PR2 | Arabidopsis | SA-responsive defense genes | m6A-modified; degraded by ECT1 to suppress overactive immunity | P. syringae DC3000 |
| ECT9 | Arabidopsis | Reader | Redundant with ECT1; suppresses ETI | Psm ES4326 (AvrRpt2) |
| ECT1/9 | Arabidopsis | Readers | Double mutant shows enhanced ETI, reduced pathogen growth, increased cell death | Psm ES4326 (AvrRpt2) |
| EDS5 | Arabidopsis | SA pathway regulator | Shows PTI-associated transcript-specific m6A remodeling | elf18 treatment |
| WRKY27 | Arabidopsis | TF (immune regulator) | Target of m6A remodeling during PTI | elf18 treatment |
| ECT2 | Arabidopsis | Reader | Promotes translation of immune transcripts (e.g., WRKY27, EDS5) | elf18 treatment, P. syringae |
| ECT3 | Arabidopsis | Reader | Stabilizes immune transcripts | elf18 treatment |
| ECT4 | Arabidopsis | Reader | Works with ECT2/3 in cooperative PTI regulation | elf18 treatment |
| VIRAL INFECTION | ||||
| TraesCS1B02G175900 | T. aestivum | Defense kinase | Differentially methylated; cysteine-rich receptor-like protein kinase in defense signaling | WYMV |
| TraesCS7B02G446900 | T. aestivum | Defense chaperone | GRP94 homolog; methylation linked to defense regulation | WYMV |
| TraesCS7A02G267400 | T. aestivum | Defense kinase | PTI1-like kinase; participates in PTI signaling, m6A-modulated | WYMV |
| TaFIP37-1 | T. aestivum | Writer | Regulates methylation of defense genes during infection | WYMV |
| TaALKBH29B | T. aestivum | Eraser | Modulates methylation of immune genes | WYMV |
| ClALKBH4B | Watermelon | Eraser | Upregulated in resistant genotype; linked to reduced m6A, enhancing immunity | CGMMV |
| OsAGO18 | Rice (Oryza sativa) | Antiviral gene | Expression correlates with m6A enrichment; regulates antiviral RNA silencing | RSV, RBSDV |
| OsSLRL1 | O. sativa | Defense regulator | Expression linked to m6A changes; modulates immunity | RSV, RBSDV |
| OsMAT3/4 | O. sativa | Writer | Upregulated during infection; enhances m6A deposition | RSV |
| OsALKBH10 | O. sativa | Eraser | Downregulated during infection; linked to viral exploitation of host m6A | RSV, RBSDV |
| CuMTA, CuMTB, CuHAKAI | Cucumber | Writers | Upregulated; alter global m6A during infection | PNRSV |
| CuSAM2a, CuSAM4 | Cucumber | SAM synthases | Provide methyl donors; upregulated in infection | PNRSV |
| CuALKBH10B | Cucumber | Eraser | Downregulated; loss of demethylation contributes to infection | PNRSV |
| CuECT2, CuECT4a/4b | Cucumber | Readers | Silencing increases viral RNA accumulation | PNRSV |
| CuPAL | Cucumber | Defense gene | Hyper-methylated and transcriptionally induced; silencing increases susceptibility | PNRSV |
| TaMTB | Wheat | Writer | Promotes infection by stabilizing WYMV RNA1; natural SNP variant increases susceptibility | WYMV |
| NbMETTL1, NbMETTL2 | N. benthamiana | Writers (METTL-like) | Overexpression reduces PPV accumulation (antiviral) | PPV |
| PvMTA | Common bean | Writer | Overexpression inhibits BCMV; silencing enhances susceptibility | BCMV |
| AtALKBH9B | Arabidopsis | Eraser | Promotes AMV infection by demethylating viral RNA; interacts with viral coat protein | AMV |
| ECT2, ECT3, ECT5 | Arabidopsis | Readers (YTH-domain) | Restrict AMV infection by recognizing methylated viral RNAs | AMV |
| SlHAKAI | Tomato | Writer | Targeted for autophagic degradation by PepMV to suppress host m6A immunity | PepMV |
| NbECT2A/2B/2C | N. benthamiana | Readers | Promote viral RNA degradation with NMD components | PepMV |
| NbUPF3, NbSMG7 | N. benthamiana | NMD pathway genes | Cooperate with m6A readers to degrade viral RNAs | PepMV |
| BjALKBH9B | Brassica juncea | Eraser | Interacts with TuMV and eIF2Bβ; regulates viral RNA methylation | TuMV |
| BjeIF2Bβ | B. juncea | Translation factor | Genome editing enhances resistance; targeted by BjALKBH9B–TuMV complex | TuMV |
| LsMETTL3, LsMETTL14 | Planthopper vector | Writers | Silencing increases RBSDV accumulation; antiviral role in insect vector | RBSDV |
| INSECT AND NEMATODES INFESTATION | ||||
| Trypsin protease inhibitor genes | O. sativa | Defense protein; JA-mediated | Strong m6A enrichment and upregulation; inhibits insect digestive enzymes, enhancing JA-mediated insect resistance | Striped stem borer (RSB) |
| MAPK cascade genes | O. sativa | Signal transduction | Enriched in resistant cultivar; m6A promotes activation of MAPK signaling, reinforcing immune responses | Striped stem borer (RSB) |
| Jasmonate biosynthesis genes | O. sativa | Hormone signaling | m6A enrichment enhances expression, boosting JA-mediated defense | Striped stem borer (RSB) |
| Terpenoid metabolism genes | O. sativa | Secondary metabolism | Enriched in resistant cultivar; supports defense via secondary metabolite accumulation | Striped stem borer (RSB) |
| PmALKBH3, PmYTHDF5, PmHAKAI1 | Pinus massoniana | Eraser, reader, writer (susceptible clones downregulated) | Downregulation associated with susceptibility to insect infestation | Monochamus alternatus |
| PmMTA, PmMTB, PmHAKAI2, PmYTHDF1–3 | P. massoniana | Writers/Readers (resistant clones upregulated) | Upregulation linked to enhanced resistance; context-specific m6A regulatory roles | M. alternatus |
| MYB TF (regulating coumestrol, psoralidin) | Soybean (Glycine max) | TF; metabolite regulation | m6A modification drives flavonoid accumulation, strengthening defense | M. alternatus |
| BBE-like 28, POD47 | G. max | ROS-related enzymes | Hypomethylated and upregulated; enhance ROS production for basal defense near nematode feeding sites | Meloidogyne incognita |
| WRKY70, HSF A7a, MYB114, MYB124, ZFP, ERF60 | G. max | TFs | Mostly inverse relation between m6A and expression; regulate immune responses | M. incognita |
| Lectin & LRR receptor kinases | G. max | Receptor proteins | Differentially expressed; involved in immune perception and signaling | M. incognita |
| Cytochrome P450s | G. max | Secondary metabolism enzymes | Modulated by m6A; support production of defense metabolites | M. incognita |
| Ubiquitin–proteasome system components | G. max | Protein degradation machinery | Differential regulation under infection; involved in immune regulation | M. incognita |
| Resistance genes, receptor-like kinases, TFs | G. max | Defense regulators (upregulated in resistant genotype) | Reduced m6A methylation enables derepression and immune activation | Heterodera glycines |
| MLO-like proteins, negative regulators | G. max | Susceptibility genes (upregulated in susceptible genotype) | Hypomethylated, allowing expression of susceptibility pathways | H. glycines |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Srivastava, R.; Lodhi, N. Role of m6A mRNA Methylation in Plant Defense. Epigenomes 2025, 9, 42. https://doi.org/10.3390/epigenomes9040042
Srivastava R, Lodhi N. Role of m6A mRNA Methylation in Plant Defense. Epigenomes. 2025; 9(4):42. https://doi.org/10.3390/epigenomes9040042
Chicago/Turabian StyleSrivastava, Rakesh, and Niraj Lodhi. 2025. "Role of m6A mRNA Methylation in Plant Defense" Epigenomes 9, no. 4: 42. https://doi.org/10.3390/epigenomes9040042
APA StyleSrivastava, R., & Lodhi, N. (2025). Role of m6A mRNA Methylation in Plant Defense. Epigenomes, 9(4), 42. https://doi.org/10.3390/epigenomes9040042







