Integrated Multimodal Omics and Dietary Approaches for the Management of Neurodegeneration
Abstract
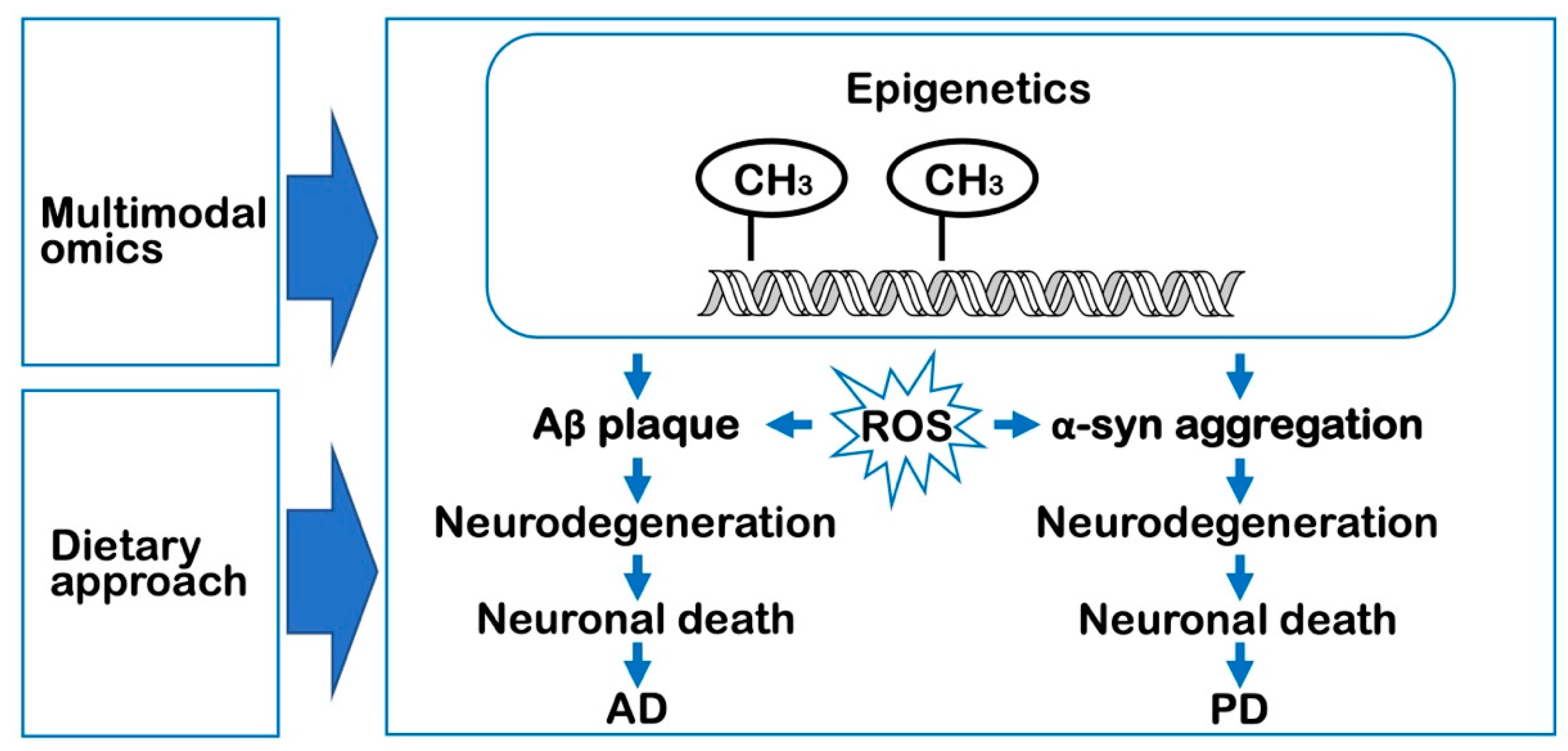
:1. Introduction
2. Epigenetics and Alzheimer’s Disease
3. Epigenetics and Parkinson’s Disease
4. Multimodal Omics Studies for the Management of Neurodegenerative Diseases
5. Dietary Approaches for the Management of Neurodegeneration
6. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Aβ | amyloid-β |
| AD | Alzheimer’s disease |
| AI | artificial intelligence |
| APOE | apolipoprotein E |
| ATAC-seq | Assay for Transposase-Accessible Chromatin sequencing |
| CITE-seq | Cellular Indexing of Transcriptomes and Epitopes by sequencing |
| CSF | cerebrospinal fluid |
| HDAC | histone deacetylase |
| lncRNA | long non-coding RNA |
| MAPT | microtubule-associated protein tau |
| miRNA | microRNA |
| ncRNA | non-coding RNA |
| PD | Parkinson’s disease |
| REAP-seq | RNA Expression and Protein sequencing |
| ROS | reactive oxygen species |
| scRNA-seq | single-cell RNA sequencing |
| scATAC-seq | Single-Cell Assay for Transposase-Accessible Chromatin sequencing |
| SN | substantia nigra |
| α-syn | α-synuclein |
| t-SNE | t-stochastic neighbor embedding |
| UTR | untranslated region |
References
- Bossy-Wetzel, E.; Schwarzenbacher, R.; Lipton, S.A. Molecular pathways to neurodegeneration. Nat. Med. 2004, 10, S2–S9. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Dan, X.; Babbar, M.; Wei, Y.; Hasselbalch, S.G.; Croteau, D.L.; Bohr, V.A. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 2019, 15, 565–581. [Google Scholar] [CrossRef] [PubMed]
- Hansson, O. Biomarkers for neurodegenerative diseases. Nat. Med. 2021, 27, 954–963. [Google Scholar] [CrossRef] [PubMed]
- Teunissen, C.E.; Verberk, I.M.W.; Thijssen, E.H.; Vermunt, L.; Hansson, O.; Zetterberg, H.; van der Flier, W.M.; Mielke, M.M.; Del Campo, M. Blood-based biomarkers for Alzheimer’s disease: Towards clinical implementation. Lancet Neurol. 2022, 21, 66–77. [Google Scholar] [CrossRef]
- Dubois, B.; Villain, N.; Frisoni, G.B.; Rabinovici, G.D.; Sabbagh, M.; Cappa, S.; Bejanin, A.; Bombois, S.; Epelbaum, S.; Teichmann, M.; et al. Clinical diagnosis of Alzheimer’s disease: Recommendations of the International Working Group. Lancet Neurol. 2021, 20, 484–496. [Google Scholar] [CrossRef]
- Berger, S.L.; Kouzarides, T.; Shiekhattar, R.; Shilatifard, A. An operational definition of epigenetics. Genes Dev. 2009, 23, 781–783. [Google Scholar] [CrossRef]
- Sen, P.; Shah, P.P.; Nativio, R.; Berger, S.L. Epigenetic mechanisms of longevity and aging. Cell 2016, 166, 822–839. [Google Scholar] [CrossRef]
- Wilson, D.M., 3rd; Cookson, M.R.; Van Den Bosch, L.; Zetterberg, H.; Holtzman, D.M.; Dewachter, I. Hallmarks of neurodegenerative diseases. Cell 2023, 186, 693–714. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- Migliore, L.; Coppedè, F. Gene-environment interactions in Alzheimer disease: The emerging role of epigenetics. Nat. Rev. Neurol. 2022, 18, 643–660. [Google Scholar] [CrossRef]
- Berson, A.; Nativio, R.; Berger, S.L.; Bonini, N.M. Epigenetic regulation in neurodegenerative diseases. Trends Neurosci. 2018, 41, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Athanasopoulos, D.; Karagiannis, G.; Tsolaki, M. Recent findings in Alzheimer disease and nutrition focusing on epigenetics. Adv. Nutr. 2016, 7, 917–927. [Google Scholar] [CrossRef] [PubMed]
- Spillantini, M.G.; Goedert, M. Tau pathology and neurodegeneration. Lancet Neurol. 2013, 12, 609–622. [Google Scholar] [CrossRef]
- Leng, F.; Edison, P. Neuroinflammation and microglial activation in Alzheimer disease: Where do we go from here? Nat. Rev. Neurol. 2021, 17, 157–172. [Google Scholar] [CrossRef]
- Matsuda, S.; Nakagawa, Y.; Tsuji, A.; Kitagishi, Y.; Nakanishi, A.; Murai, T. Implications of PI3K/AKT/PTEN signaling on superoxide dismutases expression and in the pathogenesis of Alzheimer’s disease. Diseases 2018, 6, E28. [Google Scholar] [CrossRef] [PubMed]
- Murai, T.; Matsuda, S. Therapeutic implications of probiotics in the gut microbe-modulated neuroinflammation and progression of Alzheimer’s disease. Life 2023, 13, 1466. [Google Scholar] [CrossRef]
- Halliwell, B. Oxidative stress and neurodegeneration: Where are we now? J. Neurochem. 2006, 97, 1634–1658. [Google Scholar] [CrossRef]
- Contestabile, A. Oxidative stress in neurodegeneration: Mechanisms and therapeutic perspectives. Curr. Top. Med. Chem. 2001, 1, 553–568. [Google Scholar] [CrossRef]
- Murai, T.; Matsuda, S. Pleiotropic signaling by reactive oxygen species concerted with dietary phytochemicals and microbial-derived metabolites as potent therapeutic regulators of the tumor microenvironment. Antioxidants 2023, 12, 1056. [Google Scholar] [CrossRef]
- Ionescu-Tucker, A.; Cotman, C.W. Emerging roles of oxidative stress in brain aging and Alzheimer’s disease. Neurobiol. Aging 2021, 107, 86–95. [Google Scholar] [CrossRef]
- Baker, D.J.; Petersen, R.C. Cellular senescence in brain aging and neurodegenerative diseases: Evidence and perspectives. J. Clin. Investig. 2018, 128, 1208–1216. [Google Scholar] [CrossRef]
- Zawia, N.H.; Lahiri, D.K.; Cardozo-Pelaez, F. Epigenetics, oxidative stress, and Alzheimer disease. Free Radic. Biol. Med. 2009, 46, 1241–1249. [Google Scholar] [CrossRef] [PubMed]
- Dauer, W.; Przedborski, S. Parkinson’s disease: Mechanisms and models. Neuron 2003, 39, 889–909. [Google Scholar] [CrossRef] [PubMed]
- Borsche, M.; Pereira, S.L.; Klein, C.; Grünewald, A. Mitochondria and Parkinson’s disease: Clinical, molecular, and translational aspects. J. Parkinsons Dis. 2021, 11, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Kang, X.; Hu, J.; Zhang, D.; Liang, Z.; Meng, F.; Zhang, X.; Xue, Y.; Maimon, R.; Dowdy, S.F.; et al. Reversing a model of Parkinson’s disease with in situ converted nigral neurons. Nature 2020, 582, 550–556. [Google Scholar] [CrossRef]
- Nakano, N.; Matsuda, S.; Ichimura, M.; Minami, A.; Ogino, M.; Murai, T.; Kitagishi, Y. PI3K/AKT signaling mediated by G,protein-coupled receptors is involved in neurodegenerative Parkinson’s disease. Int. J. Mol. Med. 2017, 39, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Elkouzi, A.; Vedam-Mai, V.; Eisinger, R.S.; Okun, M.S. Emerging therapies in Parkinson disease—Repurposed drugs and new approaches. Nat. Rev. Neurol. 2019, 15, 204–223. [Google Scholar] [CrossRef]
- Labbé, C.; Lorenzo-Betancor, O.; Ross, O.A. Epigenetic regulation in Parkinson’s disease. Acta Neuropathol. 2016, 132, 515–530. [Google Scholar] [CrossRef]
- Pavlou, M.A.S.; Outeiro, T.F. Epigenetics in Parkinson’s Disease. Adv. Exp. Med. Biol. 2017, 978, 363–390. [Google Scholar]
- Masliah, E.; Dumaop, W.; Galasko, D.; Desplats, P. Distinctive patterns of DNA methylation associated with Parkinson disease: Identification of concordant epigenetic changes in brain and peripheral blood leukocytes. Epigenetics 2013, 8, 1030–1038. [Google Scholar] [CrossRef]
- Espay, A.J.; Brundin, P.; Lang, A.E. Precision medicine for disease modification in Parkinson disease. Nat. Rev. Neurol. 2017, 13, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Nativio, R.; Donahue, G.; Berson, A.; Lan, Y.; Amlie-Wolf, A.; Tuzer, F.; Toledo, J.B.; Gosai, S.J.; Gregory, B.D.; Torres, C.; et al. Dysregulation of the epigenetic landscape of normal aging in Alzheimer’s disease. Nat. Neurosci. 2018, 21, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Sun, Z.; Yin, H.; Liu, X.; Wang, J.; Zhao, H.; Pang, C.H.; Wu, T.; Li, S.; Yin, Z.; et al. Data-driven materials innovation and applications. Adv. Mater. 2022, 34, e2104113. [Google Scholar] [CrossRef]
- Steponaitis, G.; Stakaitis, R.; Valiulyte, I.; Krusnauskas, R.; Dragunaite, R.; Urbanavičiūtė, R.; Tamasauskas, A.; Skiriute, D. Transcriptome-wide analysis of glioma stem cell specific m6A modifications in long-non-coding RNAs. Sci. Rep. 2022, 12, 5431. [Google Scholar] [CrossRef]
- Buenrostro, J.D.; Wu, B.; Litzenburger, U.M.; Ruff, D.; Gonzales, M.L.; Snyder, M.P.; Chang, H.Y.; Greenleaf, W.J. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 2015, 523, 486–490. [Google Scholar] [CrossRef]
- Narayanan, H.; Dingfelder, F.; Butté, A.; Lorenzen, N.; Sokolov, M.; Arosio, P. Machine learning for biologics: Opportunities for protein engineering, developability, and formulation. Trends Pharmacol. Sci. 2021, 42, 151–165. [Google Scholar] [CrossRef]
- Stoeckius, M.; Hafemeister, C.; Stephenson, W.; Houck-Loomis, B.; Chattopadhyay, P.K.; Swerdlow, H.; Satija, R.; Smibert, P. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 2017, 14, 865–868. [Google Scholar] [CrossRef] [PubMed]
- Peterson, V.M.; Zhang, K.X.; Kumar, N.; Wong, J.; Li, L.; Wilson, D.C.; Moore, R.; McClanahan, T.K.; Sadekova, S.; Klappenbach, J.A. Multiplexed quantification of proteins and transcripts in single cells. Nat. Biotechnol. 2017, 35, 936–939. [Google Scholar] [CrossRef]
- Cheung, P.; Vallania, F.; Warsinske, H.C.; Donato, M.; Schaffert, S.; Chang, S.E.; Dvorak, M.; Dekker, C.L.; Davis, M.M.; Utz, P.J.; et al. Single-cell chromatin modification profiling reveals increased epigenetic variations with aging. Cell 2018, 173, 1385–1397.e14. [Google Scholar] [CrossRef]
- Hudson, W.H.; Wieland, A. Technology meets TILs: Deciphering T cell function in the -omics era. Cancer Cell 2023, 41, 41–57. [Google Scholar] [CrossRef]
- Ahmad, A.; Sala, F.; Paiè, P.; Candeo, A.; D’Annunzio, S.; Zippo, A.; Frindel, C.; Osellame, R.; Bragheri, F.; Bassi, A.; et al. On the robustness of machine learning algorithms toward microfluidic distortions for cell classification via on-chip fluorescence microscopy. Lab Chip 2022, 22, 3453–3463. [Google Scholar] [CrossRef] [PubMed]
- Myszczynska, M.A.; Ojamies, P.N.; Lacoste, A.M.B.; Neil, D.; Saffari, A.; Mead, R.; Hautbergue, G.M.; Holbrook, J.D.; Ferraiuolo, L. Applications of machine learning to diagnosis and treatment of neurodegenerative diseases. Nat. Rev. Neurol. 2020, 16, 440–456. [Google Scholar] [CrossRef] [PubMed]
- Efremova, M.; Teichmann, S.A. Computational methods for single-cell omics across modalities. Nat. Methods 2020, 17, 14–17. [Google Scholar] [CrossRef] [PubMed]
- Corces, M.R.; Shcherbina, A.; Kundu, S.; Gloudemans, M.J.; Frésard, L.; Granja, J.M.; Louie, B.H.; Eulalio, T.; Shams, S.; Bagdatli, S.T.; et al. Single-cell epigenomic analyses implicate candidate causal variants at inherited risk loci for Alzheimer’s and Parkinson’s diseases. Nat. Genet. 2020, 52, 1158–1168. [Google Scholar] [CrossRef] [PubMed]
- Nativio, R.; Lan, Y.; Donahue, G.; Sidoli, S.; Berson, A.; Srinivasan, A.R.; Shcherbakova, O.; Amlie-Wolf, A.; Nie, J.; Cui, X.; et al. An integrated multi-omics approach identifies epigenetic alterations associated with Alzheimer’s disease. Nat Genet. 2020, 52, 1024–1035. [Google Scholar] [CrossRef] [PubMed]
- Zare Harofte, S.; Soltani, M.; Siavashy, S.; Raahemifar, K. Recent advances of utilizing artificial intelligence in lab on a chip for diagnosis and treatment. Small 2022, 18, e2203169. [Google Scholar] [CrossRef]
- Pratapa, A.; Doron, M.; Caicedo, J.C. Image-based cell phenotyping with deep learning. Curr. Opin. Chem. Biol. 2021, 65, 9–17. [Google Scholar] [CrossRef]
- Thaung Zaw, J.J.; Howe, P.R.C.; Wong, R.H.X. Sustained cerebrovascular and cognitive benefits of resveratrol in postmenopausal women. Nutrients 2020, 12, 828. [Google Scholar] [CrossRef]
- Turner, R.S.; Thomas, R.G.; Craft, S.; van Dyck, C.H.; Mintzer, J.; Reynolds, B.A.; Brewer, J.B.; Rissman, R.A.; Raman, R.; Aisen, P.S.; et al. A randomized, double-blind, placebo-controlled trial of resveratrol for Alzheimer disease. Neurology 2015, 85, 1383–1391. [Google Scholar] [CrossRef]
- Izquierdo, V.; Palomera-Ávalos, V.; Pallàs, M.; Griñán-Ferré, C. Resveratrol supplementation attenuates cognitive and molecular alterations under maternal high-fat diet intake: Epigenetic inheritance over generations. Int. J. Mol. Sci. 2021, 22, 1453. [Google Scholar] [CrossRef]
- Ma, H.; Johnson, S.L.; Liu, W.; DaSilva, N.A.; Meschwitz, S.; Dain, J.A.; Seeram, N.P. Evaluation of polyphenol anthocyanin-enriched extracts of blackberry, black raspberry, blueberry, cranberry, red raspberry, and strawberry for free radical scavenging, reactive carbonyl species trapping, anti-glycation, anti-β-amyloid aggregation, and microglial neuroprotective effects. Int. J. Mol. Sci. 2018, 19, 461. [Google Scholar]
- Pérez-Barrón, G.; Montes, S.; Aguirre-Vidal, Y.; Santiago, M.; Gallardo, E.; Espartero, J.L.; Ríos, C.; Monroy-Noyola, A. Antioxidant effect of hydroxytyrosol, hydroxytyrosol acetate and nitrohydroxytyrosol in a rat MPP+ model of Parkinson’s disease. Neurochem. Res. 2021, 46, 2923–2935. [Google Scholar] [CrossRef] [PubMed]
- Ardah, M.T.; Paleologou, K.E.; Lv, G.; Menon, S.A.; Abul Khair, S.B.; Lu, J.H.; Safieh-Garabedian, B.; Al-Hayani, A.A.; Eliezer, D.; Li, M.; et al. Ginsenoside Rb1 inhibits fibrillation and toxicity of alpha-synuclein and disaggregates preformed fibrils. Neurobiol. Dis. 2015, 74, 89–101. [Google Scholar] [CrossRef]
- Sun, X.; Cao, Y.B.; Hu, L.F.; Yang, Y.P.; Li, J.; Wang, F.; Liu, C.F. ASICs mediate the modulatory effect by paeoniflorin on α-synuclein autophagic degradation. Brain Res. 2011, 1396, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Rahimmi, A.; Tozandehjani, S.; Daraei, M.; Khademerfan, M. The neuroprotective roles of dietary micronutrients on Parkinson’s disease: A review. Mol. Biol. Rep. 2022, 49, 8051–8060. [Google Scholar] [CrossRef]
- Tomeva, E.; Krammer, U.D.B.; Switzeny, O.J.; Haslberger, A.G.; Hippe, B. Sex-specific miRNA differences in liquid biopsies from subjects with solid tumors and healthy controls. Epigenomes 2023, 7, 2. [Google Scholar] [CrossRef]
- Krammer, U.D.B.; Tschida, S.; Berner, J.; Lilja, S.; Switzeny, O.J.; Hippe, B.; Rust, P.; Haslberger, A.G. MiRNA-based “fitness score” to assess the individual response to diet, metabolism, and exercise. J. Int. Soc. Sports Nutr. 2022, 19, 455–473. [Google Scholar] [CrossRef] [PubMed]
- Liesinger, A.M.; Graff-Radford, N.R.; Duara, R.; Carter, R.E.; Hanna Al-Shaikh, F.S.; Koga, S.; Hinkle, K.M.; DiLello, S.K.; Johnson, M.F.; Aziz, A.; et al. Sex and age interact to determine clinicopathologic differences in Alzheimer’s disease. Acta Neuropathol. 2018, 136, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Gillies, G.E.; Pienaar, I.S.; Vohra, S.; Qamhawi, Z. Sex differences in Parkinson’s disease. Front. Neuroendocrinol. 2014, 35, 370–384. [Google Scholar] [CrossRef]
- López-Cerdán, A.; Andreu, Z.; Hidalgo, M.R.; Grillo-Risco, R.; Català-Senent, J.F.; Soler-Sáez, I.; Neva-Alejo, A.; Gordillo, F.; de la Iglesia-Vayá, M.; García-García, F. Unveiling sex-based differences in Parkinson’s disease: A comprehensive meta-analysis of transcriptomic studies. Biol. Sex Differ. 2022, 13, 68. [Google Scholar] [CrossRef]
- Sharma, V.K.; Mehta, V.; Singh, T.G. Alzheimer’s disorder: Epigenetic connection and associated risk factors. Curr. Neuropharmacol. 2020, 18, 740–753. [Google Scholar] [CrossRef] [PubMed]

| Methods | Targets | References |
|---|---|---|
| scATAC-seq 1 | Epigenome | [35] |
| CITE-seq 2 | Transcriptome and proteome | [37] |
| REAP-seq 3 | Transcriptome and proteome | [38] |
| EpiTOF 4 | Epigenome | [39] |
| Phytochemicals | Effects | References |
|---|---|---|
| Resveratrol | Enhance cognition | [48] |
| Resveratrol | Decrease in Aβ level | [49] |
| Resveratrol | Epigenetics | [50] |
| Anthocyanin | Inhibit Aβ fibrillation | [51] |
| Hydroxytyrosol | Antioxidant effect in PD | [52] |
| Ginsenoside | Decrease in α-syn fibrillation | [53] |
| Paeoniflorin | Decrease in α-syn accumulation | [54] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murai, T.; Matsuda, S. Integrated Multimodal Omics and Dietary Approaches for the Management of Neurodegeneration. Epigenomes 2023, 7, 20. https://doi.org/10.3390/epigenomes7030020
Murai T, Matsuda S. Integrated Multimodal Omics and Dietary Approaches for the Management of Neurodegeneration. Epigenomes. 2023; 7(3):20. https://doi.org/10.3390/epigenomes7030020
Chicago/Turabian StyleMurai, Toshiyuki, and Satoru Matsuda. 2023. "Integrated Multimodal Omics and Dietary Approaches for the Management of Neurodegeneration" Epigenomes 7, no. 3: 20. https://doi.org/10.3390/epigenomes7030020
APA StyleMurai, T., & Matsuda, S. (2023). Integrated Multimodal Omics and Dietary Approaches for the Management of Neurodegeneration. Epigenomes, 7(3), 20. https://doi.org/10.3390/epigenomes7030020





