The Role of Polycomb Proteins in Cell Lineage Commitment and Embryonic Development
Abstract
1. Introduction
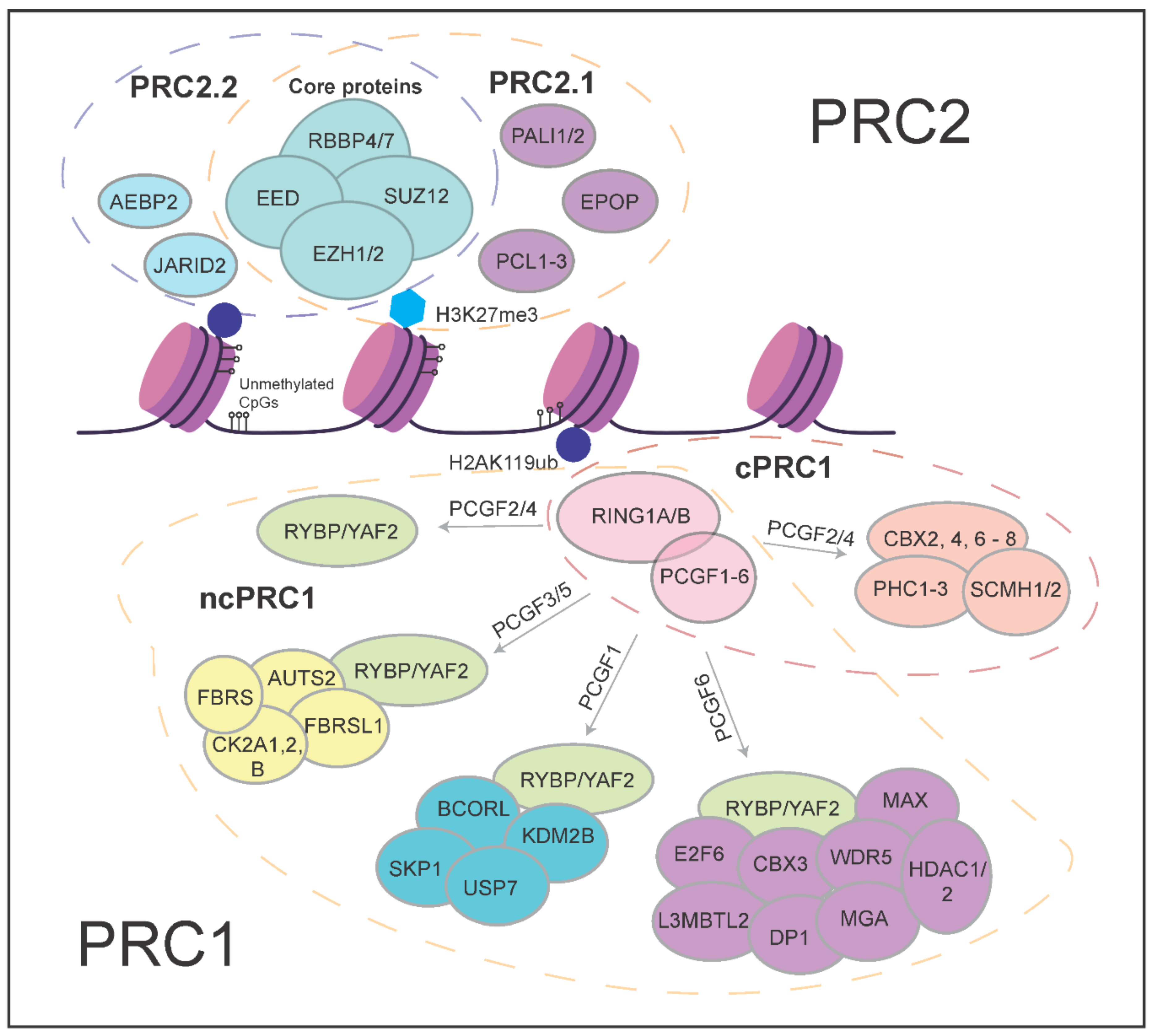
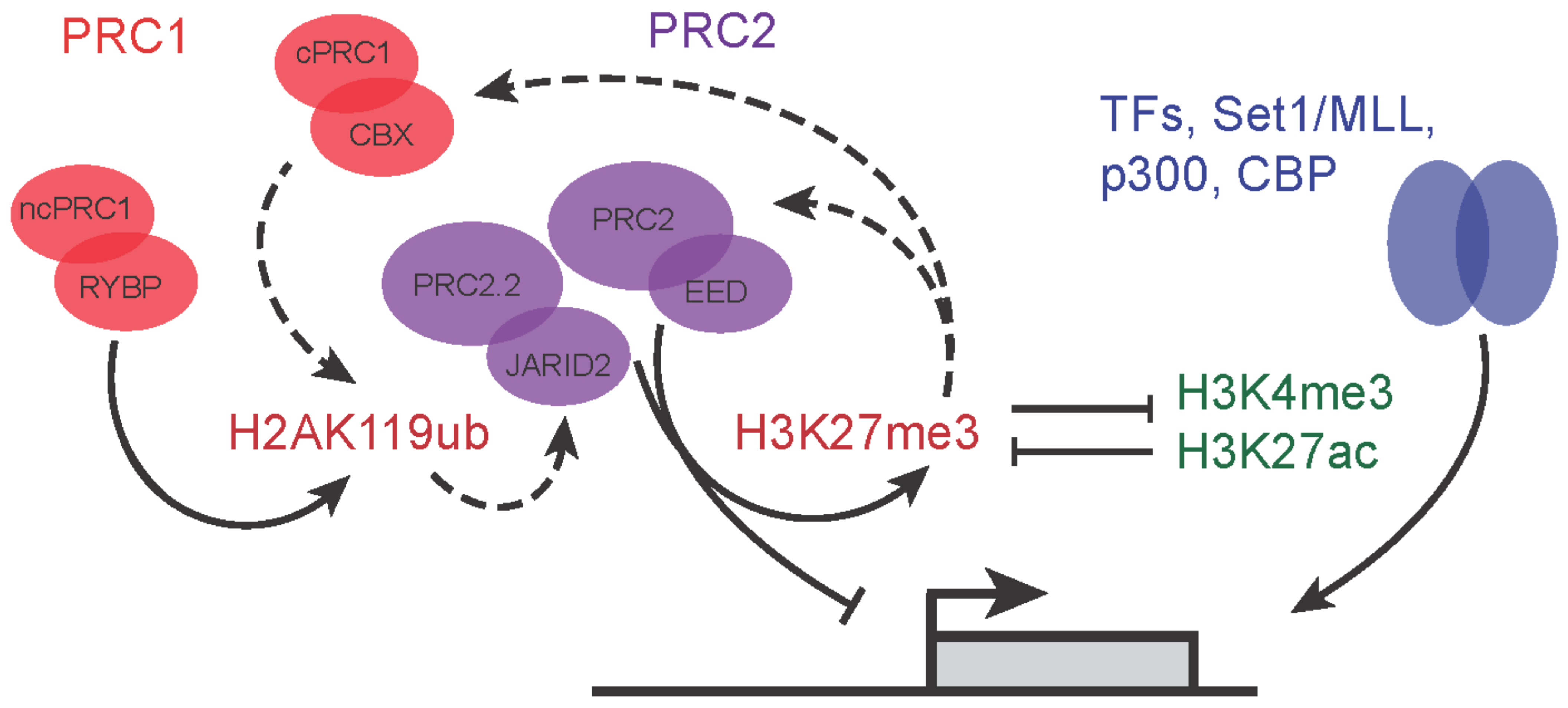
2. More Than the Sum of Its Parts—Polycomb Proteins and Their Molecular Functions
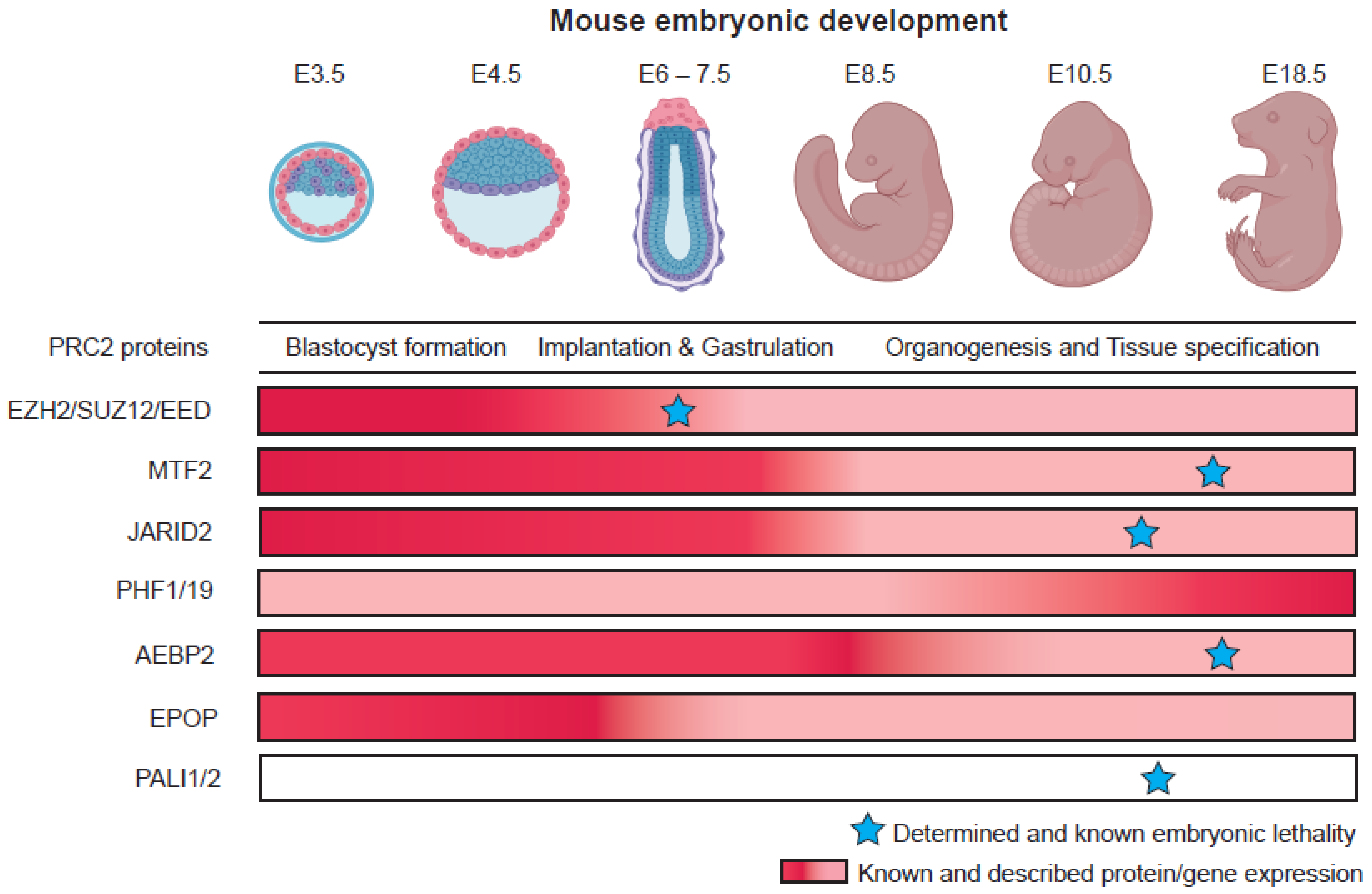
3. Roles of Polycomb Group Proteins in Pluripotency and Early Development
3.1. PRC2 Variant Complexes and Their Roles in Embryonic Stem Cells
3.2. PRC1 Variant Complexes and Their Roles in Embryonic Stem Cells
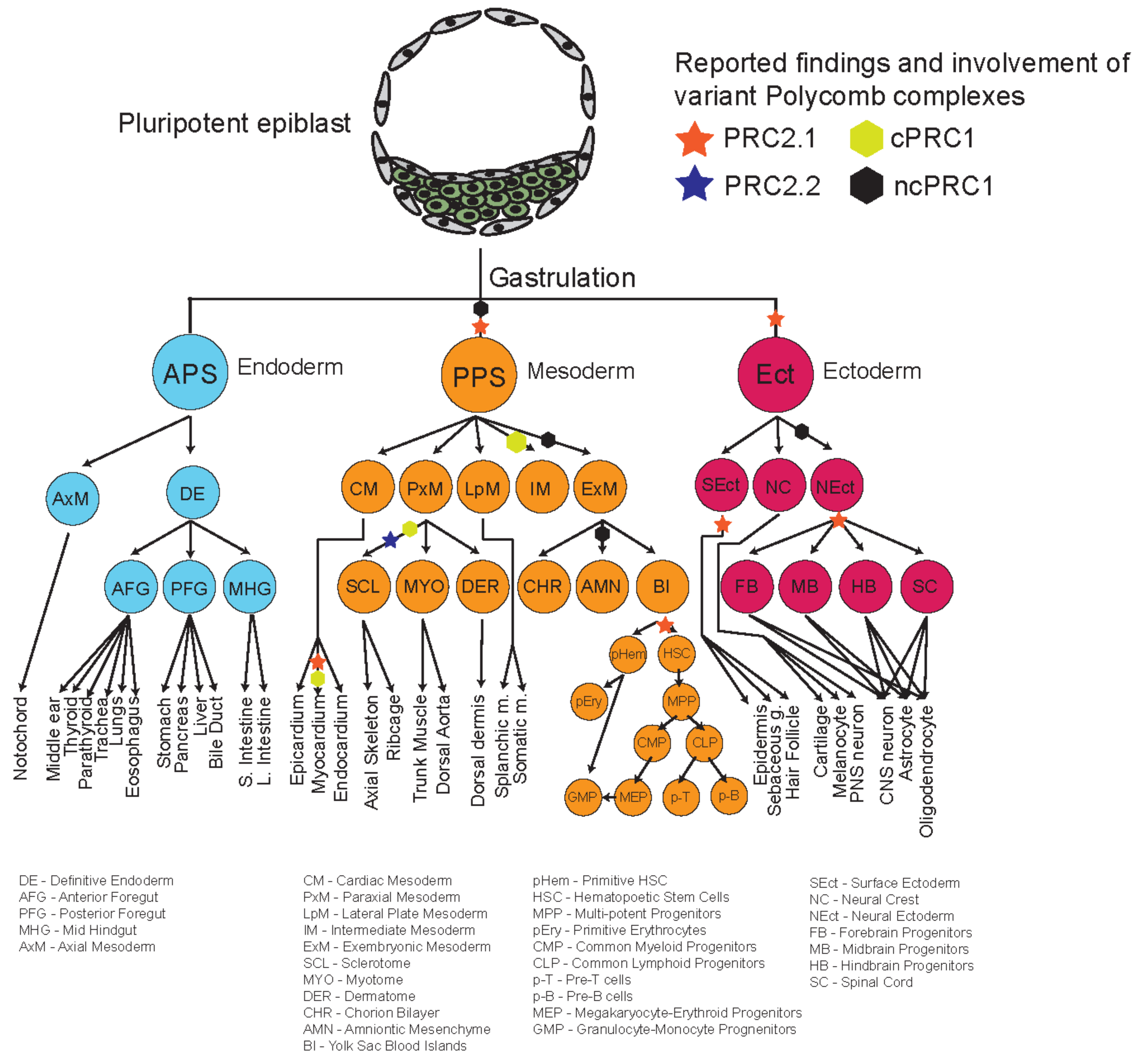
4. Roles of Polycomb Complexes in Developmental Cell Lineages
4.1. Dynamic Functions of PRC2 Variant Complexes
4.2. Dynamic Functions of PRC1 Variant Complexes
5. Towards a Systematic Analysis of Polycomb Function in Development
5.1. In Vitro 2D and 3D Culturing Systems to Study Polycomb Functions
5.2. Untangling the Functions of Polycomb Proteins Using Single-Cell Omics Technologies
6. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004, 431, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Schuettengruber, B.; Cavalli, G. Recruitment of Polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 2009, 136, 3531–3542. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.A.; Kingston, R.E. Mechanisms of Polycomb gene silencing: Knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009, 10, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Van Mierlo, G.; Veenstra, G.J.C.; Vermeulen, M.; Marks, H. The Complexity of PRC2 Subcomplexes. Trends Cell Biol. 2019, 29, 660–671. [Google Scholar] [CrossRef]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science 2002, 298, 1039–1043. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; Hart, C.M.; Francis, N.J.; Vargas, M.L.; Sengupta, A.; Wild, B.; Miller, E.L.; O’Connor, M.B.; Kingston, R.E.; Simon, J.A. Histone Methyltransferase Activity of a Drosophila Polycomb Group Repressor Complex. Cell 2002, 111, 197–208. [Google Scholar] [CrossRef]
- Blackledge, N.P.; Fursova, N.A.; Kelley, J.R.; Huseyin, M.K.; Feldmann, A.; Klose, R.J. PRC1 Catalytic Activity Is Central to Polycomb System Function. Mol. Cell 2020, 77, 857–874. [Google Scholar] [CrossRef] [PubMed]
- Loh, C.H.; van Genesen, S.; Perino, M.; Bark, M.R.; Veenstra, G.J.C. Loss of PRC2 subunits primes lineage choice during exit of pluripotency. Nat. Commun. 2021, 12, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Cooper, S.; Grijzenhout, A.; Underwood, E.; Ancelin, K.; Zhang, T.; Nesterova, T.B.; Anil-Kirmizitas, B.; Bassett, A.; Kooistra, S.M.; Agger, K.; et al. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat. Commun. 2016, 7, 13661. [Google Scholar] [CrossRef] [PubMed]
- Kalb, R.; Latwiel, S.; Baymaz, H.I.; Jansen, P.W.; Müller, C.W.; Vermeulen, M.; Müller, J. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat. Struct. Mol. Biol. 2014, 21, 569–571. [Google Scholar] [CrossRef] [PubMed]
- Tamburri, S.; Lavarone, E.; Fernández-Pérez, D.; Conway, E.; Zanotti, M.; Manganaro, D.; Pasini, D. Histone H2AK119 Mono-Ubiquitination Is Essential for Polycomb-Mediated Transcriptional Repression. Mol. Cell 2020, 77, 840–856. [Google Scholar] [CrossRef] [PubMed]
- Tavares, L.; Dimitrova, E.; Oxley, D.; Webster, J.; Poot, R.; Demmers, J.; Bezstarosti, K.; Taylor, S.; Ura, H.; Koide, H.; et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 2012, 148, 664–678. [Google Scholar] [CrossRef] [PubMed]
- Morey, L.; Helin, K. Polycomb group protein-mediated repression of transcription. Trends Biochem. Sci. 2010, 35, 323–332. [Google Scholar] [CrossRef]
- Piunti, A.; Shilatifard, A. Epigenetic balance of gene expression by polycomb and compass families. Science 2016, 352, aad9780. [Google Scholar] [CrossRef] [PubMed]
- Boulet, A.M.; Capecchi, M.R. Signaling by FGF4 and FGF8 is required for axial elongation of the mouse embryo. Dev. Biol. 2012, 371, 235–245. [Google Scholar] [CrossRef]
- Pasini, D.; Bracken, A.P.; Jensen, M.R.; Denchi, E.L.; Helin, K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004, 23, 4061–4071. [Google Scholar] [CrossRef]
- O’Carroll, D.; Erhardt, S.; Pagani, M.; Barton, S.C.; Surani, M.A.; Jenuwein, T. The Polycomb-Group Gene Ezh2 Is Required for Early Mouse Development. Mol. Cell. Biol. 2001, 21, 4330–4336. [Google Scholar] [CrossRef]
- Rothberg, J.L.M.; Maganti, H.B.; Jrade, H.; Porter, C.J.; Palidwor, G.A.; Cafariello, C.; Battaion, H.L.; Khan, S.T.; Perkins, T.J.; Paulson, R.F.; et al. Mtf2-PRC2 control of canonical Wnt signaling is required for definitive erythropoiesis. Cell Discov. 2018, 4, 21. [Google Scholar] [CrossRef]
- Conway, E.; Healy, E.; Bracken, A.P. PRC2 mediated H3K27 methylations in cellular identity and cancer. Curr. Opin. Cell Biol. 2015, 37, 42–48. [Google Scholar] [CrossRef]
- Deevy, O.; Bracken, A.P. PRC2 functions in development and congenital disorders. Dev. Camb 2019, 146, dev181354. [Google Scholar] [CrossRef]
- Li, X.; Isono, K.; Yamada, D.; Endo, T.A.; Endoh, M.; Shinga, J.; Mizutani-Koseki, Y.; Otte, A.P.; Casanova, M.; Kitamura, H.; et al. Mammalian Polycomb-Like Pcl2/Mtf2 Is a Novel Regulatory Component of PRC2 That Can Differentially Modulate Polycomb Activity both at the Hox Gene Cluster and at Cdkn2a Genes. Mol. Cell. Biol. 2011, 31, 351–364. [Google Scholar] [CrossRef] [PubMed]
- Grijzenhout, A.; Godwin, J.; Koseki, H.; Gdula, M.R.; Szumska, D.; McGouran, J.F.; Bhattacharya, S.; Kessler, B.M.; Brockdorff, N.; Cooper, S. Functional analysis of AEBP2, a PRC2 polycomb protein, reveals a trithorax phenotype in embryonic development and in ESCs. Dev. Camb. 2016, 143, 2716–2723. [Google Scholar]
- Kim, H.; Kang, K.; Ekram, M.B.; Roh, T.Y.; Kim, J. Aebp2 as an epigenetic regulator for neural crest cells. PLoS ONE 2011, 6, e25174. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Taylor, A.B.; Leal, B.Z.; Chadwell, L.V.; Ilangovan, U.; Robinson, A.K.; Schirf, V.; Hart, P.J.; Lafer, E.M.; Demeler, B.; et al. Polycomb group targeting through different binding partners of RING1B C-terminal domain. Structure 2010, 18, 966–975. [Google Scholar] [CrossRef] [PubMed]
- Messmer, S.; Franke, A.; Paro, R. Analysis of the functional role of the Polycomb chromo domain in Drosophila melanogaster. Genes Dev. 1992, 6, 1241–1254. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Levine, S.S.; Weiss, A.; Erdjument-Bromage, H.; Shao, Z.; Tempst, P.; Kingston, R.E. The Core of the Polycomb Repressive Complex Is Compositionally and Functionally Conserved in Flies and Humans. Mol. Cell. Biol. 2002, 22, 6070–6078. [Google Scholar] [CrossRef]
- Scelfo, A.; Fernández-Pérez, D.; Tamburri, S.; Zanotti, M.; Lavarone, E.; Soldi, M.; Bonaldi, T.; Ferrari, K.J.; Pasini, D. Functional Landscape of PCGF Proteins Reveals Both RING1A/B-Dependent-and RING1A/B-Independent-Specific Activities. Mol. Cell 2019, 74, 1037–1052. [Google Scholar] [CrossRef]
- Fursova, N.A.; Blackledge, N.P.; Nakayama, M.; Ito, S.; Koseki, Y.; Farcas, A.M.; King, H.W.; Koseki, H.; Klose, R.J. Synergy between Variant PRC1 Complexes Defines Polycomb-Mediated Gene Repression. Mol. Cell 2019, 74, 1020–1036. [Google Scholar] [CrossRef]
- Garcia, E.; Marcos-Gutiérrez, C.; Del Mar Lorente, M.; Moreno, J.C.; Vidal, M. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J. 1999, 18, 3404–3418. [Google Scholar] [CrossRef]
- Arrigoni, R.; Alam, S.L.; Wamstad, J.A.; Bardwell, V.J.; Sundquist, W.I.; Schreiber-Agus, N. The Polycomb-associated protein Rybp is a ubiquitin binding protein. FEBS Lett. 2006, 580, 6233–6241. [Google Scholar] [CrossRef]
- Morey, L.; Aloia, L.; Cozzuto, L.; Benitah, S.A.; Di Croce, L. RYBP and Cbx7 Define Specific Biological Functions of Polycomb Complexes in Mouse Embryonic Stem Cells. Cell Rep. 2013, 3, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Laugesen, A.; Højfeldt, J.W.; Helin, K. Molecular Mechanisms Directing PRC2 Recruitment and H3K27 Methylation. Mol. Cell 2019, 74, 8–18. [Google Scholar] [CrossRef]
- Rose, N.R.; King, H.W.; Blackledge, N.P.; Fursova, N.A.; Ember, K.J.; Fischer, R.; Kessler, B.M.; Klose, R.J. RYBP stimulates PRC1 to shape chromatin-based communication between polycomb repressive complexes. eLife 2016, 5, e18591. [Google Scholar] [CrossRef]
- Czermin, B.; Melfi, R.; McCabe, D.; Seitz, V.; Imhof, A.; Pirrotta, V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 2002, 111, 185–196. [Google Scholar] [CrossRef]
- Kuzmichev, A.; Nishioka, K.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D. Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of zeste protein. Genes Dev. 2002, 16, 2893–2905. [Google Scholar] [CrossRef] [PubMed]
- Perino, M.; van Mierlo, G.; Karemaker, I.D.; van Genesen, S.; Vermeulen, M.; Marks, H.; van Heeringen, S.J.; Veenstra, G.J.C. MTF2 recruits Polycomb Repressive Complex 2 by helical-shape-selective DNA binding. Nat. Genet. 2018, 50, 1002–1010. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liefke, R.; Jiang, J.; Kurland, J.V.; Tian, W.; Deng, P.; Zhang, W.; He, Q.; Patel, D.J.; Bulyk, M.L.; et al. Polycomb-like proteins link the PRC2 complex to CpG islands. Nature 2017, 549, 287–291. [Google Scholar] [CrossRef]
- Alekseyenko, A.A.; Gorchakov, A.A.; Kharchenko, P.V.; Kuroda, M.I. Reciprocal interactions of human C10orf12 and C17orf96 with PRC2 revealed by BioTAP-XL cross-linking and affinity purification. Proc. Natl. Acad. Sci. USA 2014, 111, 2488–2493. [Google Scholar] [CrossRef]
- Conway, E.; Jerman, E.; Healy, E.; Ito, S.; Holoch, D.; Oliviero, G.; Deevy, O.; Glancy, E.; Fitzpatrick, D.J.; Mucha, M.; et al. A Family of Vertebrate-Specific Polycombs Encoded by the LCOR/LCORL Genes Balance PRC2 Subtype Activities. Mol. Cell 2018, 70, 408–421. [Google Scholar] [CrossRef]
- Hauri, S.; Comoglio, F.; Seimiya, M.; Gerstung, M.; Glatter, T.; Hansen, K.; Aebersold, R.; Paro, R.; Gstaiger, M.; Beisel, C. A High-Density Map for Navigating the Human Polycomb Complexome. Cell Rep. 2016, 17, 583–595. [Google Scholar] [CrossRef]
- Lee, C.H.; Holder, M.; Grau, D.; Saldaña-Meyer, R.; Yu, J.R.; Ganai, R.A.; Zhang, J.; Wang, M.; LeRoy, G.; Dobenecker, M.W.; et al. Distinct Stimulatory Mechanisms Regulate the Catalytic Activity of Polycomb Repressive Complex 2. Mol. Cell 2018, 70, 435–448. [Google Scholar] [CrossRef] [PubMed]
- Kasinath, V.; Faini, M.; Poepsel, S.; Reif, D.; Feng, X.A.; Stjepanovic, G.; Aebersold, R.; Nogales, E. Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science 2018, 359, 940–944. [Google Scholar] [CrossRef] [PubMed]
- Sanulli, S.; Justin, N.; Teissandier, A.; Ancelin, K.; Portoso, M.; Caron, M.; Michaud, A.; Lombard, B.; da Rocha, S.T.; Offer, J.; et al. Jarid2 Methylation via the PRC2 Complex Regulates H3K27me3 Deposition during Cell Differentiation. Mol. Cell 2015, 57, 769–783. [Google Scholar] [CrossRef] [PubMed]
- Son, J.; Shen, S.S.; Margueron, R.; Reinberg, D. Nucleosome-binding activities within JARID2 and EZH1 regulate the function of PRC2 on chromatin. Genes Dev. 2013, 27, 2663–2677. [Google Scholar] [CrossRef] [PubMed]
- Healy, E.; Mucha, M.; Glancy, E.; Fitzpatrick, D.J.; Conway, E.; Neikes, H.K.; Monger, C.; Van Mierlo, G.; Baltissen, M.P.; Koseki, Y.; et al. PRC2.1 and PRC2.2 Synergize to Coordinate H3K27 Trimethylation. Mol. Cell 2019, 76, 437–452. [Google Scholar] [CrossRef] [PubMed]
- Perino, M.; van Mierlo, G.; Loh, C.; Wardle, S.M.T.; Zijlmans, D.W.; Marks, H.; Veenstra, G.J.C. Two Functional Axes of Feedback-Enforced PRC2 Recruitment in Mouse Embryonic Stem Cells. Stem Cell Reports 2020, 15, 1287–1300. [Google Scholar] [CrossRef]
- Vermeulen, M.; Mulder, K.W.; Denissov, S.; Pijnappel, W.W.; van Schaik, F.M.; Varier, R.A.; Baltissen, M.P.; Stunnenberg, H.G.; Mann, M.; Timmers, H.T. Selective Anchoring of TFIID to Nucleosomes by Trimethylation of Histone H3 Lysine 4. Cell 2007, 131, 58–69. [Google Scholar] [CrossRef]
- Ng, H.H.; Robert, F.; Young, R.A.; Struhl, K. Targeted Recruitment of Set1 Histone Methylase by Elongating Pol II Provides a Localized Mark and Memory of Recent Transcriptional Activity. Mol. Cell 2003, 11, 709–719. [Google Scholar] [CrossRef]
- Reinig, J.; Ruge, F.; Howard, M.; Ringrose, L. A theoretical model of Polycomb/Trithorax action unites stable epigenetic memory and dynamic regulation. Nat. Commun. 2020, 11, 4782. [Google Scholar] [CrossRef]
- Schwartz, Y.B.; Pirrotta, V. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 2007, 8, 9–22. [Google Scholar] [CrossRef]
- Grimaud, C.; Nègre, N.; Cavalli, G. From genetics to epigenetics: The tale of Polycomb group and trithorax group genes. Chromosome Res. 2006, 14, 363–375. [Google Scholar] [CrossRef]
- Woo, C.J.; Kharchenko, P.V.; Daheron, L.; Park, P.J.; Kingston, R.E. A Region of the Human HOXD Cluster that Confers Polycomb-Group Responsiveness. Cell 2010, 140, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.; Chiang, A.; Bender, W.; Shimell, M.J.; O’connor, M. Elements of the Drosophila Bithorax Complex That Mediate Repression by Polycomb Group Products. Dev. Biol. 1993, 158, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Van der Lugt, N.M.T.; Alkema, M.; Berns, A.; Deschamps, J. The Polycomb-group homolog Bmi-1 is a regulator of murine Hox gene expression. Mech. Dev. 1996, 58, 153–164. [Google Scholar] [CrossRef]
- Pasini, D.; Bracken, A.P.; Hansen, J.B.; Capillo, M.; Helin, K. The Polycomb Group Protein Suz12 Is Required for Embryonic Stem Cell Differentiation. Mol. Cell. Biol. 2007, 27, 3769–3779. [Google Scholar] [CrossRef] [PubMed]
- Faust, C.; Lawson, K.A.; Schork, N.J.; Thiel, B.; Magnuson, T. The Polycomb-group gene eed is required for normal morphogenetic movements during gastrulation in the mouse embryo. Development 1998, 125, 4495–4506. [Google Scholar] [CrossRef]
- Kumar, B.; Navarro, C.; Winblad, N.; Schell, J.P.; Zhao, C.; Weltner, J.; Baqué-Vidal, L.; Salazar Mantero, A.; Petropoulos, S.; Lanner, F.; et al. Polycomb repressive complex 2 shields naïve human pluripotent cells from trophectoderm differentiation. Nat. Cell Biol. 2022, 24, 845–857. [Google Scholar] [CrossRef]
- Zijlmans, D.W.; Talon, I.; Verhelst, S.; Bendall, A.; Van Nerum, K.; Javali, A.; Malcolm, A.A.; van Knippenberg, S.S.F.A.; Biggins, L.; To, S.K.; et al. Integrated multi-omics reveal polycomb repressive complex 2 restricts human trophoblast induction. Nat. Cell Biol. 2022, 24, 858–871. [Google Scholar] [CrossRef]
- Del Mar Lorente, M.; Marcos-Gutiérrez, C.; Pérez, C.; Schoorlemmer, J.; Ramírez, A.; Magin, T.; Vidal, M. Loss- and gain-of-function mutations show a polycomb group function for Ring1A in mice. Development 2000, 127, 5093–5100. [Google Scholar] [CrossRef]
- Voncken, J.W.; Roelen, B.A.; Roefs, M.; de Vries, S.; Verhoeven, E.; Marino, S.; Deschamps, J.; van Lohuizen, M. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc. Natl. Acad. Sci. USA 2003, 100, 2468–2473. [Google Scholar] [CrossRef]
- Takeuchi, T.; Yamazaki, Y.; Katoh-Fukui, Y.; Tsuchiya, R.; Kondo, S.; Motoyama, J.; Higashinakagawa, T. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 1995, 9, 1211–1222. [Google Scholar] [CrossRef] [PubMed]
- Højfeldt, J.W.; Hedehus, L.; Laugesen, A.; Tatar, T.; Wiehle, L.; Helin, K. Non-core Subunits of the PRC2 Complex Are Collectively Required for Its Target-Site Specificity. Mol. Cell 2019, 76, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Kloet, S.L.; Makowski, M.M.; Baymaz, H.I.; van Voorthuijsen, L.; Karemaker, I.D.; Santanach, A.; Jansen, P.W.T.C.; Di Croce, L.; Vermeulen, M. The dynamic interactome and genomic targets of Polycomb complexes during stem-cell differentiation. Nat. Struct. Mol. Biol. 2016, 23, 682–690. [Google Scholar] [CrossRef]
- Beringer, M.; Pisano, P.; Di Carlo, V.; Blanco, E.; Chammas, P.; Vizán, P.; Gutiérrez, A.; Aranda, S.; Payer, B.; Wierer, M.; et al. EPOP Functionally Links Elongin and Polycomb in Pluripotent Stem Cells. Mol. Cell 2016, 64, 645–658. [Google Scholar] [CrossRef]
- Zhen, C.Y.; Tatavosian, R.; Huynh, T.N.; Duc, H.N.; Das, R.; Kokotovic, M.; Grimm, J.B.; Lavis, L.D.; Lee, J.; Mejia, F.J.; et al. Live-cell single-molecule tracking reveals co-recognition of H3K27me3 and DNA targets polycomb Cbx7-PRC1 to chromatin. Elife 2016, 5, e17667. [Google Scholar] [CrossRef]
- Tatavosian, R.; Kent, S.; Brown, K.; Yao, T.; Duc, H.N.; Huynh, T.N.; Zhen, C.Y.; Ma, B.; Wang, H.; Ren, X. Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. J. Biol. Chem. 2019, 294, 1451–1463. [Google Scholar] [CrossRef] [PubMed]
- Plys, A.J.; Davis, C.P.; Kim, J.; Rizki, G.; Keenen, M.M.; Marr, S.K.; Kingston, R.E. Phase separation of polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev. 2019, 33, 799–813. [Google Scholar] [CrossRef]
- Blackledge, N.P.; Farcas, A.M.; Kondo, T.; King, H.W.; McGouran, J.F.; Hanssen, L.L.P.; Ito, S.; Cooper, S.; Kondo, K.; Koseki, Y.; et al. Variant PRC1 Complex-Dependent H2A Ubiquitylation Drives PRC2 Recruitment and Polycomb Domain Formation. Cell 2014, 157, 1445–1459. [Google Scholar] [CrossRef]
- Cao, R.; Tsukada, Y.I.; Zhang, Y. Role of Bmi-1 and Ring1A in H2A Ubiquitylation and Hox Gene Silencing. Mol. Cell 2005, 20, 845–854. [Google Scholar] [CrossRef]
- Wu, X.; Johansen, J.V.; Helin, K. Fbxl10/Kdm2b Recruits Polycomb Repressive Complex 1 to CpG Islands and Regulates H2A Ubiquitylation. Mol. Cell 2013, 49, 1134–1146. [Google Scholar] [CrossRef]
- He, J.; Shen, L.; Wan, M.; Taranova, O.; Wu, H.; Zhang, Y. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat. Cell Biol. 2013, 15, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Farcas, A.M.; Blackledge, N.P.; Sudbery, I.; Long, H.K.; McGouran, J.F.; Rose, N.R.; Lee, S.; Sims, D.; Cerase, A.; Sheahan, T.W. KDM2B links the polycomb repressive complex 1 (PRC1) to recognition of CpG islands. eLife 2012, 1, e00205. [Google Scholar] [CrossRef] [PubMed]
- Oliviero, G.; Brien, G.L.; Waston, A.; Streubel, G.; Jerman, E.; Andrews, D.; Doyle, B.; Munawar, N.; Wynne, K.; Crean, J. Dynamic Protein Interactions of the Polycomb Repressive Complex 2 during Differentiation of Pluripotent Cells. Mol. Cell. Proteom. 2016, 15, 3450–3460. [Google Scholar] [CrossRef] [PubMed]
- Gatchalian, J.; Kingsley, M.C.; Moslet, S.D.; Ospina, R.D.R.; Kutateladze, T.G. An aromatic cage is required but not sufficient for binding of Tudor domains of the Polycomblike protein family to H3K36me3. Epigenetics 2015, 10, 467–473. [Google Scholar] [CrossRef]
- Cai, L.; Rothbart, S.B.; Lu, R.; Xu, B.; Chen, W.Y.; Tripathy, A.; Rockowitz, S.; Zheng, D.; Patel, D.J.; Allis, C.D.; et al. An H3K36 Methylation-Engaging Tudor Motif of Polycomb-like Proteins Mediates PRC2 Complex Targeting. Mol. Cell 2013, 49, 571–582. [Google Scholar] [CrossRef] [PubMed]
- Musselman, C.A.; Avvakumov, N.; Watanabe, R.; Abraham, C.G.; Lalonde, M.E.; Hong, Z.; Allen, C.; Roy, S.; Nuñez, J.K.; Nickoloff, J.; et al. Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nat. Struct. Mol. Biol. 2012, 19, 1266–1272. [Google Scholar] [CrossRef]
- Ballaré, C.; Lange, M.; Lapinaite, A.; Martin, G.M.; Morey, L.; Pascual, G.; Liefke, R.; Simon, B.; Shi, Y.; Gozani, O.; et al. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat. Struct. Mol. Biol. 2012, 19, 1257–1265. [Google Scholar] [CrossRef]
- Brien, G.L.; Gambero, G.; O’Connell, D.J.; Jerman, E.; Turner, S.A.; Egan, C.M.; Dunne, E.J.; Jurgens, M.C.; Wynne, K.; Piao, L.; et al. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat. Struct. Mol. Biol. 2012, 19, 1273–1281. [Google Scholar] [CrossRef]
- Petracovici, A.; Bonasio, R. Distinct PRC2 subunits regulate maintenance and establishment of Polycomb repression during differentiation. Mol. Cell 2021, 81, 2625–2639. [Google Scholar] [CrossRef]
- García-Montolio, M.; Ballaré, C.; Blanco, E.; Gutiérrez, A.; Aranda, S.; Gómez, A.; Kok, C.H.; Yeung, D.T.; Hughes, T.P.; Vizán, P.; et al. Polycomb Factor PHF19 Controls Cell Growth and Differentiation Toward Erythroid Pathway in Chronic Myeloid Leukemia Cells. Front. Cell Dev. Biol. 2021, 9, 865. [Google Scholar] [CrossRef]
- Katoh-Fukui, Y.; Tsuchiya, R.; Shiroishi, T.; Nakahara, Y.; Hashimoto, N.; Noguchi, K.; Higashinakagawa, T. Male-to-female sex reversal in M33 mutant mice. Nature 1998, 393, 688–692. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, M.E.; Flenniken, A.M.; Ji, X.; Teboul, L.; Wong, M.D.; White, J.K.; Meehan, T.F.; Weninger, W.J.; Westerberg, H.; Adissu, H.; et al. High-throughput discovery of novel developmental phenotypes. Nature 2016, 537, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Hu, B.; Song, M.; Ding, Z.; Dang, Y.; Liu, Z.; Zhang, W.; Ji, Q.; Ren, R.; Ding, J.; et al. Maintenance of Nucleolar Homeostasis by CBX4 Alleviates Senescence and Osteoarthritis. Cell Rep. 2019, 26, 3643–3656. [Google Scholar] [CrossRef] [PubMed]
- Forzati, F.; Federico, A.; Pallante, P.; Abbate, A.; Esposito, F.; Malapelle, U.; Sepe, R.; Palma, G.; Troncone, G.; Scarfò, M.; et al. CBX7 is a tumor suppressor in mice and humans. J. Clin. Investig. 2012, 122, 612–623. [Google Scholar] [CrossRef]
- Isono, K.; Fujimura, Y.; Shinga, J.; Yamaki, M.; Wang, J.O.; Takihara, Y.; Murahashi, Y.; Takada, Y.; Mizutani-Koseki, Y.; Koseki, H. Mammalian Polyhomeotic Homologues Phc2 and Phc1 Act in Synergy to Mediate Polycomb Repression of Hox Genes. Mol. Cell. Biol. 2005, 25, 6694–6706. [Google Scholar] [CrossRef]
- Van der Lugt, N.M.; Domen, J.; Linders, K.; van Roon, M.; Robanus-Maandag, E.; te Riele, H.; van der Valk, M.; Deschamps, J.; Sofroniew, M.; van Lohuizen, M.; et al. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 1994, 8, 757–769. [Google Scholar] [CrossRef]
- Alkema, M.J.; van der Lugt, N.M.T.; Bobeldijk, R.C.; Berns, A.; van Lohuizen, M. Transformation of axial skeleton due to overexpression of bmi-1 in transgenic mice. Nature 1995, 374, 724–727. [Google Scholar] [CrossRef]
- Akasaka, T.; van Lohuizen, M.; van der Lugt, N.; Mizutani-Koseki, Y.; Kanno, M.; Taniguchi, M.; Vidal, M.; Alkema, M.; Berns, A.; Koseki, H. Mice doubly deficient for the Polycomb Group genes Mel18 and Bmi1 reveal synergy and requirement for maintenance but not initiation of Hox gene expression. Development 2001, 128, 1587–1597. [Google Scholar] [CrossRef]
- Pirity, M.K.; Locker, J.; Schreiber-Agus, N. Rybp/DEDAF Is Required for Early Postimplantation and for Central Nervous System Development. Mol. Cell. Biol. 2005, 25, 7193–7202. [Google Scholar] [CrossRef]
- Boulard, M.; Edwards, J.R.; Bestor, T.H. FBXL10 protects Polycomb-bound genes from hypermethylation. Nat. Genet. 2015, 47, 479–485. [Google Scholar] [CrossRef]
- Yan, Y.; Zhao, W.; Huang, Y.; Tong, H.; Xia, Y.; Jiang, Q.; Qin, J. Loss of Polycomb Group Protein Pcgf1 Severely Compromises Proper Differentiation of Embryonic Stem Cells. Sci. Rep. 2017, 7, 46276. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Lee, P.; Stafford, J.M.; von Schimmelmann, M.; Schaefer, A.; Reinberg, D. An AUTS2–Polycomb complex activates gene expression in the CNS. Nature 2014, 516, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Abe, K.; Naruse, C.; Kato, T.; Nishiuchi, T.; Saitou, M.; Asano, M. Loss of Heterochromatin Protein 1 Gamma Reduces the Number of Primordial Germ Cells via Impaired Cell Cycle Progression in Mice. Biol. Reprod. 2011, 85, 1013–1024. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brown, J.P.; Bullwinkel, J.; Baron-Lühr, B.; Billur, M.; Schneider, P.; Winking, H.; Singh, P.B. HP1γ function is required for male germ cell survival and spermatogenesis. Epigenetics Chromatin 2010, 3, 9. [Google Scholar] [CrossRef]
- Yesbolatova, A.; Saito, Y.; Kitamoto, N.; Makino-Itou, H.; Ajima, R.; Nakano, R.; Nakaoka, H.; Fukui, K.; Gamo, K.; Tominari, Y.; et al. The auxin-inducible degron 2 technology provides sharp degradation control in yeast, mammalian cells, and mice. Nat. Commun. 2020, 11, 5701. [Google Scholar] [CrossRef]
- Li, S.; Prasanna, X.; Salo, V.T.; Vattulainen, I.; Ikonen, E. An efficient auxin-inducible degron system with low basal degradation in human cells. Nat. Methods 2019, 16, 866–869. [Google Scholar] [CrossRef]
- Van den Brink, S.C.; van Oudenaarden, A. 3D gastruloids: A novel frontier in stem cell-based in vitro modeling of mammalian gastrulation. Trends Cell Biol. 2021, 31, 747–759. [Google Scholar] [CrossRef]
- Kagawa, H.; Javali, A.; Khoei, H.H.; Sommer, T.M.; Sestini, G.; Novatchkova, M.; Scholte op Reimer, Y.; Castel, G.; Bruneau, A.; Maenhoudt, N.; et al. Human blastoids model blastocyst development and implantation. Nature 2021, 601, 600–605. [Google Scholar] [CrossRef]
- Ma, A.; McDermaid, A.; Xu, J.; Chang, Y.; Ma, Q. Integrative Methods and Practical Challenges for Single-Cell Multi-omics. Trends Biotechnol. 2020, 38, 1007–1022. [Google Scholar] [CrossRef]

| Complex | Variant Complex | Non Core Subunit | In Vivo Developmental Phenotype | In Vitro Differentiation Phenotype | Cell Type or Tissue Specificity | References |
|---|---|---|---|---|---|---|
| PRC2 | PRC2.1 | MTF2 | Embryonic lethal due to severe anemia around embryonic day 15.5 | Mtf2 null cells result in faster differentiation towards all cell types in EBs | ESC/Gastrula | Faust et al., 1995: Hojfeldt et al., 2018; Perino et al.,2018; Loh et al., 2021; Rothberg et al., 2018 |
| PHF1 | ND | Increased stoichometry relative to EED during NPC differentiation | Neuronal Precursors | Kloet et al., 2016 | ||
| PHF19 | ND | Enhanced erythrocyte differentiation | Mesodermal | García-Montolio et al., 2021 | ||
| EPOP | ND | ND | ND | |||
| PALI1/2 | Lethal between E11.5 and perinatal lethality | Deregulation and reduction of H3K27me3 on PRC2 target genes | ND | Conway, E. et al., 2018 | ||
| PRC2.2 | JARID2 | Lethal between E10.5 - E18.5 | Stagnated differentiation towards all germ layers | ND | Loh et al., 2021 | |
| AEBP2 | Late-embryonic and perinatal lethality; skeletal transformations | Deregulation and increase of H3K27me3 on PRC2 target genes | ND | Grijzenhout, A. et al., 2016; Kim, H. et al., 2011 | ||
| cPRC1 | CBX2 | Male to female sex reversal | Activator of the testis - determining gene Sry | Testes, Sex Organ | Katoh-Fukui, Y. et al., 1998 | |
| CBX4 | Pre-weaning lethality | ND | ND | |||
| CBX6 | Several transformational defects in late embryonic development | ND | Enriched in NPC differentiation | Dickinson, M. E. et al., 2016 | ||
| CBX7 | ND | Enriched in early ESC stages | ||||
| CBX8 | ND | ND | Enriched in NPC differentiation | Morey, L. et al. 2012 | ||
| PHC1 | Perinatal lethality | Destabilized pluripotency by disruption of Nanog long-range interactions | ND | Isono K. et al., 2005 | ||
| PHC2 | Posterior transformation; Hox gene deregulation; Viable birth | ND | ND | Isono K. et al., 2005 | ||
| PHC3 | Phc3 null animals are born but exhibit cardiac abnormalities | ND | ND | Dickinson, M. E. et al., 2016 | ||
| PCGF2 | Posterior transformation; Hox gene deregulation; Viable birth | ND | ND | Akasaka, T. et al., 1996 | ||
| PCGF4 | Perinatal lethality, ataxia, severe haematopoietic defects | ND | ND | van der Lugt, N. M. et al., 1994 | ||
| ncPRC1 | CBX3 | Defects in germ cell development | Inhibition results in neural differentiation impairment and diverting towards mesendodermal differentiion | Neural Precursors | Abe, K. et al., 2011 | |
| PCGF1 | E12.5 lethality | Promotes ectoderm and mesoderm differentiation | Ectoderm and Mesoderm lineages | Yan Y et al., 2017 | ||
| PCGF3 | Double Pcgf3;Pcgf5 deletion results in female-specific embryonic lethality | ND | ND | Almeida, M. et al., 2017 | ||
| PCGF5 | ||||||
| PCGF6 | defects in pre-implantation and periimplantation and in placenta development | Ablation of Pcgf6 in ESCs leads to robust de-repression of such germ cell-related genes, in turn affecting cell growth and viability. | pre- and peri-implantation mouse embryo | Dickinson, M. E. et al., 2016 | ||
| KDM2B | Mid-gestation lethality; posterior transformation of the axial skeleton | Maintain pluripotency by recruitment of PRC1 to CpGs | ND | Boulard, M. et al., 2015 | ||
| RYBP | E6.5 Lethality; impaired cell proliferation | Depletion of Rybp inhibits proliferation and promotes Neuronal differentiation of embryonic neural progenitor cells (eNPCs) | Neural Precursors | Pirity, M. K. et al., 2015 | ||
| AUTS2 | Die before weaning with defects in the nervous, cardiovascular and biliary systems | ND | Neural Ectoderm | Gao, Z. et al., 2014 | ||
| L3MBTL2 | E6.5 lethality; gastrulation defects | Compromised proliferation and abnormal differentiation of L3mbtl2(-/) embryonic stem (ES) cells | ESC | Qin, J. et al., 2012 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loh, C.H.; Veenstra, G.J.C. The Role of Polycomb Proteins in Cell Lineage Commitment and Embryonic Development. Epigenomes 2022, 6, 23. https://doi.org/10.3390/epigenomes6030023
Loh CH, Veenstra GJC. The Role of Polycomb Proteins in Cell Lineage Commitment and Embryonic Development. Epigenomes. 2022; 6(3):23. https://doi.org/10.3390/epigenomes6030023
Chicago/Turabian StyleLoh, Chet H., and Gert Jan C. Veenstra. 2022. "The Role of Polycomb Proteins in Cell Lineage Commitment and Embryonic Development" Epigenomes 6, no. 3: 23. https://doi.org/10.3390/epigenomes6030023
APA StyleLoh, C. H., & Veenstra, G. J. C. (2022). The Role of Polycomb Proteins in Cell Lineage Commitment and Embryonic Development. Epigenomes, 6(3), 23. https://doi.org/10.3390/epigenomes6030023






