Quantifying Genomic Imprinting at Tissue and Cell Resolution in the Brain
Abstract
1. Genomic Imprinting: A Prototype of Epigenetic Regulation
2. The Brain Imprintome: How Many Imprinted Genes in the Brain? Transcriptome-Wide Identification of Imprinted Genes with Bulk RNA-seq
3. Monoallelic Expression of Imprinted Genes at Single-Cell Resolution: Insights from Molecular Probes, Genetic Reporters and scRNA-seq
3.1. Probes
3.2. Reporters
3.3. Single Cell RNA-seq
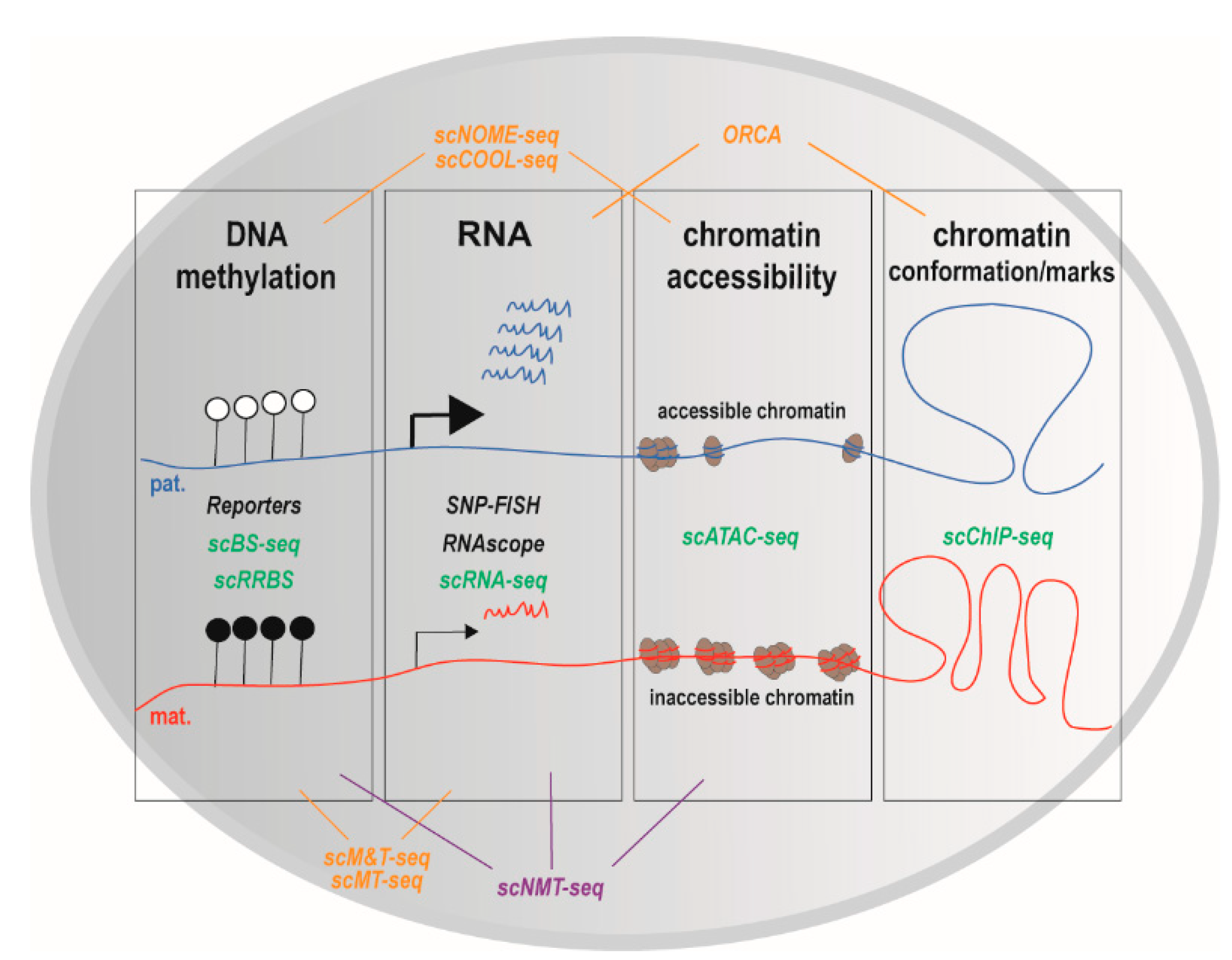
4. Single-Cell Epigenomics and Single-Cell Multi-Omics: Promising Approaches to Comprehend Genomic Imprinting at Cell Resolution
4.1. DNA Methylation
4.2. Chromatin Modifications and Conformation
4.3. Multi-Omics
5. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chess, A. Mechanisms and consequences of widespread random monoallelic expression. Nat. Rev. Genet. 2012, 13, 421–428. [Google Scholar] [CrossRef]
- Gendrel, A.-V.; Marion-Poll, L.; Katoh, K.; Heard, E. Random monoallelic expression of genes on autosomes: Parallels with X-chromosome inactivation. Sem. Cell Dev. Biol. 2016, 56, 100–110. [Google Scholar] [CrossRef]
- Khamlichi, A.A.; Feil, R. Parallels between mammalian mechanisms of monoallelic gene expression. Trends Genet. 2018, 34, 954–971. [Google Scholar] [CrossRef] [PubMed]
- Ferguson-Smith, A.C. Genomic imprinting: The emergence of an epigenetic paradigm. Nat. Rev. Genet. 2011, 12, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Hanna, C.W.; Kelsey, G. The specification of imprints in mammals. Heredity 2014, 113, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Peters, J. The role of genomic imprinting in biology and disease: An expanding view. Nat. Rev. Genet. 2014, 15, 517–530. [Google Scholar] [CrossRef]
- Davies, W.; Isles, A.R.; Burgoyne, P.S.; Wilkinson, L.S. X-linked imprinting: Effects on brain and behaviour. Bioessays 2006, 28, 35–44. [Google Scholar] [CrossRef]
- Girardot, M.; Cavaillé, J.; Feil, R. Small regulatory RNAs controlled by genomic imprinting and their contribution to human disease. Epigenetics 2012, 7, 1341–1348. [Google Scholar] [CrossRef]
- Jirtle, R.L. Available online: http://www.geneimprint.org/site/home (accessed on 10 August 2020).
- Morison, I.M.; Ramsay, J.P.; Spencer, H.G. A census of mammalian imprinting. Trends Genet. 2005, 21, 457–465. [Google Scholar] [CrossRef]
- Williamson, C.; Blake, A.; Thomas, S.; Beechey, C.; Hancock, J.; Cattanach, B.; Peters, J. Mouse Imprinting Data and References; MRC Harwell: Oxfordshire, UK, 2013; Available online: http://www.har.mrc.ac.uk/research/genomic_imprinting/ (accessed on 10 August 2020).
- Inoue, A.; Jiang, L.; Lu, F.; Suzuki, T.; Zhang, Y. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 2017, 547, 419–424. [Google Scholar] [CrossRef]
- Koerner, M.V.; Pauler, F.M.; Huang, R.; Barlow, D.P. The function of non-coding RNAs in genomic imprinting. Development 2009, 136, 1771–1783. [Google Scholar] [CrossRef] [PubMed]
- Kota, S.K.; Lleres, D.; Bouschet, T.; Hirasawa, R.; Marchand, A.; Begon-Pescia, C.; Sanli, I.; Arnaud, P.; Journot, L.; Girardot, M.; et al. ICR noncoding RNA expression controls imprinting and DNA replication at the Dlk1-Dio3 domain. Dev. Cell 2014, 31, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Tomizawa, S.; Mitsuya, K.; Totoki, Y.; Yamamoto, Y.; Kuramochi-Miyagawa, S.; Iida, N.; Hoki, Y.; Murphy, P.J.; Toyoda, A.; et al. Role for piRNAs and noncoding RNA in de Novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science 2011, 332, 848–852. [Google Scholar] [CrossRef]
- Llères, D.; Moindrot, B.; Pathak, R.; Piras, V.; Matelot, M.; Pignard, B.; Marchand, A.; Poncelet, M.; Perrin, A.; Tellier, V.; et al. CTCF modulates allele-specific sub-TAD organization and imprinted gene activity at the mouse Dlk1-Dio3 and Igf2-H19 domains. Genome Biol. 2019, 20, 272. [Google Scholar] [CrossRef]
- Hikichi, T.; Kohda, T.; Kaneko-Ishino, T.; Ishino, F. Imprinting regulation of the murine Meg1 / Grb10 and human GRB10 genes; roles of brain-specific promoters and mouse-specific CTCF-binding sites. Nucl. Acids Res. 2003, 31, 1398–1406. [Google Scholar] [CrossRef] [PubMed]
- Garfield, A.S.; Cowley, M.; Smith, F.M.; Moorwood, K.; Stewart-Cox, J.E.; Gilroy, K.; Baker, S.; Xia, J.; Dalley, J.W.; Hurst, L.D.; et al. Distinct physiological and behavioural functions for parental alleles of imprinted Grb10. Nature 2011, 469, 534–538. [Google Scholar] [CrossRef] [PubMed]
- Plasschaert, R.N.; Bartolomei, M.S. Tissue-specific regulation and function of Grb10 during growth and neuronal commitment. Proc. Natl. Acad. Sci. USA 2015, 112, 6841–6847. [Google Scholar] [CrossRef]
- Arnaud, P.; Monk, D.; Hitchins, M.; Gordon, E.; Dean, W.; Beechey, C.V.; Peters, J.; Craigen, W.; Preece, M.; Stanier, P.; et al. Conserved methylation imprints in the human and mouse GRB10 genes with divergent allelic expression suggests differential reading of the same mark. Hum. Mol. Genet. 2003, 12, 1005–1019. [Google Scholar] [CrossRef]
- Sanz, L.A.; Chamberlain, S.; Sabourin, J.-C.; Henckel, A.; Magnuson, T.; Hugnot, J.-P.; Feil, R.; Arnaud, P. A mono-allelic bivalent chromatin domain controls tissue-specific imprinting at Grb10. EMBO J. 2008, 27, 2523–2532. [Google Scholar] [CrossRef]
- Court, F.; Baniol, M.; Hagege, H.; Petit, J.S.; Lelay-Taha, M.-N.; Carbonell, F.; Weber, M.; Cathala, G.; Forne, T. Long-range chromatin interactions at the mouse Igf2/H19 locus reveal a novel paternally expressed long non-coding RNA. Nucl. Acids Res. 2011, 39, 5893–5906. [Google Scholar] [CrossRef]
- Murrell, A.; Heeson, S.; Reik, W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 2004, 36, 889–893. [Google Scholar] [CrossRef] [PubMed]
- Engel, N.; Raval, A.K.; Thorvaldsen, J.L.; Bartolomei, S.M. Three-dimensional conformation at the H19/Igf2 locus supports a model of enhancer tracking. Hum. Mol. Genet. 2008, 17, 3021–3029. [Google Scholar] [CrossRef] [PubMed]
- Sanli, I.; Feil, R. Chromatin mechanisms in the developmental control of imprinted gene expression. Int. J. Biochem. Cell Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, W.A.; Mann, M.R.W. Long noncoding RNA functionality in imprinted domain regulation. PLoS Genet. 2020, 16, e1008930. [Google Scholar] [CrossRef]
- Greer, P.L.; Hanayama, R.; Bloodgood, B.L.; Mardinly, A.R.; Lipton, D.M.; Flavell, S.W.; Kim, T.-K.; Griffith, E.C.; Waldon, Z.; Maehr, R.; et al. The Angelman syndrome protein Ube3A regulates synapse development by Ubiquitinating arc. Cell 2010, 140, 704–716. [Google Scholar] [CrossRef]
- Lopez, S.J.; Laufer, B.I.; Beitnere, U.; Berg, E.L.; Silverman, J.L.; O’Geen, H.; Segal, D.J.; LaSalle, J.M. Imprinting effects of UBE3A loss on synaptic gene networks and Wnt signaling pathways. Hum. Mol. Genet. 2019, 28, 3842–3852. [Google Scholar] [CrossRef]
- Yashiro, K.; Riday, T.T.; Condon, K.H.; Roberts, A.C.; Bernardo, D.R.; Prakash, R.; Weinberg, R.J.; Ehlers, M.D.; Philpot, B.D. Ube3a is required for experience-dependent maturation of the neocortex. Nat. Neurosci. 2009, 12, 777–783. [Google Scholar] [CrossRef]
- Mueller, O.T.; Coovadia, A. Gene symbol: UBE3A. Disease: Angelman syndrome. Hum. Genet. 2008, 124, 304. [Google Scholar]
- Albrecht, U.; Sutcliffe, J.S.; Cattanach, B.M.; Beechey, C.V.; Armstrong, D.; Eichele, G.; Beaudet, A.L. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat. Genet. 1997, 17, 75–78. [Google Scholar] [CrossRef]
- Huang, H.S.; Allen, J.A.; Mabb, A.M.; King, I.F.; Miriyala, J.; Taylor-Blake, B.; Sciaky, N.; Dutton, J.W.; Lee, H.M.; Chen, X.; et al. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature 2011. [Google Scholar] [CrossRef]
- Powell, W.T.; Coulson, R.L.; Gonzales, M.L.; Crary, F.K.; Wong, S.S.; Adams, S.; Ach, R.A.; Tsang, P.; Yamada, N.A.; Yasui, D.H.; et al. R-loop formation at Snord116 mediates topotecan inhibition of Ube3a-antisense and allele-specific chromatin decondensation. Proc. Natl. Acad. Sci. USA 2013, 110, 13938–13943. [Google Scholar] [CrossRef] [PubMed]
- Seitz, H.; Youngson, N.; Lin, S.-P.; Dalbert, S.; Paulsen, M.; Bachellerie, J.-P.; Ferguson-Smith, A.C.; Cavaillé, J. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat. Genet. 2003, 34, 261–262. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, L.S.; Davies, W.; Isles, A.R. Genomic imprinting effects on brain development and function. Nat. Rev. Neurosci. 2007, 8, 832–843. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.-C.; Bennett, K.; Gregg, C. Epigenetic and cellular diversity in the brain through allele-specific effects. Trends Neurosci. 2018, 41, 925–937. [Google Scholar] [CrossRef] [PubMed]
- Monk, D.; Mackay, D.J.G.; Eggermann, T.; Maher, E.R.; Riccio, A. Genomic imprinting disorders: Lessons on how genome, epigenome and environment interact. Nat. Rev. Genet. 2019, 20, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Eggermann, T.; Perez de Nanclares, G.; Maher, E.R.; Temple, I.K.; Tumer, Z.; Monk, D.; Mackay, D.J.; Gronskov, K.; Riccio, A.; Linglart, A.; et al. Imprinting disorders: A group of congenital disorders with overlapping patterns of molecular changes affecting imprinted loci. Clin. Epigenet. 2015, 7, 123. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, S.J.; Lalande, M. Angelman syndrome, a genomic imprinting disorder of the brain. J. Neurosci. Off. J. Soc. Neurosci. 2010, 30, 9958–9963. [Google Scholar] [CrossRef]
- Judson, M.C.; Wallace, M.L.; Sidorov, M.S.; Burette, A.C.; Gu, B.; van Woerden, G.M.; King, I.F.; Han, J.E.; Zylka, M.J.; Elgersma, Y.; et al. GABAergic neuron-specific loss of Ube3a causes angelman syndrome-like EEG abnormalities and enhances seizure susceptibility. Neuron 2016, 90, 56–69. [Google Scholar] [CrossRef]
- Wallace, M.L.; Burette, A.C.; Weinberg, R.J.; Philpot, B.D. Maternal loss of Ube3a produces an excitatory/inhibitory imbalance through neuron type-specific synaptic defects. Neuron 2012, 74, 793–800. [Google Scholar] [CrossRef]
- Berrios, J.; Stamatakis, A.M.; Kantak, P.A.; McElligott, Z.A.; Judson, M.C.; Aita, M.; Rougie, M.; Stuber, G.D.; Philpot, B.D. Loss of UBE3A from TH-expressing neurons suppresses GABA co-release and enhances VTA-NAc optical self-stimulation. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef]
- Ferron, S.R.; Charalambous, M.; Radford, E.; McEwen, K.; Wildner, H.; Hind, E.; Morante-Redolat, J.M.; Laborda, J.; Guillemot, F.; Bauer, S.R.; et al. Postnatal loss of Dlk1 imprinting in stem cells and niche astrocytes regulates neurogenesis. Nature 2011, 475, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Prasasya, R.; Grotheer, K.V.; Siracusa, L.D.; Bartolomei, M.S. Temple syndrome and Kagami-Ogata syndrome: Clinical presentations, genotypes, models and mechanisms. Hum. Mol. Genet. 2020. [Google Scholar] [CrossRef] [PubMed]
- Hatada, I.; Ohashi, H.; Fukushima, Y.; Kaneko, Y.; Inoue, M.; Komoto, Y.; Okada, A.; Ohishi, S.; Nabetani, A.; Morisaki, H.; et al. An imprinted gene p57 KIP2 is mutated in Beckwith–Wiedemann syndrome. Nat. Genet. 1996, 14, 171–173. [Google Scholar] [CrossRef] [PubMed]
- Brioude, F.; Kalish, J.M.; Mussa, A.; Foster, A.C.; Bliek, J.; Ferrero, G.B.; Boonen, S.E.; Cole, T.; Baker, R.; Bertoletti, M.; et al. Clinical and molecular diagnosis, screening and management of Beckwith–Wiedemann syndrome: An international consensus statement. Nat. Rev. Endocrinol. 2018, 14, 229–249. [Google Scholar] [CrossRef]
- Gardiner, K.; Chitayat, D.; Choufani, S.; Shuman, C.; Blaser, S.; Terespolsky, D.; Farrell, S.; Reiss, R.; Wodak, S.; Pu, S.; et al. Brain abnormalities in patients with Beckwith–Wiedemann syndrome. Am. J. Med. Genet. Part A 2012, 158A, 1388–1394. [Google Scholar] [CrossRef]
- Bouschet, T.; Dubois, E.; Reynes, C.; Kota, S.K.; Rialle, S.; Maupetit-Mehouas, S.; Pezet, M.; Le Digarcher, A.; Nidelet, S.; Demolombe, V.; et al. In Vitro corticogenesis from embryonic stem cells recapitulates the In Vivo epigenetic control of imprinted gene expression. Cereb. Cortex 2017, 27, 2418–2433. [Google Scholar] [CrossRef]
- Imaizumi, Y.; Furutachi, S.; Watanabe, T.; Miya, H.; Kawaguchi, D.; Gotoh, Y. Role of the imprinted allele of the Cdkn1c gene in mouse neocortical development. Sci. Rep. 2020, 10, 1884. [Google Scholar] [CrossRef]
- Laukoter, S.; Beattie, R.; Pauler, F.M.; Amberg, N.; Nakayama, K.I.; Hippenmeyer, S. Imprinted Cdkn1c genomic locus cell-autonomously promotes cell survival in cerebral cortex development. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef]
- Reik, W.; Walter, J. Genomic imprinting: Parental influence on the genome. Nat. Rev. Genet. 2001, 2, 21–32. [Google Scholar] [CrossRef]
- Pearce, G.P.; Spencer, H.G. Population genetic models of genomic imprinting. Genetics 1992, 130, 899–907. [Google Scholar]
- Yu, S.; Yu, D.; Lee, E.; Eckhaus, M.; Lee, R.; Corria, Z.; Accili, D.; Westphal, H.; Weinstein, L.S. Variable and tissue-specific hormone resistance in heterotrimeric Gs protein α-subunit (Gsα) knockout mice is due to tissue-specific imprinting of the Gsα gene. Proc. Natl. Acad. Sci. USA 1998, 95, 8715–8720. [Google Scholar] [CrossRef] [PubMed]
- Gould, T.D.; Pfeifer, K. Imprinting of mouse Kvlqt1 is developmentally regulated. Hum. Mol. Genet. 1998, 7, 483–487. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rougeulle, C.; Glatt, H.; Lalande, M. The Angelman syndrome candidate gene, UBE3AIE6-AP, is imprinted in brain. Nat. Genet. 1997, 17, 14–15. [Google Scholar] [CrossRef] [PubMed]
- Babak, T.; Deveale, B.; Armour, C.; Raymond, C.; Cleary, M.A.; van der Kooy, D.; Johnson, J.M.; Lim, L.P. Global survey of genomic imprinting by transcriptome sequencing. Curr. Biol. 2008, 18, 1735–1741. [Google Scholar] [CrossRef]
- Wang, X.; Sun, Q.; McGrath, S.D.; Mardis, E.R.; Soloway, P.D.; Clark, A.G. Transcriptome-wide identification of novel imprinted genes in neonatal mouse brain. PLoS ONE 2008, 3, e3839. [Google Scholar] [CrossRef]
- Wang, Q.; Li, K.; Zhang, D.; Li, J.; Xu, G.; Zheng, J.; Yang, N.; Qu, L. Next-generation sequencing techniques reveal that genomic imprinting is absent in day-old gallus gallus domesticus brains. PLoS ONE 2015, 10, e0132345. [Google Scholar] [CrossRef]
- Nodine, M.D.; Bartel, D.P. Maternal and paternal genomes contribute equally to the transcriptome of early plant embryos. Nature 2012, 482, 94–97. [Google Scholar] [CrossRef]
- Andergassen, D.; Dotter, C.P.; Wenzel, D.; Sigl, V.; Bammer, P.C.; Muckenhuber, M.; Mayer, D.; Kulinski, T.M.; Theussl, H.-C.; Penninger, J.M.; et al. Mapping the mouse Allelome reveals tissue-specific regulation of allelic expression. eLife 2017, 6. [Google Scholar] [CrossRef]
- Babak, T.; DeVeale, B.; Tsang, E.K.; Zhou, Y.; Li, X.; Smith, K.S.; Kukurba, K.R.; Zhang, R.; Li, J.B.; van der Kooy, D.; et al. Genetic conflict reflected in tissue-specific maps of genomic imprinting in human and mouse. Nat. Genet. 2015, 47, 544–549. [Google Scholar] [CrossRef]
- Gulyás-Kovács, A.; Keydar, I.; Xia, E.; Fromer, M.; Hoffman, G.; Ruderfer, D.; Sachidanandam, R.; Chess, A. Unperturbed expression bias of imprinted genes in schizophrenia. Nat. Commun. 2018, 9, 2914. [Google Scholar] [CrossRef]
- DeVeale, B.; van der Kooy, D.; Babak, T. Critical evaluation of imprinted gene expression by RNA-Seq: A new perspective. PLoS Genet. 2012, 8, e1002600. [Google Scholar] [CrossRef] [PubMed]
- Kelsey, G.; Bartolomei, M.S. Imprinted genes and the number is? PLoS Genet. 2012, 8. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, B.; Monajemi, R.; Gagalova, K.K.; Ho, D.; Draisma, H.H.M.; van de Wiel, M.A.; Franke, L.; Heijmans, B.T.; van Meurs, J.; Jansen, R.; et al. RNA-Seq in 296 phased trios provides a high-resolution map of genomic imprinting. BMC Biol. 2019, 17, 50. [Google Scholar] [CrossRef] [PubMed]
- Perez, J.D.; Rubinstein, N.D.; Fernandez, D.E.; Santoro, S.W.; Needleman, L.A.; Ho-Shing, O.; Choi, J.J.; Zirlinger, M.; Chen, S.K.; Liu, J.S.; et al. Quantitative and functional interrogation of parent-of-origin allelic expression biases in the brain. eLife 2015, 4. [Google Scholar] [CrossRef]
- Tran, D.A.; Bai, A.Y.; Singh, P.; Wu, X.; Szabo, P.E. Characterization of the imprinting signature of mouse embryo fibroblasts by RNA deep sequencing. Nucl. Acids Res. 2014, 42, 1772–1783. [Google Scholar] [CrossRef]
- Wang, X.; Soloway, P.D.; Clark, A.G. A survey for novel imprinted genes in the mouse placenta by mRNA-seq. Genetics 2011, 189, 109–122. [Google Scholar] [CrossRef][Green Version]
- Bonthuis, P.J.; Huang, W.C.; Stacher Horndli, C.N.; Ferris, E.; Cheng, T.; Gregg, C. Noncanonical genomic imprinting effects in offspring. Cell Rep. 2015, 12, 979–991. [Google Scholar] [CrossRef] [PubMed]
- Gregg, C.; Zhang, J.; Weissbourd, B.; Luo, S.; Schroth, G.P.; Haig, D.; Dulac, C. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science 2010, 329, 643–648. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Huang, S.-C.; Tung, C.-C.; Chou, C.-H.; Gau, S.S.-F.; Huang, H.-S. Analysis of genome-wide monoallelic expression patterns in three major cell types of mouse visual cortex using laser capture microdissection. PLoS ONE 2016, 11, e0163663. [Google Scholar] [CrossRef]
- Lorenc, A.; Linnenbrink, M.; Montero, I.; Schilhabel, M.B.; Tautz, D. Genetic differentiation of hypothalamus parentally biased transcripts in populations of the house mouse implicate the prader–willi syndrome imprinted region as a possible source of behavioral divergence. Mol. Biol. Evol. 2014, 31, 3240–3249. [Google Scholar] [CrossRef]
- Wang, X.; Clark, A.G. Using next-generation RNA sequencing to identify imprinted genes. Heredity (Edinb) 2014, 113, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Reynes, C.; Kister, G.; Rohmer, M.; Bouschet, T.; Varrault, A.; Dubois, E.; Rialle, S.; Journot, L.; Sabatier, R. ISoLDE: A data-driven statistical method for the inference of allelic imbalance in datasets with reciprocal crosses. Bioinformatics 2019. [Google Scholar] [CrossRef] [PubMed]
- Degner, J.F.; Marioni, J.C.; Pai, A.A.; Pickrell, J.K.; Nkadori, E.; Gilad, Y.; Pritchard, J.K. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics 2009, 25, 3207–3212. [Google Scholar] [CrossRef]
- Luo, S.; Valencia, C.A.; Zhang, J.; Lee, N.-C.; Slone, J.; Gui, B.; Wang, X.; Li, Z.; Dell, S.; Brown, J.; et al. Biparental Inheritance of Mitochondrial DNA in Humans. Proc. Natl. Acad. Sci. USA 2018, 115, 13039. [Google Scholar] [CrossRef] [PubMed]
- Santoni, F.A.; Stamoulis, G.; Garieri, M.; Falconnet, E.; Ribaux, P.; Borel, C.; Antonarakis, S.E. Detection of imprinted genes by single-cell allele-specific gene expression. Am. J. Hum. Genet. 2017, 100, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Zink, F.; Magnusdottir, D.N.; Magnusson, O.T.; Walker, N.J.; Morris, T.J.; Sigurdsson, A.; Halldorsson, G.H.; Gudjonsson, S.A.; Melsted, P.; Ingimundardottir, H.; et al. Insights into imprinting from parent-of-origin phased methylomes and transcriptomes. Nat. Genet. 2018, 50, 1542–1552. [Google Scholar] [CrossRef]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931. [Google Scholar] [CrossRef]
- Levesque, M.J.; Ginart, P.; Wei, Y.; Raj, A. Visualizing SNVs to quantify allele-specific expression in single cells. Nat. Methods 2013, 10, 865–867. [Google Scholar] [CrossRef]
- Ginart, P.; Kalish, J.M.; Jiang, C.L.; Yu, A.C.; Bartolomei, M.S.; Raj, A. Visualizing allele-specific expression in single cells reveals epigenetic mosaicism in an H19 loss-of-imprinting mutant. Genes Dev. 2016, 30, 567–578. [Google Scholar] [CrossRef]
- Kaffer, C.R.; Srivastava, M.; Park, K.-Y.; Ives, E.; Hsieh, S.; Batlle, J.; Grinberg, A.; Huang, S.-P.; Pfeifer, K. A transcriptional insulator at the imprinted H19/Igf2 locus. 12. Genes Dev. 2000, 14, 1908–1919. [Google Scholar]
- Symmons, O.; Chang, M.; Mellis, I.A.; Kalish, J.M.; Park, J.; Suszták, K.; Bartolomei, M.S.; Raj, A. Allele-specific RNA imaging shows that allelic imbalances can arise in tissues through transcriptional bursting. PLoS Genet. 2019, 15, e1007874. [Google Scholar] [CrossRef] [PubMed]
- Larsson, A.J.M.; Johnsson, P.; Hagemann-Jensen, M.; Hartmanis, L.; Faridani, O.R.; Reinius, B.; Segerstolpe, Å.; Rivera, C.M.; Ren, B.; Sandberg, R. Genomic encoding of transcriptional burst kinetics. Nature 2019, 565, 251–254. [Google Scholar] [CrossRef]
- Swanzey, E.; Stadtfeld, M. A reporter model to visualize imprinting stability at the Dlk1 locus during mouse development and in pluripotent cells. Development 2016. [Google Scholar] [CrossRef] [PubMed]
- Bonthuis, P.J.; Steinwand, S.; Huang, W.-C.; Horndli, C.N.S.; Emery, J.; Kravitz, S.; Ferris, E.; Gregg, C. Dopa decarboxylase is a genetic hub of parental control over offspring behavior. bioRxiv 2020. [Google Scholar] [CrossRef]
- Deng, Q.; Ramsköld, D.; Reinius, B.; Sandberg, R. Single-cell RNA-Seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 2014, 343, 193–196. [Google Scholar] [CrossRef]
- Saraiva, L.R.; Ibarra-Soria, X.; Khan, M.; Omura, M.; Scialdone, A.; Mombaerts, P.; Marioni, J.C.; Logan, D.W. Hierarchical deconstruction of mouse olfactory sensory neurons: From whole mucosa to single-cell RNA-seq. Sci. Rep. 2015, 5, 1–17. [Google Scholar] [CrossRef]
- Chen, R.; Wu, X.; Jiang, L.; Zhang, Y. Single-cell RNA-Seq reveals hypothalamic cell diversity. Cell Rep. 2017, 18, 3227–3241. [Google Scholar] [CrossRef]
- Laukoter, S.; Pauler, F.M.; Beattie, R.; Amberg, N.; Hansen, A.H.; Streicher, C.; Penz, T.; Bock, C.; Hippenmeyer, S. Cell-type specificity of genomic imprinting in cerebral cortex. Neuron 2020. [Google Scholar] [CrossRef]
- Picelli, S. Full-Length Single-cell RNA sequencing with smart-seq2. In Single Cell Methods: Sequencing and Proteomics; Proserpio, V., Ed.; Springer: New York, NY, USA, 2019; pp. 25–44. ISBN 978-1-4939-9240-9. [Google Scholar]
- Xu, J.; Carter, A.C.; Gendrel, A.-V.; Attia, M.; Loftus, J.; Greenleaf, W.J.; Tibshirani, R.; Heard, E.; Chang, H.Y. Landscape of monoallelic DNA accessibility in mouse embryonic stem cells and neural progenitor cells. Nat. Genet. 2017, 49, 377–386. [Google Scholar] [CrossRef]
- Gendrel, A.-V.; Attia, M.; Chen, C.-J.; Diabangouaya, P.; Servant, N.; Barillot, E.; Heard, E. Developmental dynamics and disease potential of random monoallelic gene expression. Dev. Cell 2014, 28, 366–380. [Google Scholar] [CrossRef]
- Moffitt, J.R.; Bambah-Mukku, D.; Eichhorn, S.W.; Vaughn, E.; Shekhar, K.; Perez, J.D.; Rubinstein, N.D.; Hao, J.; Regev, A.; Dulac, C.; et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 2018, 362. [Google Scholar] [CrossRef] [PubMed]
- Kelsey, G.; Stegle, O.; Reik, W. Single-cell epigenomics: Recording the past and predicting the future. Science 2017, 358, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, Y.; Shivalila, C.S.; Soldner, F.; Markoulaki, S.; Jaenisch, R. Tracing dynamic changes of DNA methylation at single-cell resolution. Cell 2015, 163, 218–229. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, Y.; Wu, H.; Song, Y.; Shivalila, C.S.; Markoulaki, S.; Jaenisch, R. Parent-of-origin DNA methylation dynamics during mouse development. Cell Rep. 2016, 16, 3167–3180. [Google Scholar] [CrossRef]
- Pott, S. Simultaneous measurement of chromatin accessibility, DNA methylation, and nucleosome phasing in single cells. eLife 2017, 6, e23203. [Google Scholar] [CrossRef]
- Kelly, T.K.; Liu, Y.; Lay, F.D.; Liang, G.; Berman, B.P.; Jones, P.A. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res. 2012, 22, 2497–2506. [Google Scholar] [CrossRef]
- Smallwood, S.A.; Lee, H.J.; Angermueller, C.; Krueger, F.; Saadeh, H.; Peat, J.; Andrews, S.R.; Stegle, O.; Reik, W.; Kelsey, G. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods 2014, 11, 817–820. [Google Scholar] [CrossRef]
- Guo, H.; Zhu, P.; Wu, X.; Li, X.; Wen, L.; Tang, F. Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res. 2013, 23, 2126–2135. [Google Scholar] [CrossRef]
- Zhu, C.; Preissl, S.; Ren, B. Single-cell multimodal omics: The power of many. Nat. Methods 2020, 17, 11–14. [Google Scholar] [CrossRef]
- Rotem, A.; Ram, O.; Shoresh, N.; Sperling, R.A.; Goren, A.; Weitz, D.A.; Bernstein, B.E. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat. Biotechnol. 2015, 33, 1165–1172. [Google Scholar] [CrossRef]
- Mateo, L.J.; Murphy, S.E.; Hafner, A.; Cinquini, I.S.; Walker, C.A.; Boettiger, A.N. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 2019, 568, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Angermueller, C.; Clark, S.J.; Lee, H.J.; Macaulay, I.C.; Teng, M.J.; Hu, T.X.; Krueger, F.; Smallwood, S.A.; Ponting, C.P.; Voet, T.; et al. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat. Methods 2016, 13, 229–232. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Huang, K.; An, Q.; Du, G.; Hu, G.; Xue, J.; Zhu, X.; Wang, C.-Y.; Xue, Z.; Fan, G. Simultaneous profiling of transcriptome and DNA methylome from a single cell. Genome Biol. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Li, L.; Li, J.; Wu, X.; Hu, B.; Zhu, P.; Wen, L.; Tang, F. Single-cell multi-omics sequencing of mouse early embryos and embryonic stem cells. Cell Res. 2017, 27, 967–988. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.J.; Argelaguet, R.; Kapourani, C.-A.; Stubbs, T.M.; Lee, H.J.; Alda-Catalinas, C.; Krueger, F.; Sanguinetti, G.; Kelsey, G.; Marioni, J.C.; et al. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat. Commun. 2018, 9, 781. [Google Scholar] [CrossRef]
- Argelaguet, R.; Clark, S.J.; Mohammed, H.; Stapel, L.C.; Krueger, C.; Kapourani, C.-A.; Imaz-Rosshandler, I.; Lohoff, T.; Xiang, Y.; Hanna, C.W.; et al. Multi-omics profiling of mouse gastrulation at single-cell resolution. Nature 2019, 576, 487–491. [Google Scholar] [CrossRef]
- Hemberger, M.; Dean, W.; Reik, W. Epigenetic dynamics of stem cells and cell lineage commitment: Digging Waddington’s canal. Nat. Rev. Mol. Cell Biol. 2009, 10, 526–537. [Google Scholar] [CrossRef]
- Dixit, A.; Parnas, O.; Li, B.; Chen, J.; Fulco, C.P.; Jerby-Arnon, L.; Marjanovic, N.D.; Dionne, D.; Burks, T.; Raychndhury, R.; et al. Perturb-seq: Dissecting molecular circuits with scalable single cell RNA profiling of pooled genetic screens. Cell 2016, 167, 1853–1866. [Google Scholar] [CrossRef]
- Proserpio, V. Methods in Molecular Biology. In Single Cell Methods: Sequencing and Proteomics; Proserpio, V., Ed.; Springer: New York, NY, USA, 2019; Volume 1979, ISBN 978-1-4939-9239-3. [Google Scholar]
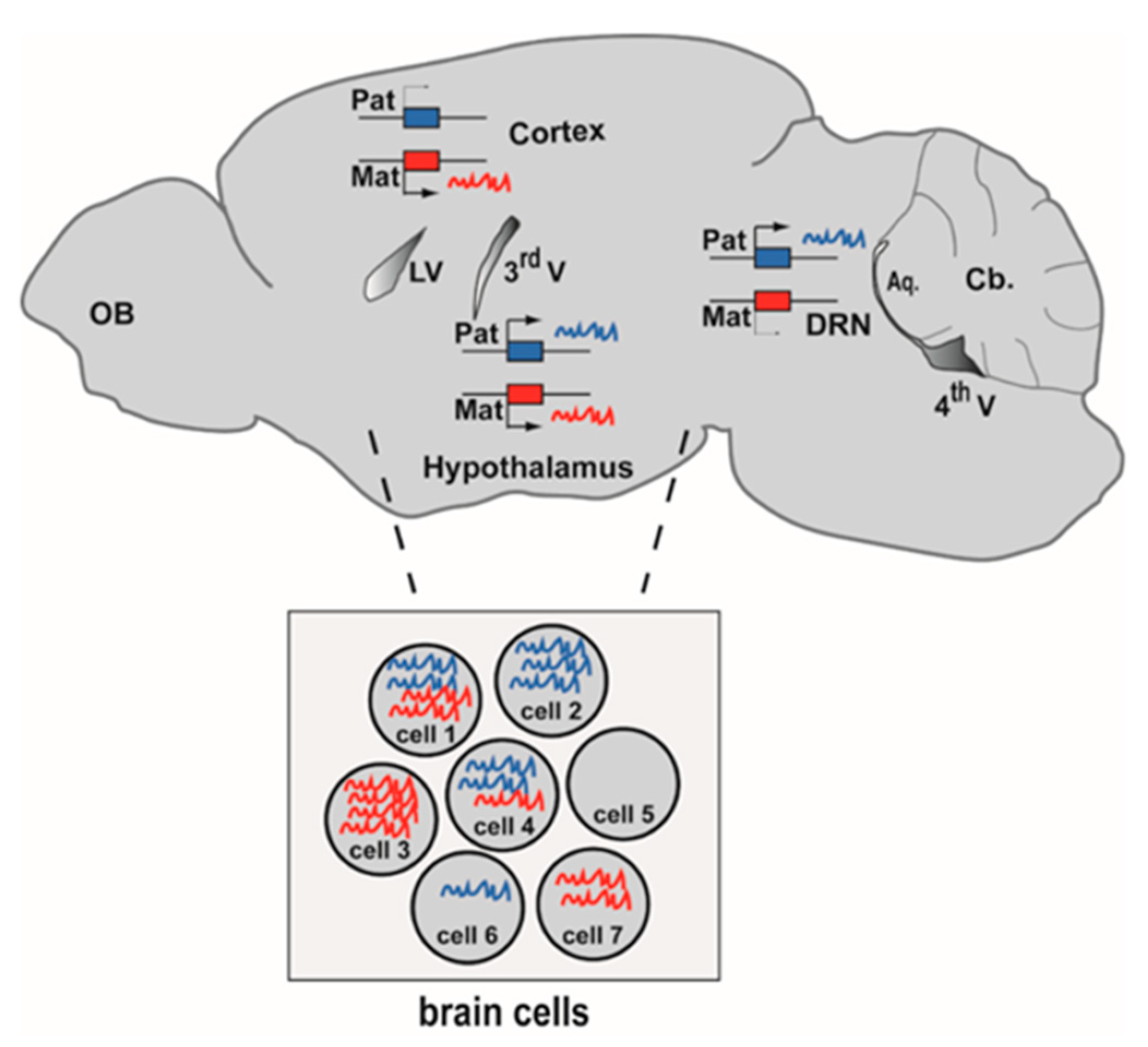

| Study: First Author Name, Reference | Brain Region | Mouse Strains | Reciprocal Crosses | Replicates | Concordance of SNPs | Strand Specific RNA-seq | Validation by Other Methods | Additional Epigenetic Marks |
|---|---|---|---|---|---|---|---|---|
| Andergassen, [60] | whole brain | FVB × Cast | Yes | Yes | Yes | Yes | No | allele-specific ChIP (H3K27ac) |
| Babak, [61] | 13 brain parts | C57BL/6 × Cast | Yes | Yes | Yes | Yes | Pyrosequencing | DNA Methylation |
| Bonthuis, [69] | arcuate and dorsal raphe | C57BL/6 × Cast | Yes | Yes | Yes | No | Pyrosequencing, RNAscope | allele-specific ChIP (H3K9ac and H3K9me3) |
| Bouschet, [48] | cerebral cortex | C57BL/6 × JF1 | Yes | Yes | No | Yes | Sanger/RFLP | DNA Methylation |
| DeVeale, [63] | whole brain | C57BL/6 × Cast | Yes | Yes | Yes | Yes | Pyrosequencing | No |
| Gregg, [70] | Cortex and hypothalamus | C57BL/6 × Cast | Yes | Yes | No | No | iPLEX Sequenom | No |
| Lin, [71] | 3 cell types of visual cortex | C57BL/6 × Cast | Yes | No | No | No | Sanger | No |
| Lorenc, [72] | hypothalamus | WSB × PWD | Yes | Yes | Yes | No | Pyrosequencing | No |
| Perez, [66] | cortex, hypothalamuscerebellum | C57BL/6 × Cast | Yes | Yes | No | No | Pyrosequencing | No |
| Wang, [57] | Whole brain | PWD × AKR | Yes | Yes | No | No | Sanger Pyrosequencing | No |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Varrault, A.; Dubois, E.; Le Digarcher, A.; Bouschet, T. Quantifying Genomic Imprinting at Tissue and Cell Resolution in the Brain. Epigenomes 2020, 4, 21. https://doi.org/10.3390/epigenomes4030021
Varrault A, Dubois E, Le Digarcher A, Bouschet T. Quantifying Genomic Imprinting at Tissue and Cell Resolution in the Brain. Epigenomes. 2020; 4(3):21. https://doi.org/10.3390/epigenomes4030021
Chicago/Turabian StyleVarrault, Annie, Emeric Dubois, Anne Le Digarcher, and Tristan Bouschet. 2020. "Quantifying Genomic Imprinting at Tissue and Cell Resolution in the Brain" Epigenomes 4, no. 3: 21. https://doi.org/10.3390/epigenomes4030021
APA StyleVarrault, A., Dubois, E., Le Digarcher, A., & Bouschet, T. (2020). Quantifying Genomic Imprinting at Tissue and Cell Resolution in the Brain. Epigenomes, 4(3), 21. https://doi.org/10.3390/epigenomes4030021





