Deciphering the Heterogeneity of Pancreatic Cancer: DNA Methylation-Based Cell Type Deconvolution Unveils Distinct Subgroups and Immune Landscapes
Abstract
1. Introduction
2. Results
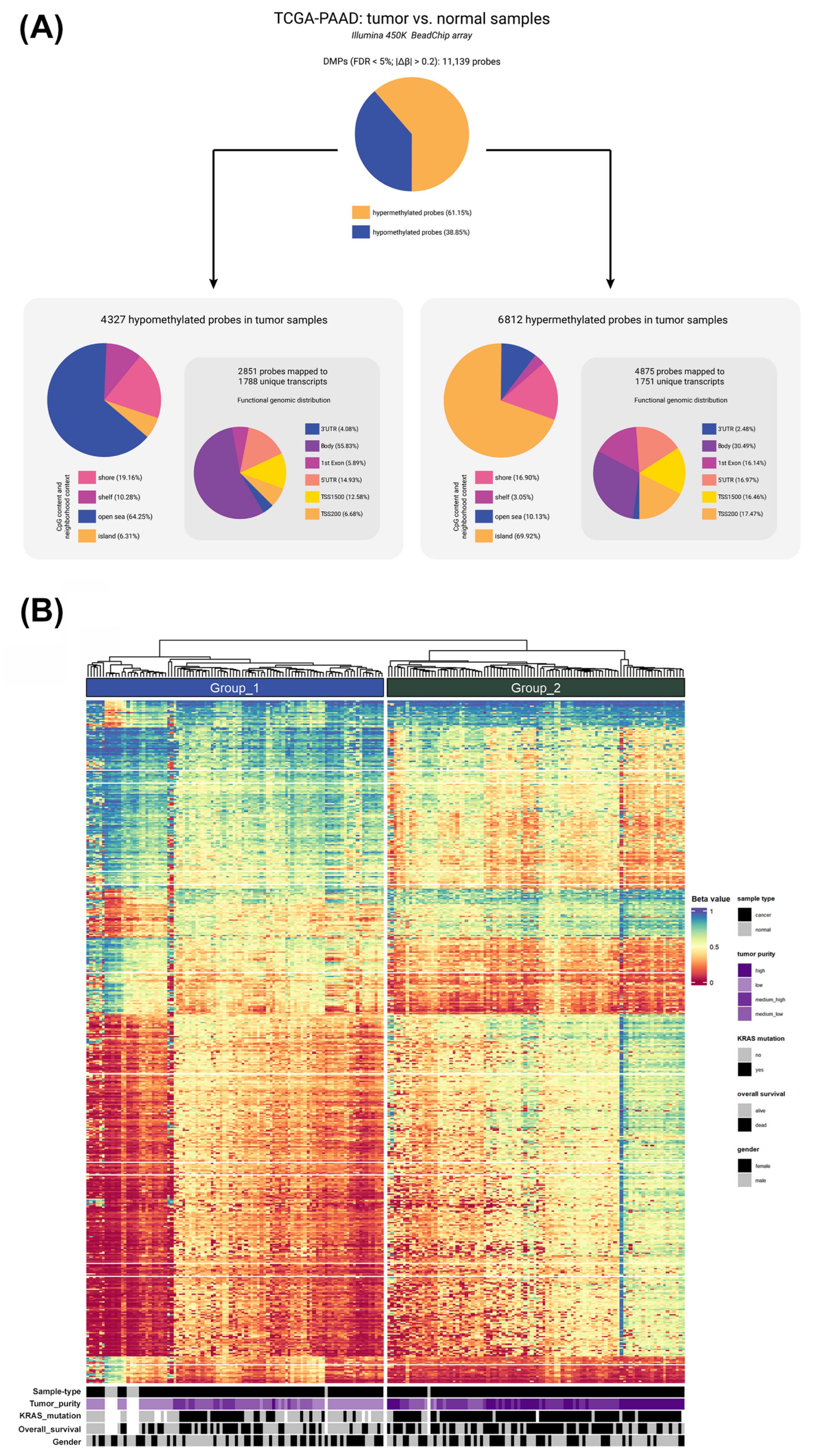
2.1. Global DNA Methylation Profiling Identifies Distinct Patterns in Pancreatic Cancer
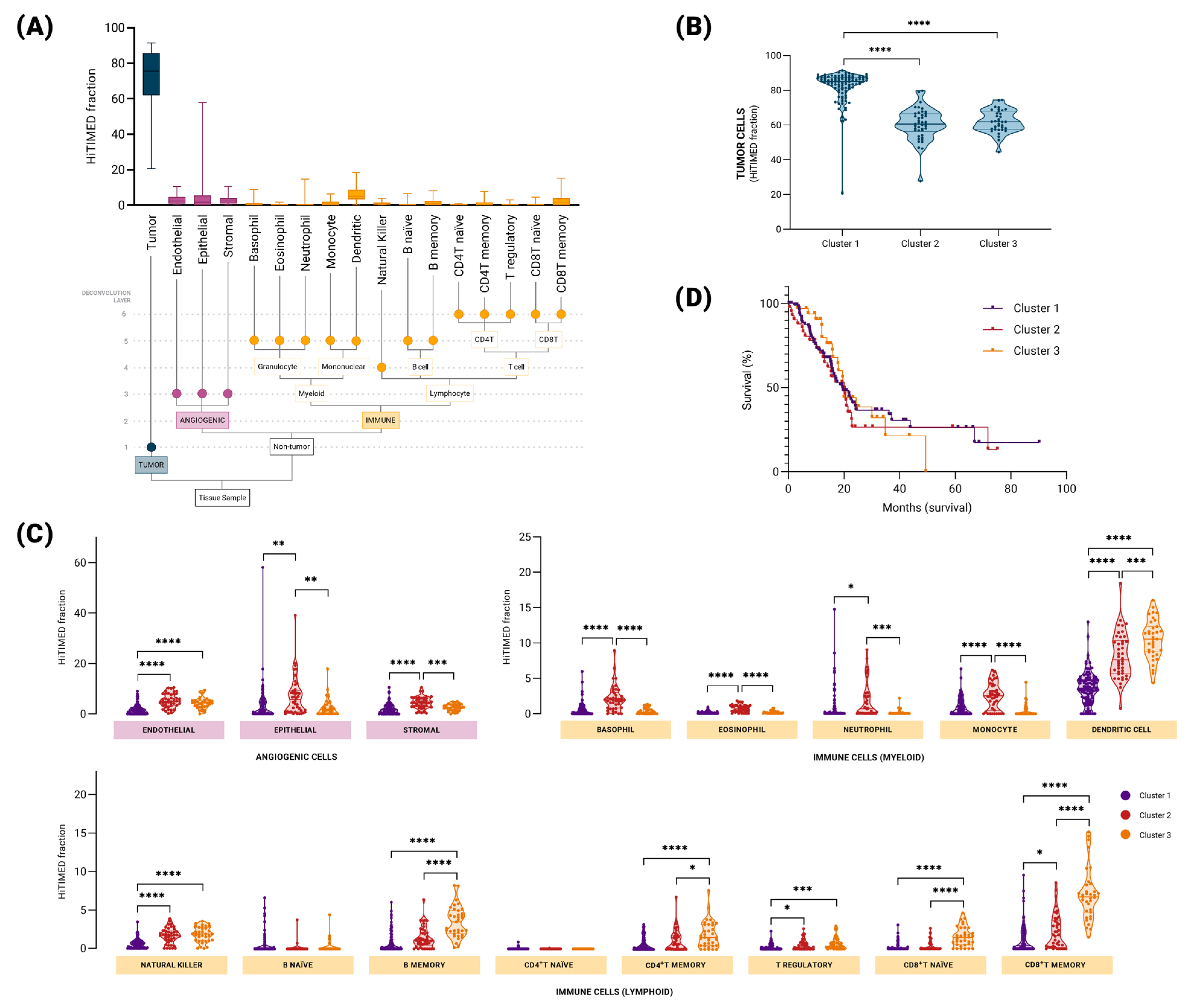
2.2. Hierarchical DNA Methylation-Based Tumor Deconvolution Reveals Distinct Patterns of Tumor Immune Microenvironment in Pancreatic Cancer
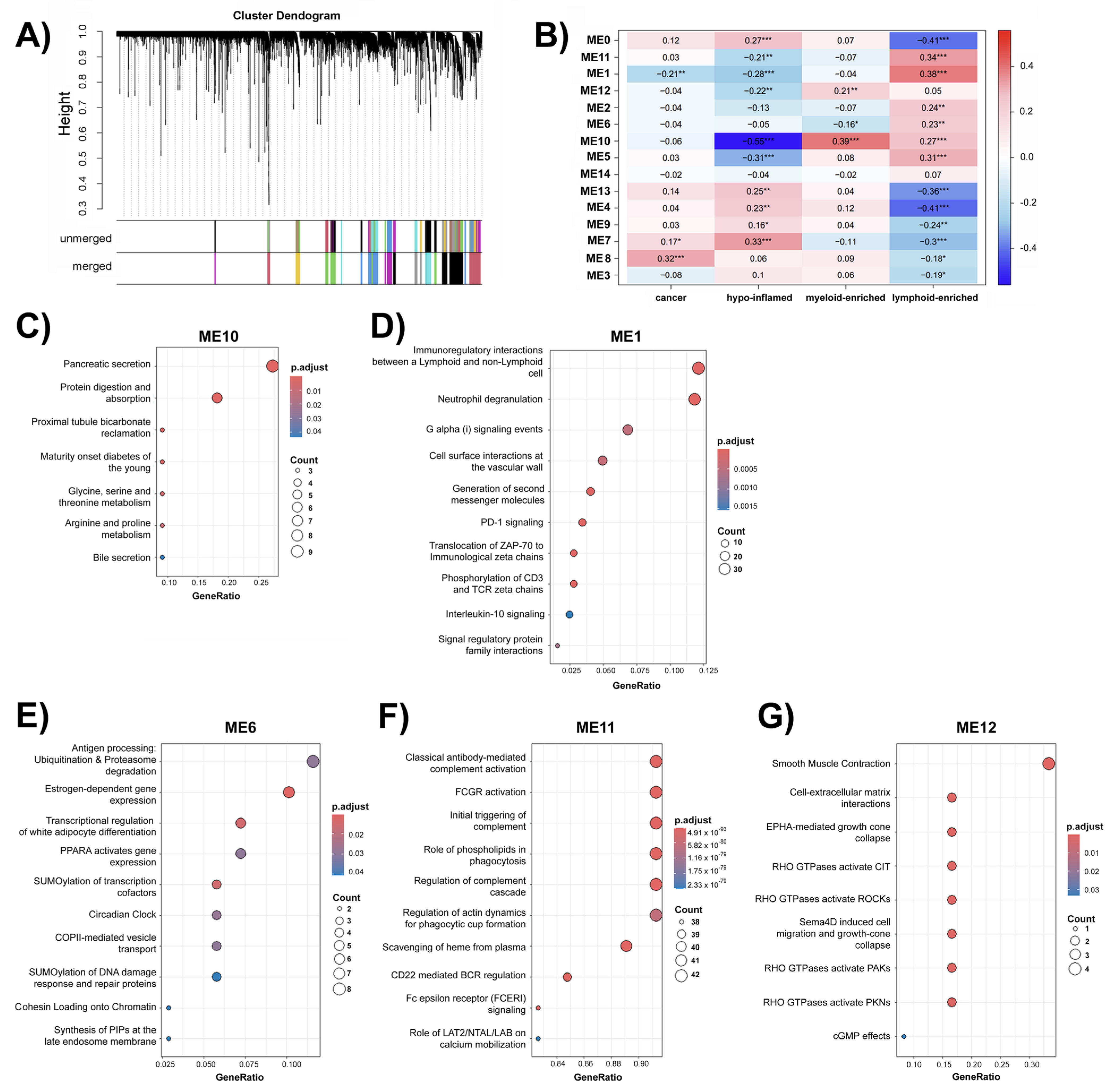
2.3. Weighted Correlation Network Analysis (WGCNA) Uncovers Group of Genes Associated with Immune Groups in Pancreatic Cancer
3. Discussion
4. Materials and Methods
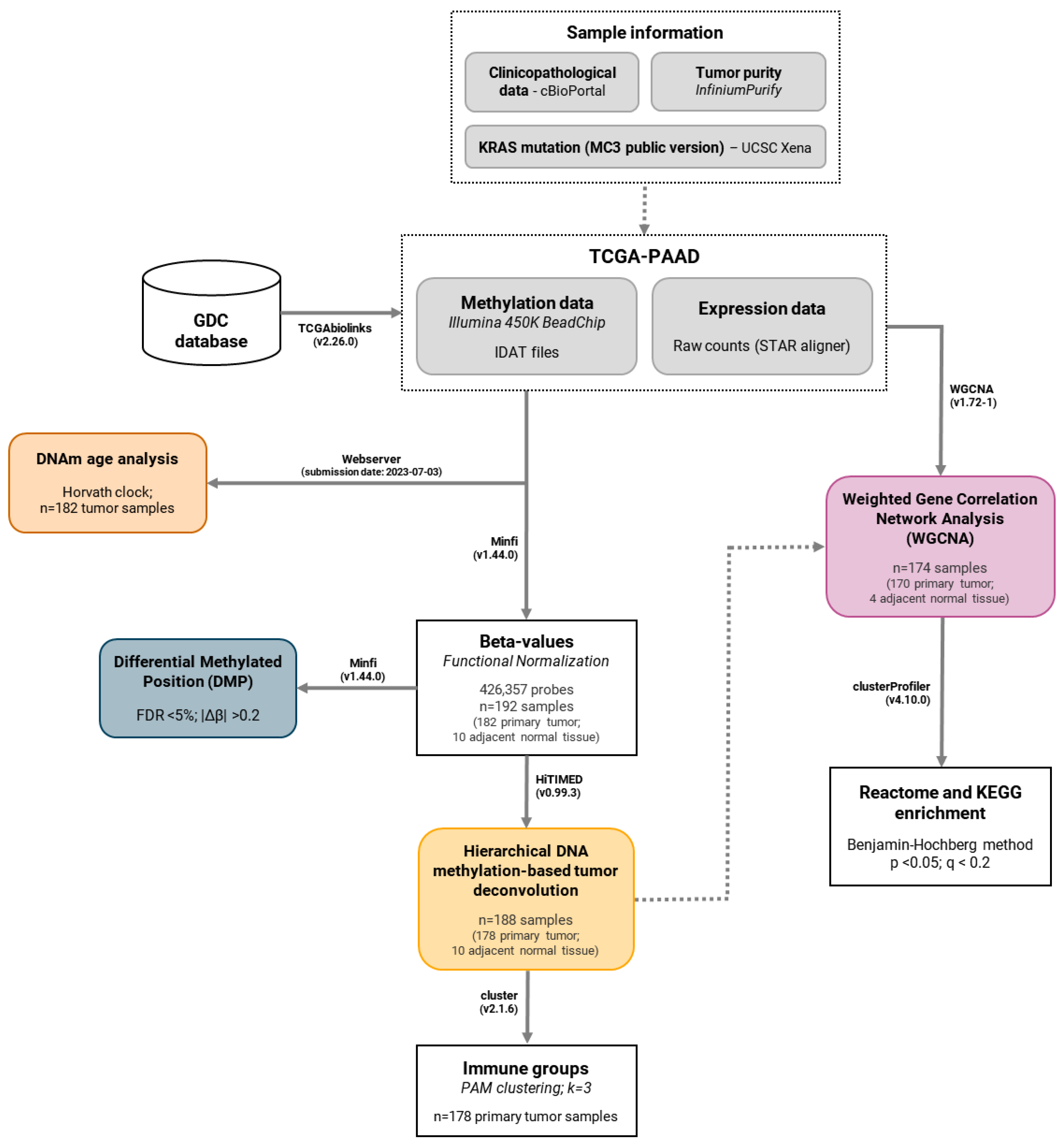
4.1. DNA Methylation Profiling in Pancreatic Cancer
4.1.1. Data Acquisition and Pre-Processing
4.1.2. Differentially Methylated Positions Analysis
4.2. DNA Methylation Age Analysis
4.3. Hierarchical DNA Methylation-Based Tumor Deconvolution
4.4. Gene Expression Analysis Using a Network-Based Approach
4.4.1. Data Acquisition and Pre-Processing
4.4.2. Weighted Gene Correlation Network Analysis (WGCNA)
4.4.3. Enrichment Analysis of WGCNA Modules
4.5. Histopathological Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| BCR | B-cell receptor |
| CAFs | Cancer-associated fibroblasts |
| CD22 | Cluster of Differentiation 22 |
| CD247 | CD247 molecule |
| CD3 | Cluster of differentiation 3 |
| CD4+T | Cluster of differentiation 4 T lymphocyte |
| CD3D | CD3 delta subunit of T-cell receptor complex |
| CD3E | CD3 epsilon subunit of T-cell receptor complex |
| CD3G | CD3 gamma subunit of T-cell receptor complex |
| CD8A | CD8 subunit alpha |
| CD8+T | Cluster of differentiation cytotoxic T lymphocytes |
| CpG | Cytosine Guanine dinucleotide |
| DMP | Differentially methylated position |
| DNAm | DNA methylation |
| EAA | Epigenetic Age Acceleration |
| ECM | Extracellular matrix |
| FDR | False discovery rate |
| GDC | Genomic Data Commons |
| GTPase | Guanosine triphosphatases |
| HiTIMED | Hierarchical Tumor Immune Microenvironment Epigenetic Deconvolution |
| IFI27 | Interferon alpha inducible protein 27 |
| IDAT | Illumina Data (raw intensity data) |
| KLRD1 | Killer cell lectin-like receptor D1 |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KRAS | Kirsten rat sarcoma viral oncogene |
| MS4A1 | Membrane spanning 4-domains A1 |
| MC3 | Multi-Center Mutation Calling in Multiple Cancers |
| ME | Module Eigengene |
| NCAM1 | Neural cell adhesion molecule 1 |
| PAAD | Pancreatic adenocarcinoma |
| PAKs | p21-activated kinase |
| PAM | Partitioning around medoids |
| PD-1 | Programmed cell death protein 1 |
| PDAC | Pancreatic ductal adenocarcinoma |
| PKNs | Protein kinases N |
| RAS-MAPK | Ras-mitogen-activated protein kinase |
| RHO | Ras homolog |
| ROCKs | Rho-associated protein kinases |
| RREB1 | Ras-responsive element binding protein 1 |
| RRID | Research Resource Identifier |
| Sema4D | Semaphorin 4D |
| SNP | Single Nucleotide Polymorphism |
| TCGA-PAAD | The Cancer Genome Atlas—Pancreatic adenocarcinoma cohort |
| TCR | T cell receptor |
| TGFB1 | Transforming growth factor beta 1 |
| TGFB2 | Transforming growth factor beta 2 |
| TGFB3 | Transforming growth factor beta 3 |
| TIME | Tumor immune microenvironment |
| TME | Tumor microenvironment |
| TOM | Topological overlap matrix |
| TP53 | Tumor suppressor p53 gene |
| TSS | Transcription Start Site |
| UCSC | The University of California, Santa Cruz |
| UTR | Untranslated region |
| VEGF | Vascular Endothelial Growth Factor |
| WGCNA | Weighted Gene Co-expression Network Analysis |
| ZAP-70 | Zeta-chain-associated protein kinase 70 |
References
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Bożyk, A.; Wojas-Krawczyk, K.; Krawczyk, P.; Milanowski, J. Tumor Microenvironment—A Short Review of Cellular and Interaction Diversity. Biology 2022, 11, 929. [Google Scholar] [CrossRef]
- Liu, B.; Xie, Y.; Zhang, Y.; Tang, G.; Lin, J.; Yuan, Z.; Liu, X.; Wang, X.; Huang, M.; Luo, Y.; et al. Spatial deconvolution from bulk DNA methylation profiles determines intratumoral epigenetic heterogeneity. Cell Biosci. 2025, 23, 7. [Google Scholar] [CrossRef]
- Gentles, A.J.; Newman, A.M.; Liu, C.L.; Bratman, S.V.; Feng, W.; Kim, D.; Nair, V.S.; Xu, Y.; Khuong, A.; Hoang, C.D.; et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015, 21, 938–945. [Google Scholar] [CrossRef]
- Raghavan, S.; Winter, P.S.; Navia, A.W.; Williams, H.L.; DenAdel, A.; Lowder, K.E.; Galvez-Reyes, J.; Kalekar, R.L.; Mulugeta, N.; Kapner, K.S.; et al. Microenvironment drives cell state, plasticity, and drug response in pancreatic cancer. Cell 2021, 184, 6119–6137.e26. [Google Scholar] [CrossRef]
- Conway, J.W.; Braden, J.; Wilmott, J.S.; Scolyer, R.A.; Long, G.V.; Pires da Silva, I. The effect of organ-specific tumor microenvironments on response patterns to immunotherapy. Front. Immunol. 2022, 17, 1030147. [Google Scholar] [CrossRef]
- Maman, S.; Witz, I.P. A history of exploring cancer in context. Nat. Rev. Cancer 2018, 18, 359–376. [Google Scholar] [CrossRef] [PubMed]
- Xue, R.; Zhang, Q.; Cao, Q.; Kong, R.; Xiang, X.; Liu, H.; Feng, M.; Wang, F.; Cheng, J.; Li, Z.; et al. Liver tumour immune microenvironment subtypes and neutrophil heterogeneity. Nature 2022, 612, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Mun, J.Y.; Leem, S.H.; Lee, J.H.; Kim, H.S. Dual Relationship Between Stromal Cells and Immune Cells in the Tumor Microenvironment. Front. Immunol. 2022, 13, 864739. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Shen, M.; Wu, L.; Yang, H.; Yao, Y.; Yang, Q.; Du, J.; Liu, L.; Li, Y.; Bai, Y. Stromal cells in the tumor microenvironment: Accomplices of tumor progression? Cell Death Dis. 2023, 14, 587. [Google Scholar] [CrossRef]
- Wang, L.; Geng, H.; Liu, Y.; Liu, L.; Chen, Y.; Wu, F.; Liu, Z.; Ling, S.; Wang, Y.; Zhou, L. Hot and cold tumors: Immunological features and the therapeutic strategies. MedComm (2020) 2023, 26, e343. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Byrne, K.T.; Yan, F.; Yamazoe, T.; Chen, Z.; Baslan, T.; Richman, L.P.; Lin, J.H.; Sun, Y.H.; Rech, A.J.; et al. Tumor Cell-Intrinsic Factors Underlie Heterogeneity of Immune Cell Infiltration and Response to Immunotherapy. Immunity 2018, 49, 178–193.e7. [Google Scholar] [CrossRef]
- Sun, X.X.; Yu, Q. Intra-tumor heterogeneity of cancer cells and its implications for cancer treatment. Acta Pharmacol. Sin. 2015, 36, 1219–1227. [Google Scholar] [CrossRef]
- Reiter, J.G.; Baretti, M.; Gerold, J.M.; Makohon-Moore, A.P.; Daud, A.; Iacobuzio-Donahue, C.A.; Azad, N.S.; Kinzler, K.W.; Nowak, M.A.; Vogelstein, B. An analysis of genetic heterogeneity in untreated cancers. Nat. Rev. Cancer 2019, 19, 639–650. [Google Scholar] [CrossRef]
- Rawla, P.; Sunkara, T.; Gaduput, V. Epidemiology of Pancreatic Cancer: Global Trends, Etiology and Risk Factors. World J. Oncol. 2019, 10, 10–27. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2017, 32, 185–203.e13. [Google Scholar] [CrossRef]
- Ho, W.J.; Jaffee, E.M.; Zheng, L. The tumour microenvironment in pancreatic cancer—Clinical challenges and opportunities. Nat. Rev. Clin. Oncol. 2020, 17, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Falcomatà, C.; Bärthel, S.; Schneider, G.; Rad, R.; Schmidt-Supprian, M.; Saur, D. Context-Specific Determinants of the Immunosuppressive Tumor Microenvironment in Pancreatic Cancer. Cancer Discov. 2023, 13, 278–297. [Google Scholar] [CrossRef] [PubMed]
- Neoptolemos, J.P.; Kleeff, J.; Michl, P.; Costello, E.; Greenhalf, W.; Palmer, D.H. Therapeutic developments in pancreatic cancer: Current and future perspectives. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 333–348. [Google Scholar] [CrossRef]
- Yabar, C.S.; Winte, J.M. Pancreatic Cancer: A Review. Gastroenterol. Clin. N. Am. 2016, 45, 429–445. [Google Scholar] [CrossRef]
- Lanfredini, S.; Thapa, A.; O’Neill, E. RAS in pancreatic cancer. Biochem. Soc. Trans. 2019, 47, 961–972. [Google Scholar] [CrossRef]
- Bear, A.S.; Vonderheide, R.H.; O’Hara, M.H. Challenges and Opportunities for Pancreatic Cancer Immunotherapy. Cancer Cell 2020, 38, 788–802. [Google Scholar] [CrossRef]
- Guidry, K.; Vasudevaraja, V.; Labbe, K.; Mohamed, H.; Serrano, J.; Guidry, B.W.; DeLorenzo, M.; Zhang, H.; Deng, J.; Sahu, S.; et al. DNA Methylation Profiling Identifies Subgroups of Lung Adenocarcinoma with Distinct Immune Cell Composition, DNA Methylation Age, and Clinical Outcome. Clin. Cancer Res. 2022, 28, 3824–3835. [Google Scholar] [CrossRef]
- Liebner, D.A.; Huang, K.; Parvin, J.D. MMAD: Microarray microdissection with analysis of differences is a computational tool for deconvoluting cell type-specific contributions from tissue samples. Bioinformatics 2014, 30, 682–689. [Google Scholar] [CrossRef] [PubMed]
- Im, Y.; Kim, Y. A Comprehensive Overview of RNA Deconvolution Methods and Their Application. Mol. Cells 2023, 46, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Khodadoust, M.S.; Liu, C.L.; Newman, A.M.; Alizadeh, A.A. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol. Biol. 2018, 1711, 243–259. [Google Scholar] [PubMed]
- Chakravarthy, A.; Furness, A.; Joshi, K.; Ghorani, E.; Ford, K.; Ward, M.J.; King, E.V.; Lechner, M.; Marafioti, T.; Quezada, S.A.; et al. Pan-cancer deconvolution of tumour composition using DNA methylation. Nat. Commun. 2018, 9, 3220. [Google Scholar] [CrossRef]
- Liu, X.; Wang, W.; Liu, X.; Zhang, Z.; Yu, L.; Li, R.; Guo, D.; Cai, W.; Quan, X.; Wu, H.; et al. Multi-omics analysis of intra-tumoural and inter-tumoural heterogeneity in pancreatic ductal adenocarcinoma. Clin. Transl. Med. 2022, 12, e670. [Google Scholar] [CrossRef]
- Ma, Y.; Zhou, X. Spatially informed cell-type deconvolution for spatial transcriptomics. Nat. Biotechnol. 2022, 40, 1349–1359. [Google Scholar] [CrossRef]
- Hwang, W.L.; Jagadeesh, K.A.; Guo, J.A.; Hoffman, H.I.; Yadollahpour, P.; Reeves, J.W.; Mohan, R.; Drokhlyansky, E.; Van Wittenberghe, N.; Ashenberg, O.; et al. Single-nucleus and spatial transcriptome profiling of pancreatic cancer identifies multicellular dynamics associated with neoadjuvant treatment. Nat. Genet. 2022, 54, 1178–1191. [Google Scholar] [CrossRef]
- Grünwald, B.T.; Devisme, A.; Andrieux, G.; Vyas, F.; Aliar, K.; McCloskey, C.W.; Macklin, A.; Jang, G.H.; Denroche, R.; Romero, J.M.; et al. Spatially confined sub-tumor microenvironments in pancreatic cancer. Cell 2021, 184, 5577–5592.e18. [Google Scholar] [CrossRef]
- Zhou, D.C.; Jayasinghe, R.G.; Chen, S.; Herndon, J.M.; Iglesia, M.D.; Navale, P.; Wendl, M.C.; Caravan, W.; Sato, K.; Storrs, E.; et al. Spatially restricted drivers and transitional cell populations cooperate with the microenvironment in untreated and chemo-resistant pancreatic cancer. Nat. Genet. 2022, 54, 1390–1405. [Google Scholar] [CrossRef]
- Wang, J.; Fite, B.Z.; Kare, A.J.; Wu, B.; Raie, M.; Tumbale, S.K.; Zhang, N.; Davis, R.R.; Tepper, C.G.; Aviran, S.; et al. Multiomic analysis for optimization of combined focal and immunotherapy protocols in murine pancreatic cancer. Theranostics 2022, 12, 7884–7902. [Google Scholar] [CrossRef]
- Trieu, V.; Potts, M.; Myers, S.; Richardson, S.; Qazi, S. TGFB2 Gene Methylation in Tumors with Low CD8+ T-Cell Infiltration Drives Positive Prognostic Overall Survival Responses in Pancreatic Ductal Adenocarcinoma. Int. J. Mol. Sci. 2025, 10, 5567. [Google Scholar] [CrossRef] [PubMed]
- Sadozai, H.; Acharjee, A.; Eppenberger-Castori, S.; Gloor, B.; Gruber, T.; Schenk, M.; Karamitopoulou, E. Distinct Stromal and Immune Features Collectively Contribute to Long-Term Survival in Pancreatic Cancer. Front. Immunol. 2021, 19, 643529. [Google Scholar] [CrossRef] [PubMed]
- Tew, B.Y.; Durand, J.K.; Bryant, K.L.; Hayes, T.K.; Peng, S.; Tran, N.L.; Gooden, G.C.; Buckley, D.N.; Der, C.J.; Baldwin, A.S.; et al. Genome-wide DNA methylation analysis of KRAS mutant cell lines. Sci. Rep. 2020, 10, 10149. [Google Scholar] [CrossRef] [PubMed]
- Sherman, M.H.; Beatty, G.L. Tumor Microenvironment in Pancreatic Cancer Pathogenesis and Therapeutic Resistance. Annu. Rev. Pathol. 2023, 18, 123–148. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, Z.; Yang, S.; Xiao, M. Extracellular matrix remodeling in tumor progression and immune escape: From mechanisms to treatments. Mol. Cancer 2023, 22, 48. [Google Scholar] [CrossRef]
- El Baba, N.; Farran, M.; Khalil, E.A.; Jaafar, L.; Fakhoury, I.; El-Sibai, M. The Role of Rho GTPases in VEGF Signaling in Cancer Cells. Anal. Cell. Pathol. 2020, 2020, 2097214. [Google Scholar] [CrossRef]
- Sierra, J.R.; Corso, S.; Caione, L.; Cepero, V.; Conrotto, P.; Cignetti, A.; Piacibello, W.; Kumanogoh, A.; Kikutani, H.; Comoglio, P.M.; et al. Tumor angiogenesis and progression are enhanced by Sema4D produced by tumor-associated macrophages. J. Exp. Med. 2008, 205, 1673–1685. [Google Scholar]
- Spiering, D.; Hodgson, L. Dynamics of the Rho-family small GTPases in actin regulation and motility. Cell Adh. Migr. 2011, 5, 170–180. [Google Scholar] [CrossRef]
- Ridley, A.J. Rho GTPase signalling in cell migration. Curr. Opin. Cell Biol. 2015, 36, 103–112. [Google Scholar] [CrossRef]
- Brummelman, J.; Pilipow, K.; Lugli, E. The Single-Cell Phenotypic Identity of Human CD8(+) and CD4(+) T Cells. Int. Rev. Cell Mol. Biol. 2018, 341, 63–124. [Google Scholar]
- Rossy, J.; Williamson, D.J.; Benzing, C.; Gaus, K. The integration of signaling and the spatial organization of the T cell synapse. Front. Immunol. 2012, 23, 352. [Google Scholar] [CrossRef]
- Sharpe, A.H.; Pauken, K.E. The diverse functions of the PD1 inhibitory pathway. Nat. Rev. Immunol. 2018, 18, 153–167. [Google Scholar] [CrossRef]
- Riley, J.L. PD-1 signaling in primary T cells. Immunol. Rev. 2009, 229, 114–125. [Google Scholar] [CrossRef]
- Clark, E.A.; Giltiay, N.V. CD22: A Regulator of Innate and Adaptive B Cell Responses and Autoimmunity. Front. Immunol. 2018, 9, 2235. [Google Scholar] [CrossRef]
- Pishesha, N.; Harmand, T.J.; Ploegh, H.L. A guide to antigen processing and presentation. Nat. Rev. Immunol. 2022, 22, 751–764. [Google Scholar] [CrossRef] [PubMed]
- Sarma, J.V.; Ward, P.A. The complement system. Cell Tissue Res. 2011, 343, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef] [PubMed]
- Aryee, M.J.; Jaffe, A.E.; Corrada-Bravo, H.; Ladd-Acosta, C.; Feinberg, A.P.; Hansen, K.D.; Irizarry, R.A. Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014, 30, 1363–1369. [Google Scholar] [CrossRef] [PubMed]
- Fortin, J.P.; Labbe, A.; Lemire, M.; Zanke, B.W.; Hudson, T.J.; Fertig, E.J.; Greenwood, C.M.; Hansen, K.D. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 2014, 15, 503. [Google Scholar] [CrossRef]
- Chen, Y.A.; Lemire, M.; Choufani, S.; Butcher, D.T.; Grafodatskaya, D.; Zanke, B.W.; Gallinger, S.; Hudson, T.J.; Weksberg, R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 2013, 8, 203–209. [Google Scholar] [CrossRef]
- Hansen, K.D. Illumina Human Methylation 450kanno.ilmn12.hg19: Annotation for Illumina’s 450k Methylation Arrays, R package, version 0.6.1; Bioconductor: 2021. Available online: https://ropensci.org/blog/2021/11/16/how-to-cite-r-and-r-packages/ (accessed on 15 August 2025).
- Ellrott, K.; Bailey, M.H.; Saksena, G.; Covington, K.R.; Kandoth, C.; Stewart, C.; Hess, J.; Ma, S.; Chiotti, K.E.; McLellan, M.; et al. Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines. Cell Syst. 2018, 6, 271–281.e7. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Feng, H.; Chen, M.; Wu, H.; Zheng, X. InfiniumPurify: An R package for estimating and accounting for tumor purity in cancer methylation research. Genes. Dis. 2018, 5, 43–45. [Google Scholar] [CrossRef]
- Hansen, K.D.; Aryee, M. Illumina Human Methylation 450k Manifest: Annotation for Illumina’s 450k Methylation Arrays, R package, version 0.4.0; Bioconductor: 2012. Available online: https://ropensci.org/blog/2021/11/16/how-to-cite-r-and-r-packages/ (accessed on 15 August 2025).
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 2013, 14, R115. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wiencke, J.K.; Kelsey, K.T.; Koestler, D.C.; Christensen, B.C.; Salas, L.A. HiTIMED: Hierarchical tumor immune microenvironment epigenetic deconvolution for accurate cell type resolution in the tumor microenvironment using tumor-type-specific DNA methylation data. J. Transl. Med. 2022, 20, 516. [Google Scholar] [CrossRef]
- Maechler, M.; Rousseeuw, P.; Struyf, A.; Hubert, M.; Hornik, K. Cluster: Cluster Analysis Basics and Extensions, R package, version 2.1.4; Bioconductor: 2022. Available online: https://ropensci.org/blog/2021/11/16/how-to-cite-r-and-r-packages/ (accessed on 15 August 2025).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 17. [Google Scholar] [CrossRef]
- Durinck, S.; Moreau, Y.; Kasprzyk, A.; Davis, S.; De Moor, B.; Brazma, A.; Huber, W. BioMart and Bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics 2005, 21, 3439–3440. [Google Scholar] [CrossRef] [PubMed]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef] [PubMed]
- Carlson, M.; Falcon, S.; Pages, H.; Li, N. org.Hs.eg.db: Genome wide annotation for Human. R package, 2019. version 3.8.2.
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Gutman, D.A.; Cobb, J.; Somanna, D.; Park, Y.; Wang, F.; Kurc, T.; Saltz, J.H.; Brat, D.J.; Cooper, L.A. Cancer Digital Slide Archive: An informatics resource to support integrated in silico analysis of TCGA pathology data. J. Am. Med. Inform. Assoc. 2013, 20, 1091–1098. [Google Scholar] [CrossRef]
| Group 1 | Group 2 | |||||
|---|---|---|---|---|---|---|
| Variables | Samples Total | N | (%) | N | (%) | p-Value * |
| Gender | 182 | 87 | 95 | (100%) | 0.6553 | |
| Female | 41 | 41 | ||||
| Male | 46 | 54 | (100%) | |||
| Age (years) | 87 | 95 | 0.6362 | |||
| <50 | 11 | 9 | ||||
| >50 | 76 | 86 | ||||
| Overall Survival | 182 | 87 | (100%) | 95 | (100%) | 0.0046 |
| Alive | 50 | (57.5%) | 34 | (35.8%) | ||
| Dead | 37 | (42.5%) | 61 | (64.2%) | ||
| KRAS mutation | 173 | 80 | (100%) | 93 | (100%) | <0.0001 |
| Presence | 30 | (37.5%) | 84 | (90.3%) | ||
| Absence | 50 | (62.5%) | 9 | (9.7%) | ||
| Tumor purity | 182 | 87 | (100%) | 95 | (100%) | <0.0001 |
| Low | 46 | (52.9%) | 2 | (2.1%) | ||
| Medium–low | 27 | (31.0%) | 12 | (12.6%) | ||
| Medium–high | 13 | (14.9%) | 37 | (38.9%) | ||
| High | 1 | (1.1%) | 44 | (46.3%) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mitsuyasu Barbosa, B.; Todorovic Fabro, A.; da Silva Gomes, R.; Rainho, C.A. Deciphering the Heterogeneity of Pancreatic Cancer: DNA Methylation-Based Cell Type Deconvolution Unveils Distinct Subgroups and Immune Landscapes. Epigenomes 2025, 9, 34. https://doi.org/10.3390/epigenomes9030034
Mitsuyasu Barbosa B, Todorovic Fabro A, da Silva Gomes R, Rainho CA. Deciphering the Heterogeneity of Pancreatic Cancer: DNA Methylation-Based Cell Type Deconvolution Unveils Distinct Subgroups and Immune Landscapes. Epigenomes. 2025; 9(3):34. https://doi.org/10.3390/epigenomes9030034
Chicago/Turabian StyleMitsuyasu Barbosa, Barbara, Alexandre Todorovic Fabro, Roberto da Silva Gomes, and Claudia Aparecida Rainho. 2025. "Deciphering the Heterogeneity of Pancreatic Cancer: DNA Methylation-Based Cell Type Deconvolution Unveils Distinct Subgroups and Immune Landscapes" Epigenomes 9, no. 3: 34. https://doi.org/10.3390/epigenomes9030034
APA StyleMitsuyasu Barbosa, B., Todorovic Fabro, A., da Silva Gomes, R., & Rainho, C. A. (2025). Deciphering the Heterogeneity of Pancreatic Cancer: DNA Methylation-Based Cell Type Deconvolution Unveils Distinct Subgroups and Immune Landscapes. Epigenomes, 9(3), 34. https://doi.org/10.3390/epigenomes9030034







