Abstract
It is generally accepted that epigenetic modifications, such as DNA and histone methylations, affect transcription and that a gene’s transcription feeds back on its epigenetic profile. Depending on the epigenetic modification, positive and negative feedback loops have been described. Here, we study whether such interrelation are mandatory and how transcription factor networks affect it. We apply self-organizing map machine learning to a published data set on the specification and differentiation of murine intestinal stem cells in order to provide an integrative view of gene transcription and DNA, as well as histone methylation during this process. We show that, although gain/loss of H3K4me3 at a gene promoter is generally considered to be associated with its increased/decreased transcriptional activity, such an interrelation is not mandatory, i.e., changes of the modification level do not necessarily affect transcription. Similar considerations hold for H3K27me3. In addition, even strong changes in the transcription of a gene do not necessarily affect its H3K4me3 and H3K27me3 modification profile. We provide a mechanistic explanation of these phenomena that is based on a model of epigenetic regulation of transcription. Thereby, the analyzed data suggest a broad variance in gene specific regulation of histone methylation and support the assumption of an independent regulation of transcription by histone methylation and transcription factor networks. The results provide insights into basic principles of the specification of tissue stem cells and highlight open questions about a mechanistic modeling of this process.
1. Introduction
In the last decade, epigenetic profiling of cell populations and tissues has become a standard method in characterizing molecular regulation during homeostasis and tissue transformation. First studies already noted a strong correlation between epigenetic marks that are associated with a gene and the transcription of the gene itself [1,2]. Today, a strong impact, in particular of histone methylations, on transcription is commonly assumed. Nevertheless, there is still a debate on whether epigenetic modifications control the transcription level of associated genes or stabilize their transcriptional states that have been acquired in advance [3]. Moreover, the feedback of a gene’s transcription on its epigenetic profile is still not fully understood.
Several molecular biology studies of the last years have shed light on these questions. Tri-methylation of different lysines of histone 3, such as H3K4me3 and H3K27me3, have been shown to affect transcription. While H3K4me3 supports transcription initiation by recruiting RNA polymerase II [4], H3K27me3 represses transcription by preventing binding of acetyl-transferases to target genes [5]. Conversely, active transcription has been linked to the recruitment of H3K4me3 histone methyltransferases (HMTs) [6] and binding of activating transcription factors has been suggested to compete with the binding of H3K27me3 HMTs [7].
Together, these interactions define a positive feedback between H3K4me3 and transcription and a negative feedback between H3K27me3 and transcription and are in good agreement with the correlations found in whole genome profiling studies [2]. In contrast to these histone methylations, promoter DNA methylation has been described to become recruited after a particular transcriptional state has been established and, thus, is considered to affect in particular the stability of transcriptional states [8,9]. Nevertheless, DNA methylation can affect transcription indirectly by repelling H3K4me3 [10] and H3K27me3 HMTs [7]. The modifications that were set by these HMTs in turn interfere with binding of DNA methyl-transferases (DNMTs). DNMTs are repelled by H3K4me3 [11] and evidence has been provided that they are recruited by H3K27me3 [12].
Based on such experimental findings, we have established a computational model of epigenetic regulation of transcription [13,14] and applied it in order to explain gene regulatory phenomena associated with stem cell differentiation and aging [15,16,17,18]. In parallel, we have developed data analysis tools that are based on self-organizing maps (SOMs) that enable a combined analysis of transcription and epigenetic data [19,20]. After applying these tools to data sets on developmental or tissue transformation processes, we observed that strong correlations between transcription and histone methylation, although existent for important steps of regulation, do not appear to be mandatory and apply only to a well-defined subset of genes.
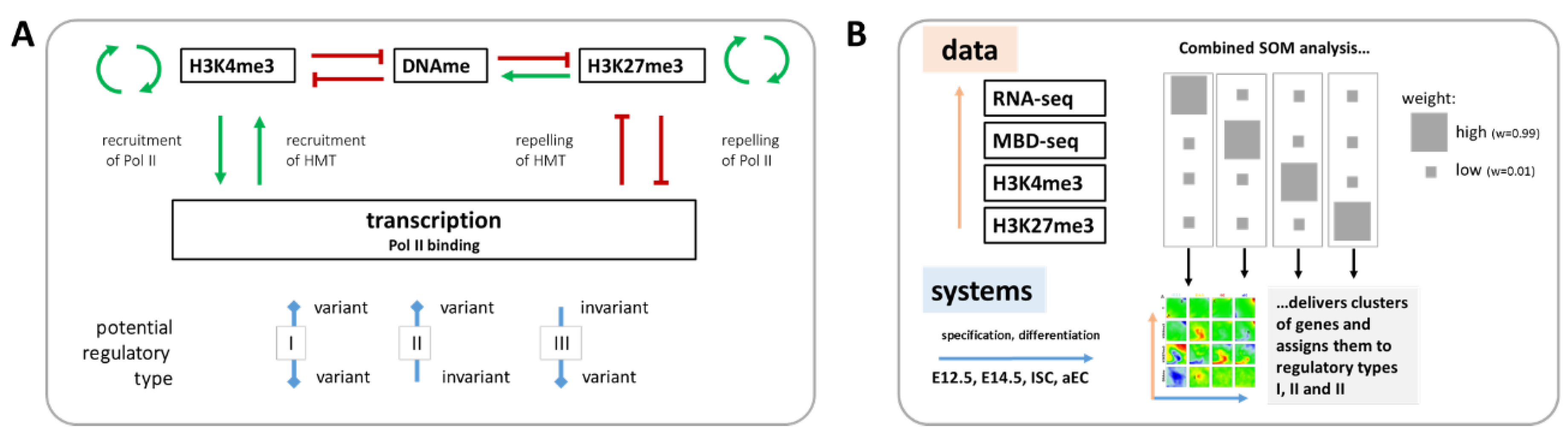
Here, we study such relations re-analyzing RNA-seq, H3K4me3, and H3K27me3 ChIP-seq as well as methyl-CpG binding domain protein (MBD)-seq data on cells of four different states of the specification and differentiation of mouse intestinal stem cells (ISCs) published by Kazakevych et al. [21]. In this study, embryonic intestinal epithelial progenitors at embryonic day E12.5 and E14.5, adult ISCs and their differentiated progeny (adult enterocytes, aECs) were isolated and compared, providing new insights into intestinal tissue formation. We focus here on regulatory principles of this process. We identify three different phenotypes of regulation (I-III) that cover correlated as well as independent regulation of transcription and epigenetic profiles. We provide a theoretical explanation for the regulatory behavior that was observed and discuss consequences for quantitative modeling of epigenetic regulation in processes, such as stem cell specification and differentiation. Figure 1 provides a schematic overview of our data analysis approach.
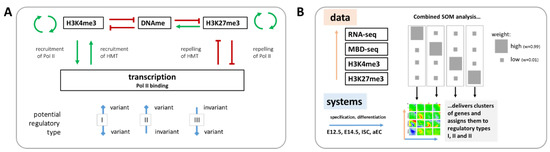
Figure 1.
Schematic overview of the study design. (A) Aim of the study: Known interactions between transcription and epigenetic states that are characterized by the histone methylations H3K4me3 and H3K27me3 and DNA-methylation (DNAme) are indicated. We ask whether mutual dependence between transcription and epigenetics is mandatory (I), or whether an independent variability between both can be observed (II and III). Pol II: RNA polymerase II, HMT: histone methyltransferase; and (B) For this purpose, published [21] transcriptome (RNA-seq) and epigenetic (H3K4me3 and H3K27me3 ChIP-seq and MBD-seq) data of four states of ISC specification and differentiation (progenitors at embryonic days E12.5 and 14.5; ISCs; aEC) were re-analyzed using a combinatorial SOM. Subsets of genes were assigned to regulation types I, II or III.
2. Results
2.1. Transcription ‘Dominated’ Self-Organizing Maps (SOMs)
In the first SOM analysis, we combined RNA-seq data with one of the epigenetic data sets on H3K4me3, H3K27me3, and promoter DNA (DNAme) methylation, respectively. For this purpose, we used a SOM that was recently developed by us [20]. For 11,404 out of the 31,591 genes with Ensembl-ID, no transcription levels were provided in the data set [21]. Thus, combined data from 20,187 genes entered our SOMs. We weighted the RNA-seq data with w = 0.999 and the epigenetic data with 0.001. Thus, according to the SOM principles [20], genes that change their transcription during ISC specification and differentiation sort to the boundaries of the SOM grid, while those being invariantly transcribed are preferentially sorted to the center in between the variant ones, as a rule of thumb. Here, the low weighted epigenetic properties that are associated with the genes induce an additional clustering. The SOM portraits obtained (averages over the replicates) from the four different cell types E12.5, E14.5, ISCs, and aECs are shown in Figure 2A. In the following, we focus on selected spots of the SOM portraits that are representative of different kinds of regulation.
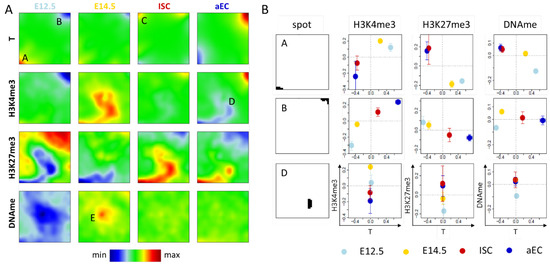
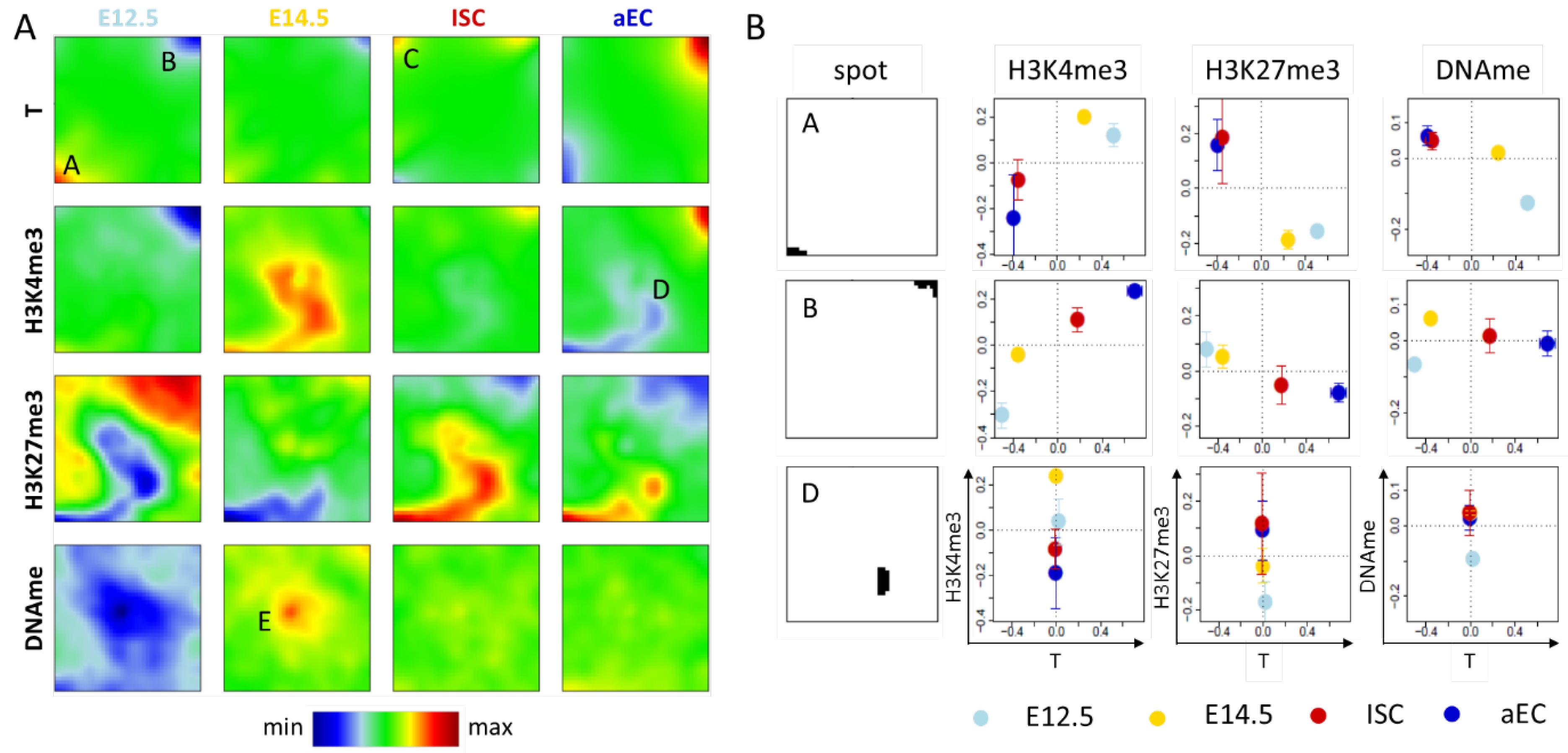
Figure 2.
Transcription ‘dominated’ SOMs. (A) Shown here are SOM portraits for transcription (T) separately combined with H3K4me3, H3K27me3, and DNA methylation (DNAme) measured for four different intestinal cell types (E12.5, E14.5, ISCs and aECs). The transcription portraits of the T-H3K4me3 SOM, the T-H3K27me3 SOM, and the T-DNAme SOM are very similar, those of the T-H3K4me3 SOM (1st row) are shown. The green area in these portraits defines the area of transcription invariance. In this area, the different methylations induce a specific clustering of genes (2nd–4th row). Spots discussed in the text are indicated by capital letters. Row-wise color-coding ranges from minimum (blue) to maximum (red); and (B) Pairwise correlations between the properties of the different cell types for selected spots of the SOMs. For the indicated properties, the average value over the spot genes is given as a function of their average transcription. Properties of spots C and E genes are provided in the Appendix A (Figure A1).
2.1.1. Correlated Changes of Epigenetic Profiles and Transcription
In all SOM portraits of the transcription states (Figure 2A, 1st row), two specific spots are present. Spot A genes are highly transcribed in both embryonic states (red), while they are repressed in ISCs and aECs (blue). Spot B shows the opposite regulation. Spot C is a cell type specific spot and the associated genes are highly transcribed only in ISCs (red spot). Beside changes in transcription, genes of spot A and B also show changes of their epigenetic profiles, as seen by color changes in the respective maps (Figure 2A 2nd-4th row and Figure 2B). Repression of transcription of spot A genes during ISC specification and differentiation is accompanied by a decrease in the abundance of H3K4me3 and an increase in H3K27me3, while parallel activation of spot B genes associates with opposite changes. Thus, the regulation of spot A and B genes is consistent with the common assumption that H3K4me3/H3K27me3 are positively/negatively correlated with transcription [2,22]. We call regulation where transcription correlates with changes of a defined epigenetic modification: regulation of type I of this modification.
While Figure 2B shows correlated changes of mean transcription and mean methylations during ISC specification and differentiation, such correlation was also calculated for the metagenes (pixels) of the individual spots. Results for the signed-covariance (SCOV) are shown in the Appendix A (Figure A1). As expected, H3K4me3 shows an exclusive positive correlation with transcription and H3K27me3 an exclusive negative correlation. Only for DNA methylation are both types of correlation with transcription observed, in agreement with the regulatory schemes that are provided in Figure 1A.
Gene ontology (GO) set enrichment analysis [23] revealed that genes of spot A are enriched in gene sets such as ‘multicellular organism development’ (biological process (BP), FDR < 1 × 10−8), i.e., they are linked to early developmental processes, while those of spot B are enriched in gene sets, such as ‘antigen processing and presentation’ (BP, FDR < 1 × 10−7) and ‘brush border’ (cellular component (CC), FDR < 1 × 10−19) documenting epithelial differentiation (the distribution of all genes of these sets across the SOM is given in Figure A2A). Thus, major steps of ISC specification and differentiation involve type I regulation. However, changes of the average fraction of H3K4me3 and H3K27me3 modified genes are in the range of 0.5 for spot A and B (Figure 2B). This indicates that only about half of these genes switch their respective modification state from 0 to 1 or vice versa. Thus, part of the genes changes transcription while the epigenome remains stable. This indicates the presence of a different type of regulation. This will be analyzed in more detail below.
2.1.2. Epigenetic Regulation without Changes of Transcription
The SOM portraits of the epigenetic states (Figure 2A) show spots D and E in the areas of virtually invariant transcription. Spot D (T-H3K4me3 SOM) is associated with genes that lose H3K4me3. These genes are not significantly enriched in GO sets. In contrast to genes following type I regulation schemes, histone modification changes of spot D genes are not paralleled by changes of transcription. We call such regulation: regulation of type II of the modification.
Applied to DNA methylation, the regulation of type II involves genes that change DNA methylation at stable transcription. An example is genes that are associated with spot E (T-DNAme SOM). Here, hypo- and hyper-methylation of genes occurs at progenitors at E12.5 and E14.5, respectively. This kind of regulation refers to several hundreds of genes. These genes are highly enriched in the set ‘G−protein coupled receptor signaling pathway’ (BP, FDR < 1 × 10−68). Interestingly, many of spot D and E genes are transcriptionally repressed as the average of their absolute transcription values is very small (Figure A2B).
In summary, although genes that are associated with major regulation processes of ISC specification and differentiation show correlated changes of transcription and histone methylation and/or DNA methylation, such correlation appears to be not mandatory. Strong epigenetic changes can occur without pronounced changes in transcription. The question arises whether our analysis also detects transcriptional changes that take place without changing epigenetic profiles.
2.2. SOMs ‘Dominated’ by Epigenetics
In a second series of SOM calculations, we again combined the RNA-seq data separately with each of the epigenetic data sets (H3K4me3, H3K27me3, DNAme). However, we down-weighted the RNA-seq data (w = 0.001) while up-weighting the epigenetic data (0.999). Thus, according to the SOM principles [20], genes that change their epigenetic state during ISC specification and differentiation sort to the boundaries of the SOM grid, while those being epigenetically invariant are sorted preferentially to the center.
2.2.1. H3K4me3 ‘Dominated’ SOMs
The SOM portraits of an H3K4me3 ‘dominated’ SOM (H3K4me3-T SOM) for the four different cell types (E12.5, E14.5, ISC, and aEC) are shown in Figure 3. The H3K4me3 portraits (Figure 3A, 1st row) show several spots at the boundaries of the SOM grid, among them are spots F and G. Genes that are associated with spot F recruit H3K4me3 during ISC specification and differentiation. In parallel, their transcription becomes upregulated. Genes associated with spot G show the opposite regulation. Consistently, both types of genes show a positive correlation between H3K4me3 and transcription (see: SCOV row). Thus, spot F and G comprise genes of regulation type I.
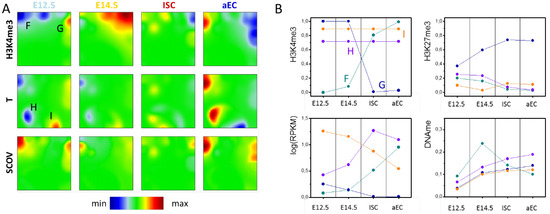
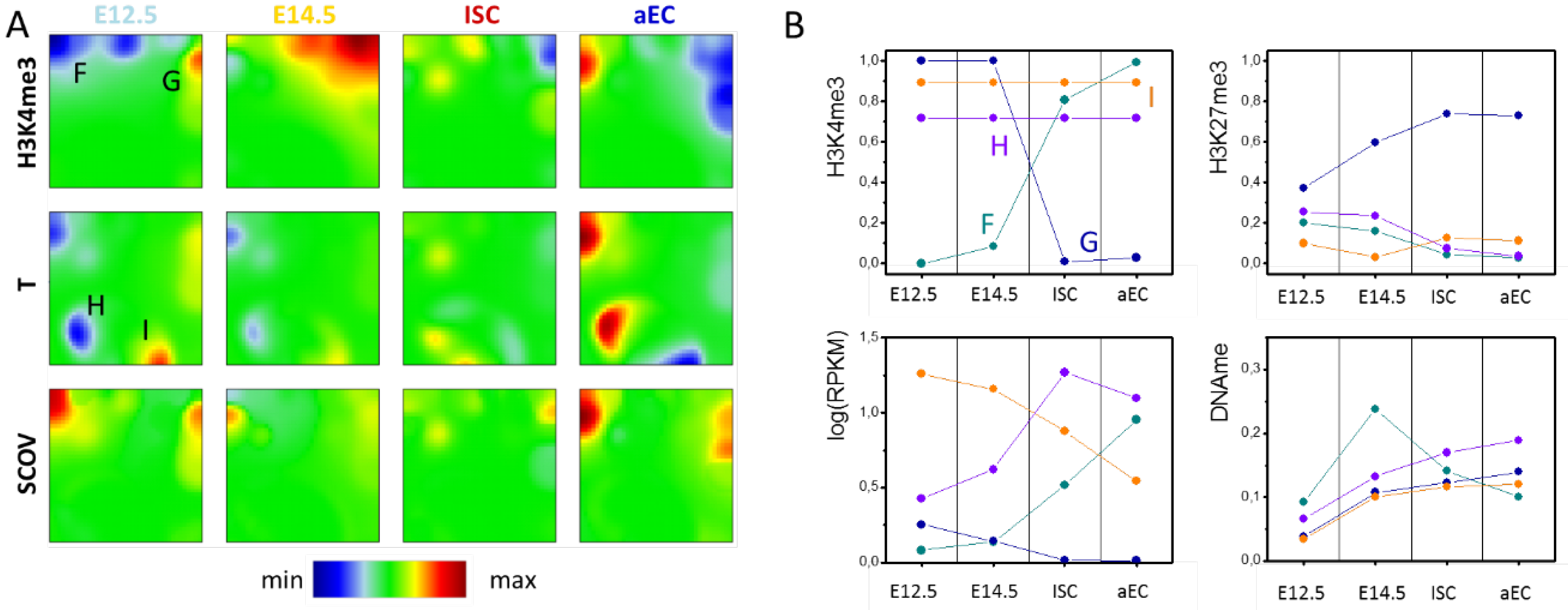
Figure 3.
H3K4me3 ‘dominated’ SOM. (A) Shown here are portraits for H3K4me3 (w = 0.999), transcription (T, w = 0.001) and the signed-covariance (SCOV) measured for progenitors at E12.5 and E14.5, ISCs and aECs. The area that is green in all H3K4me3 portraits defines the area of H3K4me3 invariance. Spots discussed in the text are indicted by capital letters; and (B) Absolute values of spot averages (T: 3 replicates; H3K4me3, H3K27me3, DNAme: 2 replicates each). Regulation of spot F: Gain of H3K4me3 and parallel loss of H3K27me3 and DNA methylation are accompanied by an increase of transcription. Regulation of spot G: Loss of H3K4me3 and gain of H3K27me3 are paralleled by a decrease of transcription. The spots H and I comprise transcribed genes with high H3K4me3 level that change transcription. Note that spot I refers to virtually invariant H3K4me3, although it is located at the border of the SOM due to the underlying effect of transcription on SOM-structure.
Similarly to genes of spot A and B of the T-H3K4me3 SOM, spot G and F genes enrich in the GO sets ‘multicellular organism development (BP, FDR < 1 × 10−6)’ and ‘brush border (CC, FDR < 1 × 10−3)’, respectively. The spot averages of the absolute transcription levels and histone and DNA methylations provide some more details on the genes’ regulation (Figure 3B). Spot F contains genes that are associated with unmodified histones in the progenitors at E12.5 and E14.5. Many of them recruit promoter DNA methylation in E14.5. During ISC specification and differentiation, these genes gain H3K4me3 in adult cells, while in parallel they lose DNA methylation. This regulation is consistent with the DNA de-methylation driven adult stem cell specification and differentiation that is described by Kazakevych et al. [21]. Repression of spot G genes affects many bivalent genes. They are transcribed on an intermediate level and become repressed following loss of H3K4me3 and gain of H3K27me3. This is consistent with histone methylation driven repression of early developmental genes [21].
In the area of virtually invariant H3K4me3 modification, the transcription portraits show two additional spots H and I (Figure 3A). Spot H genes become activated in ISCs, while those of spot I are inversely regulated. These genes undergo changes in transcription without changing their H3K4me3 level. We call this scenario regulation of type III with respect to the epigenetic modification under consideration. The genes of spot H and I represent part of the genes located in the spots B and A of the T-H3K4me3 SOM, respectively (Figure 2A). They represent those parts that do not change H3K4me3 (see above). Notably, they enrich in different GO sets. Genes of spot H enrich weakly (FDR < 0.01) in sets that are associated with different response mechanisms (BP), among them ‘response to external stimulus’, ‘response to chemical’, and ‘response to stress’. Spot I genes enrich in sets as ‘regulation of cell cycle (BP, FDR < 1 × 10−10)’ and ‘cellular metabolic process (BP, FDR < 1 × 10−14)’.
In summary, according to our SOM analyses, epigenetic marker profiles can vary without affecting the level of transcription and vice versa. Here, we demonstrated the reversal for H3K4me3 and transcription. Examples for SOM analysis combining H3K27me3 or DNA methylation with transcription are provided in the Appendix A (Figure A3).
2.3. Model Considerations
In the following, we provide explanations for the observed gene regulation scenarios. Thereby, we take advantage of the model of epigenetic regulation of transcription that was developed by our group [13,14]. The model describes the interrelations between transcription (T) and three epigenetic modifications, H3K4me3, H3K27me3, and DNA methylation. These methylation levels are quantified by the fraction of modified nucleosomes in the promoter of a given gene mK (K = 4, 27) in case of histone methylations and by the fraction of methylated CpGs in the promoter mCpG in the case of DNA methylation.
2.3.1 Regulation of Histone Modifications
The binding probabilities of the HMTs, ΘK (K = 4, 27), are calculated assuming a positive feedback between the presence of the histone mark K and the recruitment of its HMTs. Such feedback has been demonstrated for both H3K4me3 and H3K27me3 [24]. According to this assumption, the binding probability of the HMT of histone mark K depends on the fraction mK (K = 4, 27) of modified nucleosomes of the NH cooperative nucleosomes that are associated with the promoter and is given by:
Here, ε0K is the ground enthalpy per bound HMT complex and εBSK and εHMK are the free energies of HMT binding to DNA and to histone mark K, respectively. They are specific for histone mark K and they are scaled by the Boltzmann unit. The last term in the exponent describes the dependence of the histone modification level on the transcription T. The coupling constant λΚ was set (−1) for H3K4me3 and (+1) for H3K27me3.
Assuming a dynamic equilibrium between histone modification and de-modification, ΘK has to solve:
where CK is the de-modification constant CK= kde/kmod; i.e., the ratio between the de-modification rate kde and the modification rate kmod for the respective modification. The resulting self-consistent equation provides solutions for the modification level mK in dependence of T [13,14].
According to these considerations, the dependence of H3K4me3 on the transcription is described by an S-shaped curve (Figure 4, H3K27me3 by a Z-shaped curve). This shape has also been observed experimentally [2]. For low transcription, the modification level mK varies only slightly with T, and thus can be considered as being independent. The same applies to high transcription values. Thus, these regions are associated with regulation of type III. In case the transcription is increased above T2 (decreased below T1) for low (high) transcription, both transcription and the modification state change, i.e., regulation of type I takes place. In the case a gene is transcribed in the range of bi-stability (Figure 4, three solutions exist for m, the intermediate state is unstable), a temporary change of its transcription level can enforce a modification switch. After systems relaxation, only an epigenetic change will be detected and the effective regulation is classified as type II.
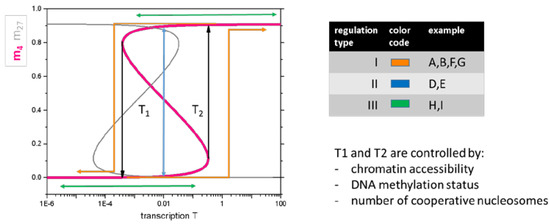
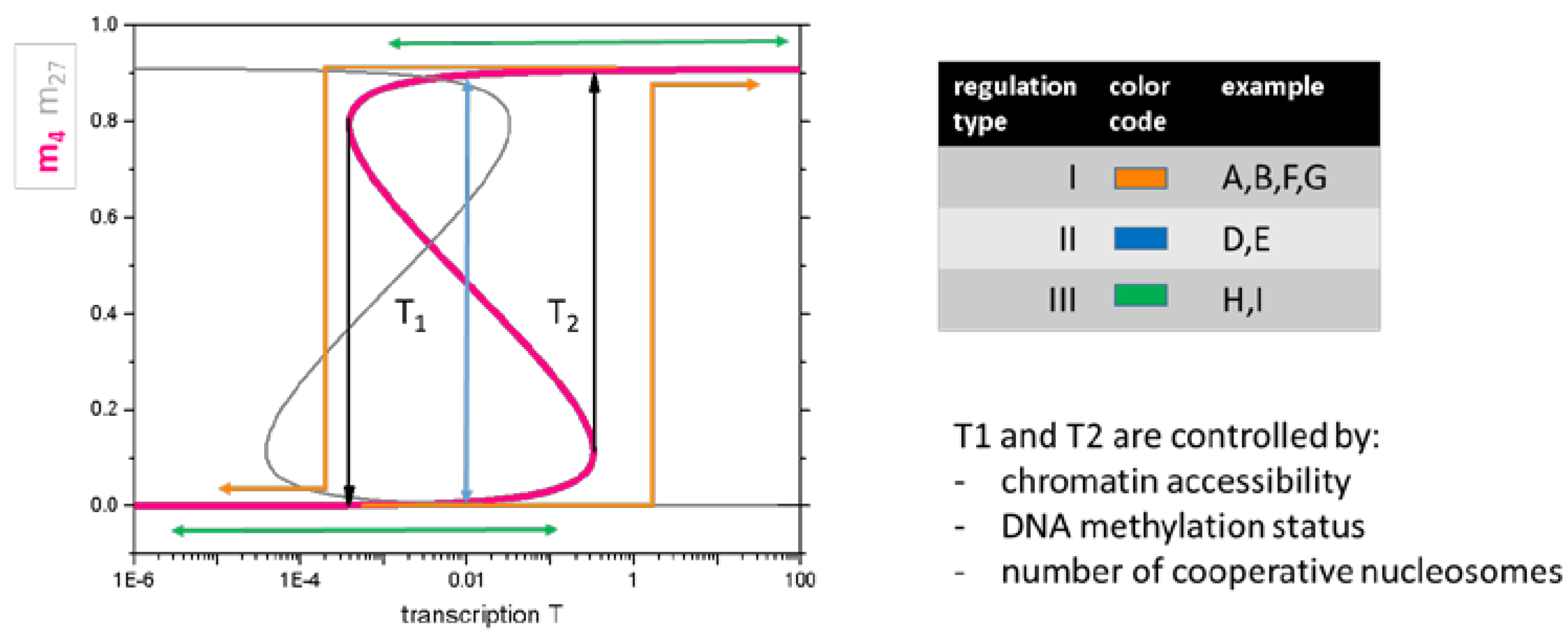
Figure 4.
Basic model of the regulation types I–III. Model of histone modification. According to Equation (1), the H3K4me3 level (pink line) increases with increasing transcription. In case of sufficient feedback between reading and writing of the modification by the HMTs, a bistable modification region is observed between the threshold transcription levels T1 and T2. Here, low and high H3K4me3 states coexist. Increasing/decreasing transcription at low/high H3K4me3 indeed only marginally affects modification as long as T2/T1 is not exceeded. This explains regulation type III. A transient increase/decrease of transcription exceeding T2/T1 enables a switch from low to high, respective high to low modification, potentially explaining regulation of type II. In case the change is permanent, the regulation refers to type I. The H3K27me3 level shows an ‘inverse’ behavior (grey line).
Thus, basic properties of the epigenetic machinery alone provide an explanation for the regulatory scenarios observed when analyzing the data of Kazakevych et al. [21]. For example, the regulatory scenarios associated with the spots of the H3K4me3 ‘dominated’ SOM (Figure 3, Spot F–I) can be interpreted as follows: the scenarios that are associated with the spots F and G represent switches of H3K4me3 based on activation and repression of the transcription above and below the thresholds T2 and T1, respectively. Genes of the spots H and I change their transcription but never cross the transcription thresholds.
2.3.2. Regulation of Transcription
The model of histone modification explains the regulation of type II as a modification switch enabled by transient changes of transcription, thereby, for a start neglecting feedback of the modification on transcription. In the model of transcription, we assume that transcription actually depends on the modification levels m4 and m27. It is calculated by solving a differential equation for transcript abundance, self-consistently with Equations (1) and (2):
Here, Pmax is the maximum promoter activity and δ the transcript degradation rate. The constant m0 << 1 ensures that genes can be transcribed even with a small rate also if the promoter is devoid of H3K4me3 (m4 = 0). FTF and Fauto are the regulation factors for the transcription factor (TF)-network and the auto-regulation of the gene, respectively. FTF is given by:
Here, εi is the decrease of the free energy of RNA polymerase II binding at the base promoter of the gene following binding of the TF i to its promoter. It is scaled by the Boltzmann unit. Ti is the transcription of this TF. The product runs over all TFs that are bound to the promoter. Changes of FTF can refer to both changes in the number of bound TFs and changes of the transcription of TFs. Fauto is defined in the same way while considering the binding of the gene to its own promoter. Details about the underlying regulatory principles, which are based on thermodynamics, can be found in [25].
Thus, according to this model, epigenetics strongly feeds back on transcription and an alternative explanation for type II regulation is required. Notably, the regulation that is found for genes associated with spot D and E of the T-H3K4me3 SOM (Figure 2) occurs at very low transcription (Figure A2). According to Equation (3), the variance of T becomes small in case of a small effective promoter activity (Peff << δ). This occurs if transcription is repressed by the TF-network. In this case, the term X = (m0 + m4)(1 − m27), which describes the dependence of transcription on the histone modification levels, becomes down-scaled and the absolute variance becomes small, i.e., becomes hardly detectable. Thus, assuming independent regulation of transcription by epigenetics and TF-networks can alternatively explain regulation type II, consistently with the regulation of the gene sets associated with spot D and E in the T-H3K4me3 SOM (Figure 2).
2.4. Epigenetic Regulation of Transcription: Transition States
2.4.1. Properties of Transition States
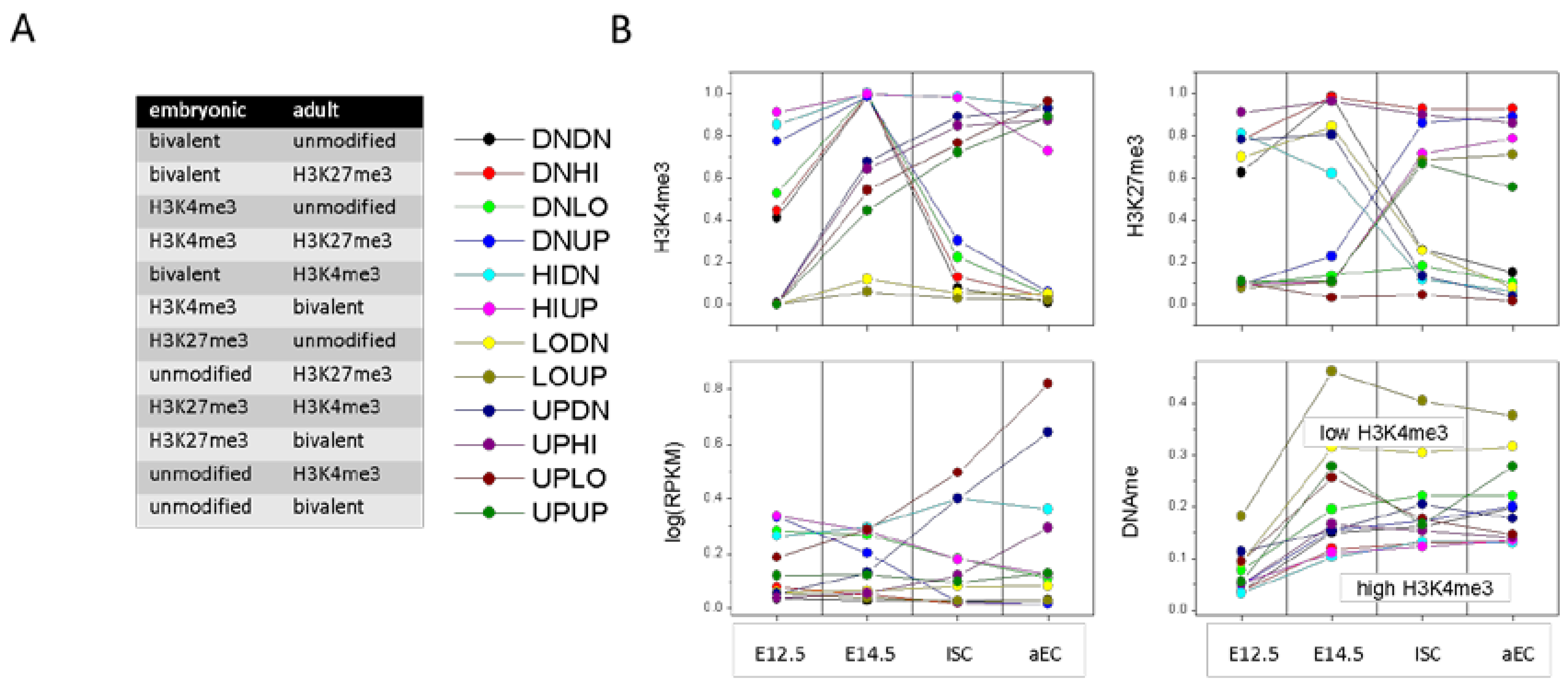
In order to test the above assumption, we identified those genes that change their histone modification profiles during ISC specification and differentiation in a defined way. For this purpose, each gene obtained a code (X,Y) with X,Y = UP, DN, HI, LO, depending on whether the histone modification level increases (UP), decreases (DN), or remains stable at high (HI) or low (LO) level during ISC specification and differentiation, respectively (see: Methods, Section 4.3). The code sequence is (H3K4me3; H3K27me3). There are 16 possible cases how a gene can behave, among them are 12 cases with changes of the histone modification profile. In the following, these cases are called transition states. We found all transition states to be well populated during ISC specification and differentiation, (>100 genes, Appendix B: Figure A4), except for the transitions from an H3K27me3 or an unmodified state into a bistable state. Thus, during ISC specification and differentiation the epigenetic transitions are realized in opposite directions with similar frequencies.
Figure 5 shows the average regulatory profiles of all transition states. Obviously, the recruitment of H3K4me3 already occurs in progenitors at E14.5, while the loss of H3K4me3 and changes of the H3K27me3 level are seen predominantly in ISCs. In agreement with the protective function of H3K4me3 against promoter DNA methylation, highest (lowest) values for this type of DNA methylation are seen for genes with stable low (high) H3K4me3. On average, the subsets of genes that carry H3K4me3 in embryonic progenitors and lose it in adult cells are lower transcribed than those of the subsets that gain H3K4me3 in the adult cells.
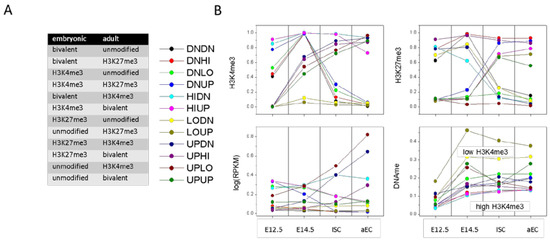
Figure 5.
Transcriptional states. (A) Modification in embryonic and adult states and the corresponding transition state (down, DN; up, UP; high, HI; low, LO); and (B) Properties of transition states. Averages are taken over all genes of the individual transition states and subsequently over the replicates. Recruitment of H3K4me3 (UP Y) occurs in progenitors at E14.5, while loss (DN Y) takes place in ISCs. The H3K4me3 level determines the promoter DNA methylation level.
Transition states that enrich in GO sets of the category BP comprise repression of bivalent genes by H3K4me3 loss (DNHI), activation of unmodified genes by H3K4me3 gain (UPLO), and a switch from the H3K27me3 to the H3K4me3 state (UPDN). Among the respective GO-sets are: ‘negative regulation of Wnt signaling pathway’ (DNHI, FDR < 1 × 10−3), ‘intestinal absorption’ (UPLO, FDR < 1 × 10−4), and ‘antigen processing and presentation’ (UPDN, FDR < 1 × 10−4).
2.4.2. Clustering of State Specific Transcription Profiles
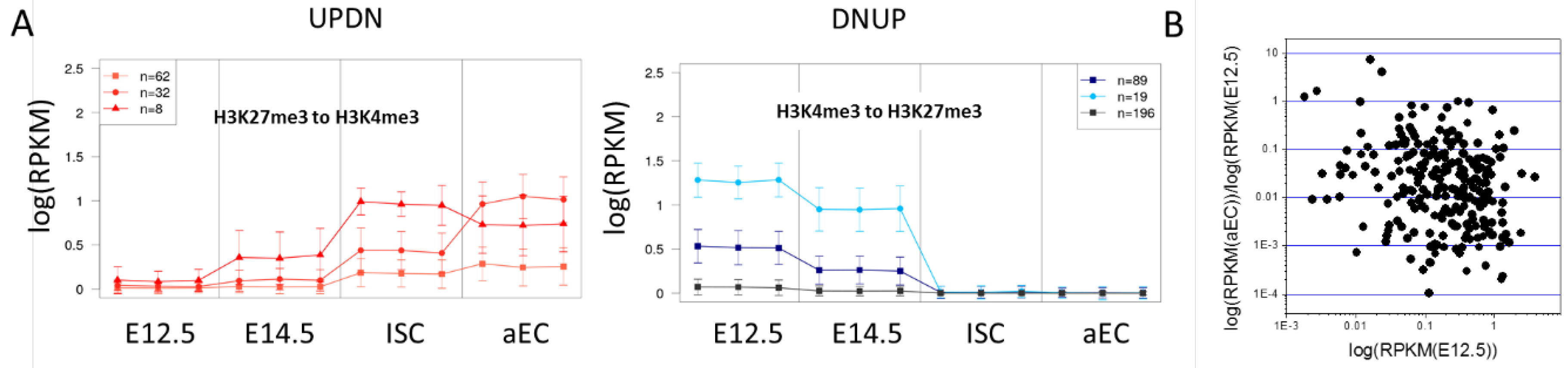
The averaged transcription values of the transition sets are somewhat misleading regarding the ongoing regulation. The transition state specific changes of the epigenetic profiles are similar for most genes. Moreover, all genes of a particular transition set change their transcription in the expected way and only a few deviate from the state specific behavior. However, the transcriptional changes take place at very different transcription levels (Figure 6A).
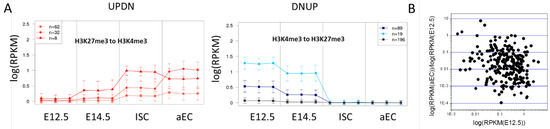
Figure 6.
Similar epigenetic regulation at different transcription levels. (A) Average transcription behavior of transition set subsets UPDN (H3K27me3 to H3K4me3) and DNUP (H3K4me3 to H3K27me3). Similar behavior was clustered, considering all three replicates independently (mean, sd). The cluster size is indicated. Higher absolute transcription changes are seen for genes with an initially higher transcription; and (B) Transcription fold changes between the adult ECs and progenitors at E12.5 are not correlated with the initial transcription in progenitors at E12.5 (DNUP set, H3K4me3 to H3K27me3).
Thereby, no correlation is detectable between the initial transcription and the fold-change that was observed during the regulation (for an example see: Figure 6B), suggesting that epigenetics enable a defined fold-change of the transcription level and the TF-networks define the level where the transition starts. The average of the fold change associated with the switch is 0.37 [0.06] for DNLO and 0.41 [0.06] for HIUP, but 0.11 [0.03] for DNUP (mean [SEM]). This indicates that H3K4me3 and H3K27me3 induce independent effects that can be described by individual factors, as suggested by Equation (3). Note that we analyzed gene repression only, thus the saturation of transcription does not affect the results.
Our results suggest that independent regulation of transcription by epigenetics and TF-networks appears to be a common regulatory mechanism during ISC specification and differentiation. Transition states comprise genes of regulation type I (for high transcription) and type II (for low transcription) regulation. Regarding transcriptional repression, genes of the regulation type II represent the vast majority (repression of repressed) genes, while gene activation frequently affects genes of regulation type I (substantial activation). This explains why, on average, the subsets of genes that carry H3K4me3 in embryonic and lose it in adult cells are less transcribed than those of the subsets that gain H3K4me3 in adult cells and indicates the presence of a hysteresis of transcriptional regulation (Figure 4).
2.5. TF-Based Regulation of Transcription: Adaptive States
2.5.1. Properties of Adaptive States
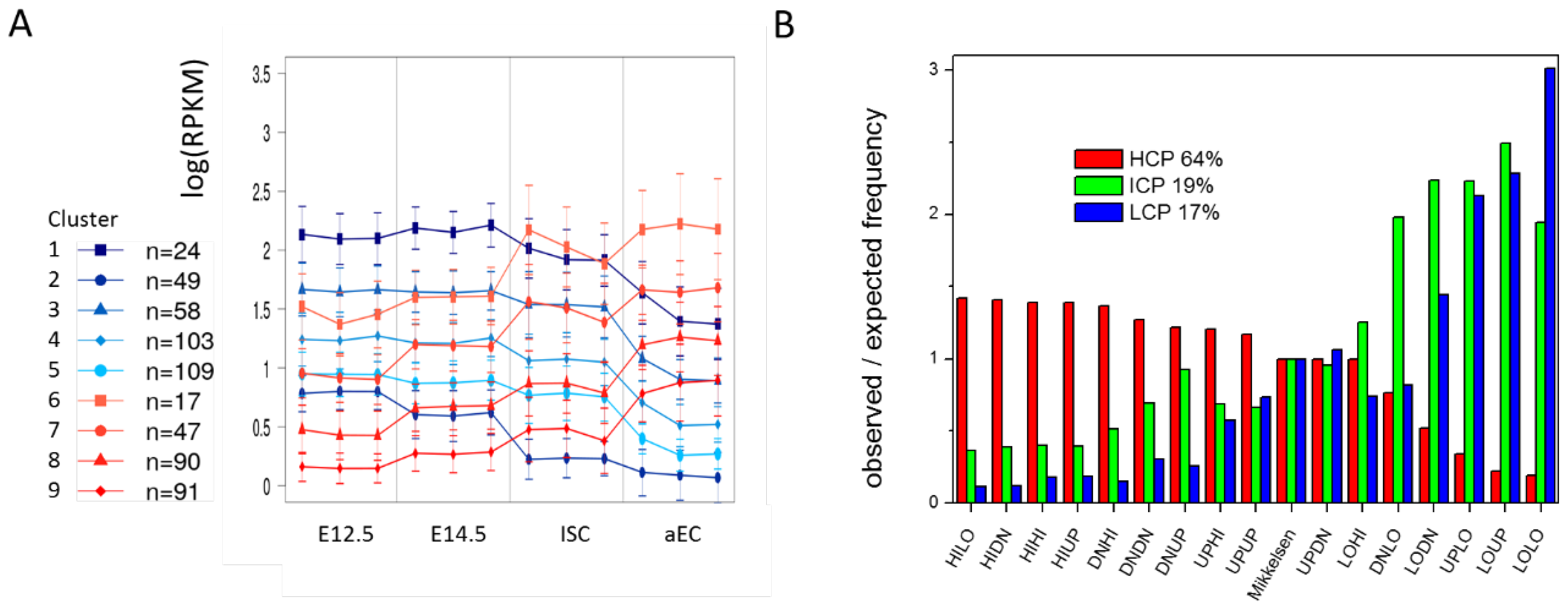
The next question was on how the transcription of genes with a stable epigenetic profile, i.e., HILO (H3K4me3), HIHI (bivalent), LOLO (unmodified), and LOHI (H3K27me3) is regulated. Actually, subsets of these genes change transcription substantiating the existence of type III regulation with respect to histone methylations. This refers to H3K4me3 modified genes (HILO, Figure 7A), but also to bivalent (HIHI) and unmodified genes (LOLO) (Appendix B: Figure A4), while H3K27me3 modified (LOHI) genes are stably repressed. Repressed HILO genes (cluster 1 and 3) show enrichment for GO terms, such as ‘regulation of gene expression’ (BP, FDR < 1 × 10−7) and ‘chromatin organization’ (BP FDR < 1 × 10−9), while activated HILO genes (cluster 6 and 7) show enrichment in terms ‘small molecule metabolic process’ (BP, FDR < 1 × 10−3) and ‘mitochondrion’ (CC, FDR < 1 × 10−7). As these genes do not qualitatively change their epigenetic profile, we call their regulatory states: adaptive states. The question remained, how this ‘adaptation’ that is representative for regulation type III, is regulated.
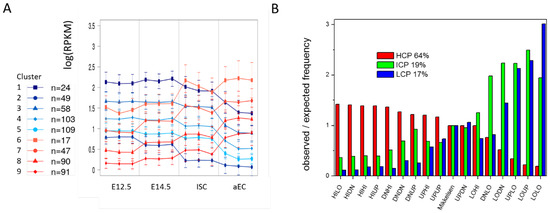
Figure 7.
Adaptive states. (A) Transcriptional profiles for genes of the HILO state (stable H3K4me3). Genes have been clustered according to their transcriptional profiles. Only genes with a minimal transcriptional standard deviation across their profile of 0.25 were considered. Cluster sizes n are indicated. Subsets are found that increase or decrease transcription similarly at different levels of transcription (errors: sd); and (B) Gene regulatory states differ in the CpG density of their promoters. Genes that are stably H3K4me3 modified are preferentially associated with high CpG density (HCP) promoters, while stably unmodified genes are preferentially associated with intermediate CpG density (ICP) and low CpG density (LCP) promoters. Average fractions of the different promoters are indicated. The classification was taken from Mikkelsen et al. [1].
We hypothesized that TF-networks are the major regulators of these genes, i.e., changes in TF-networks change the regulation factor FTF of these genes (Equation (4)), and thus their effective promoter activity Peff (Equation (3)). Analyzing the behavior of these genes in detail, we recognized an additional implication of epigenetics. During ISC specification and differentiation, LOLO genes that become transcriptionally up-regulated behave like UPLO genes; i.e., their promoters become de-methylated (Appendix B: Figure A4). This indicates that in both cases DNA de-methylation might be the driving force of transcriptional activation of the genes.
In our model, DNA methylation does not directly affect transcription, which is in accordance with experimental observations [26]. Nevertheless, the model can explain this kind of regulation, assuming that DNA methylation changes affect the binding affinity of TFs, which is in accordance with experiments [27]. According to Equation (4), TF binding at the gene promoter changes the free energy of RNA polymerase II recruitment. Consequently, if promoter DNA methylation affects TF binding, then it will change the gene’s transcription level as well.
Our results suggest that DNA de-methylation-driven adaptation of TF-networks controls part of the intestinal ISC specification and differentiation. In contrast to the interactions that were considered so far, this defines an indirect regulatory interaction between epigenetics and transcription.
2.5.2. Regulation Types Are Associated with Specific Promotor Types
Changes of DNA methylation depend on the CpG density at the promoter [1]. Depending on their CpG density, gene promoters have been classified in high (HCP), intermediate (ICP), and low (LCP) CpG density. In case of ISC specification and differentiation, we observed the well-known behavior that genes associated with stable H3K4me3 and low DNA methylation are enriched in HCG promoter genes, while genes that are associated with unmodified states and high DNA methylation are enriched in ICP and LCP promoter genes. Figure 7B provides detailed information of these differences. According to these results, LOLO (and also UPLO genes) are genes with ICP or LCP promoters. Thus, DNA methylation changes during ISC specification and differentiation control the transcription of ICP and LCP promoter genes. These genes are known to be associated with tissue specific genes [28].
3. Discussion
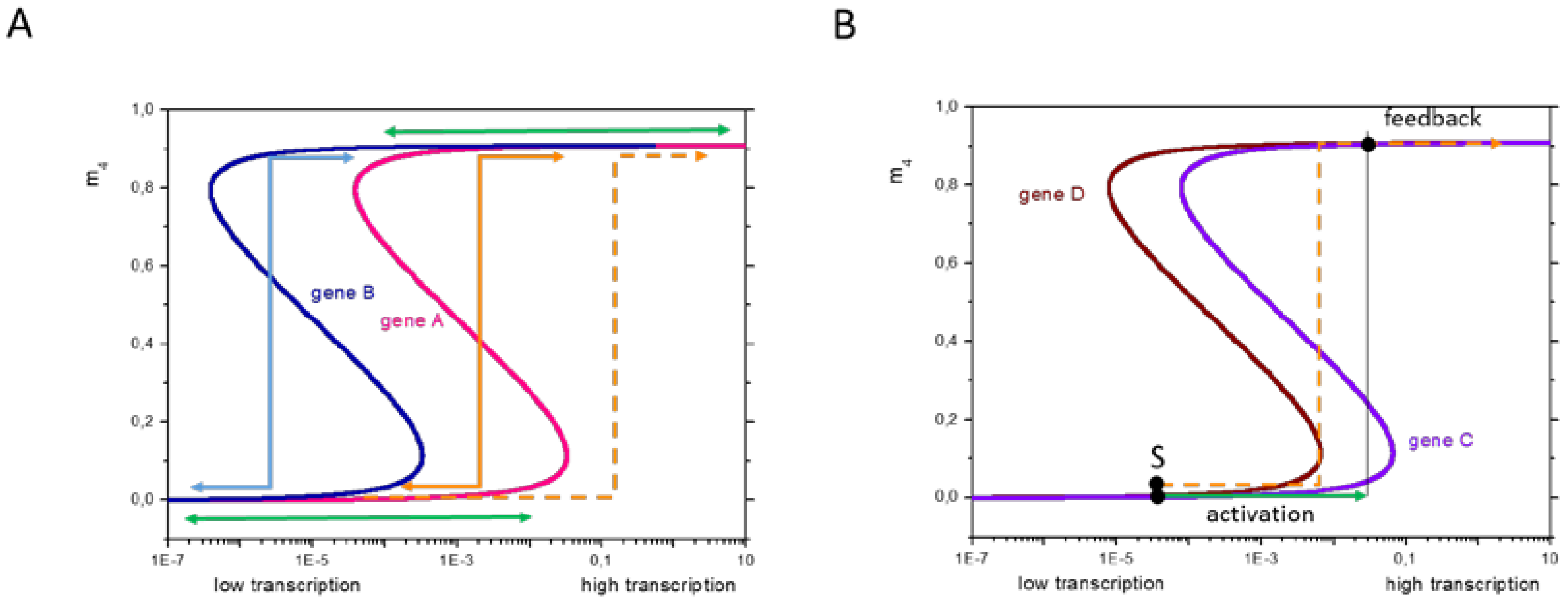
Many studies have described correlative relationships between occupancy of histone marks at distinct gene regions and transcription. Only recently have studies focused on elucidating clear functional consequences of specific histone marks during different stages of transcription [29]. Here, we analyzed whether and under which conditions such correlations are found and how (virtually) missing correlations can be mechanistically explained. Applying a combinatorial SOM, we identified three phenotypes of regulation, all being frequently observed during ISC specification and differentiation, and proposed a model to explain them. The main model conclusions are summarized in Figure 8.
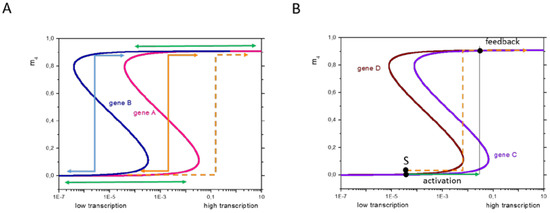
Figure 8.
Gene specific regulation of histone methylation. (A) Regulation types are a consequence of broad variance in gene specific regulation of histone methylation. Genes which change the modification state at high transcription (gene A) can show either type I or type III regulation; those that switch at low transcription (gene B) can show either type II or type III (color code as in Figure 4). Switches can occur due to transient (solid lines) or permanent (dashed lines) changes of transcription. Subsequent to the switch, the transcription changes due to feedback effects; and (B) Cooperation of epigenetics and transcription factor (TF)-networks. Changes of the epigenetic profile of a gene induces changes of TF-binding and thus changes in transcription (here: activation). These changes may (dashed orange arrow, type I) or not (green arrow, type III) be sufficient to induce further epigenetic changes associated with feedback. The initial transcription is indicated by S.
However, some basic questions remain open: (I) Our model defines a prototypic regulation for H3K4me3 (H3K27me3) that is characterized by a switch between low (high) and high (low) modification with increasing transcription at a defined threshold level of transcription. This regulation might or might not include a hysteresis. As we have detected all possible histone modification states at a broad range of transcription, this suggests that the thresholds have a broad distribution among the genes. A comprehensive validation of this hypothesis has not been provided so far; in particular, as experimental results are based on binary data, i.e., a quantitative interpretation of ChIP- and MBD-seq is still missing. (II) We propose an independent regulation of transcription by epigenetic and TF-networks. This suggestion is based on the observation that the fold-changes of transcription during a defined epigenetic switch do not depend on the initial transcription value. Here, we neglected that genes of the transition sets are regulated on top e.g., by TF-networks. However, in case repression and activation of genes are dominated by these TF-networks, one would expect that individual genes deviate from the regulation mode of their transition set; e.g. epigenetically up-regulated genes would effectively become down-regulated. We found such behavior, but only for a few genes (<10 genes per transition set).
Analyzing different states of ISC specification and differentiation, we have not addressed the question about the dynamics of transition states so far [9]. So, what induces these changes? Is it actually transcription? Alternative dynamics relate to changes in properties of the epigenetic machinery. This would easily explain the parallel changes for differentially transcribed genes. But then what does enable the paradoxical, opposite regulation of the transition states (e.g., DNUP versus UPDN)? We consider transient changes of transcription (activation and repression) due to external signals as the most likely explanation. The long term change in transcription might then result from a feedback of changing modification on transcription, either directly (Figure 8A, Equation (3)) or indirectly by modifying TF-binding.
From the mathematical point of view, there are alternative interpretations of the observed regulations. Decreasing the absolute value of the coupling parameter λK (Equation (1)) enforces regulation of type III, where the histone modification state no longer depends on the transcription. In this case, transcription would no longer affect the recruitment of HMTs. Similarly, scaling the absolute value of the modification levels in term X (mK* = mK I, with I < 1, Equation (3)) describes an induction of regulation of type II, where the transcription no longer depends on the histone modification level. In this case, histone modification would no longer affect the recruitment of RNA polymerase II. Although possible, enabling an increasing broad spectrum of regulatory states [14], both processes would impair the formation of a stable epigenome.
In agreement with previous findings [30], our study suggests that models of transcriptional regulation during ISC specification and differentiation have to account for the changes in TF-binding due to changing epigenetic profiles, including DNA methylation. While the prediction of TF-binding, based on DNA structure and epigenetic profiles, is state of the art bioinformatics [31,32,33] to the best of our knowledge, the underlying principles have not been integrated into a mechanistic model of the cooperation between epigenetics and TF-networks in gene regulation so far. We consider this step as crucial in order to come up with self-consistent regulatory models of tissue stem cell specification and differentiation. Considering the effects of distal regulatory elements, such as enhancers, is a further challenge [34]. Here, related data have been provided by Kazakevych et al. [21].
While the impact of epigenetics on transcription has been moved into the focus of epigenetic research, studies on the consequences of these interrelations and their variability for regulatory dynamics during tissue formation and transformation are still rare [35]. Our studies just provide a first insight into this field. Our results remain restricted to ISCs. However, we expect similar results for other somatic stem cell pools.
4. Materials and Methods
4.1. Data and Preprocessing for SOM
We downloaded raw sequencing data on murine ISC specification and differentiation [21] (GEO accession number GSE89684). Among the data, we selected those for transcription (three replicates), DNA methylation, H3K4me3 and H3K27me3 modification (two replicates each) in four cell types: embryonic intestinal epithelial progenitors at E12.5 and E14.5, ISCs, and aECs.
In case of RNA-seq data, raw counts were normalized to the total number of reads, resulting in one read per million (rpm) value per gene. These rpm values were normalized to the genes length (reads per kilobase million, RPKM) and transformed into log-scale. Histone and DNA methylation peaks, i.e., genomic regions, in which these modifications are enriched, have been identified by Kazakevych et al., [21] and were used as provided to identify modification peaks within the promoter region of genes (defined as transcription start side TSS +/− 1000 bases) and gene bodies (defined as region between the TSS and the last base of the gene). The Ensembl-gene reference list has been taken from UCSC Table Browser.
If a peak has a minimum overlap of 5% with a promoter region and/or a gene body, the respective gene-associated histones are considered to carry the modification in a binary present (1) or non-present manner (0). In the case of DNA methylation, we considered promoter DNA methylation only. For further analysis, normalized RNA-seq and binary modification data have been used.
4.2. Combinatorial SOM Method and Gene Annotation
For SOM training, we used data from two, randomly selected replicates of each cell type only to guarantee an equally weighted contribution from each data type. The data were transformed into a unique, ‘harmonized’ scale by normalizing them with respect to the mean absolute value averaged over the whole data entity. Afterwards, we centralized them separately for each data entity with respect to the mean value of each gene averaged over all samples. This centralization step emphasizes further analysis on differential values independent of the respective absolute transcription, DNA, and histone methylation levels. These data entered our in-house pipeline called multiSOMe [20].
The data sets, consisting of e.g., transcription and H3K4me3 data, are multiplied by weighting factors w and (1 − w), respectively, with w ϵ [0,1] and the weighted profiles are merged into combined profiles. The combined profiles are then clustered into metagene profiles, with each metagene serving as a representative of a cluster of combined profiles. We used a grid of 40 × 40 metagenes and applied default parametrization, as implemented in oposSOM [36].
After SOM training data are back transformed into their original scales for visualization and further downstream analysis. The transcription and combined modification states of the samples are visualized by color coding the metagene values. Because of the joint training of transcription and methylation data, each gene is located at the same position in both maps. Correlation between transcription and methylation was analyzed while calculating the signed square root covariance (SCOV) for each metagene and was visualized by color coding the metagenes. We calculated mean transcription, methylation, and covariance landscapes for each cell type by averaging the respective metagene values over the samples. The obtained mosaic images show a smooth texture with red and blue spot-like regions. These ‘spots’ represent clusters of co-transcribed or co-methylated genes.
4.3. Identification of Transition States and Clustering of Transcriptional Profiles
Transition states are defined by the properties of the H3K4me3 and H3K27me3 profiles of the genes. For each modification, we calculated the sum over all embryonic (S1) and all adult (S2) modification levels of all replicates, separately. We assign the state up (UP) to all genes with: S2 − S1 > 1, the state down (DN) to all genes with: S1 − S2 > 1. All other (abs(S2 − S1) <= 1) genes receive the state high (HI) if: S1 + S2 > 4 and otherwise state low (LO). The transition state than results from the combined state (X,Y) regarding H3K4me3 and H3K27me3 (with X,Y ϵ UP, DN, HI, LO).
Each gene of a particular transition state is characterized by its transcriptional profile, which is defined by the set of its transcription values in all measured samples. In order to cluster these profiles for subsets of genes, we used a modification of the k-median cluster algorithm (see Appendix B). The algorithm calculates a number of cluster centers (representative profiles), and assigns each gene to one of them using the least square method. In an iterative step, the cluster centers are recalculated depending on the transcriptional profiles of the genes that are assigned to the individual cluster and the genes are re-assigned to these new centers until the association between the genes and the centers is stable. As a result, one achieves a number of gene clusters and some outlier genes that do not show any similarity with the identified clusters.
Author Contributions
Conceptualization, J.G.; Methodology, T.T., L.H., G.A. and H.B.; Software, T.T. and L.H.; Formal Analysis, T.T., L.H., H.B. and J.G.; Resources, GEO accession number GSE89684 [21]; Data Curation, T.T. and L.H.; Writing-Original Draft Preparation: J.G.; Writing-Review & Editing, G.A., H.B. and J.G.; Visualization, T.T., L.H., H.B. and J.G.
Funding
The study was supported by the German Federal Ministry of Education and Science (BMBF) grants INDRA and CAPSYS (grant numbers: 031A312, 01ZX1304A).
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A. Additional SOM Analysis
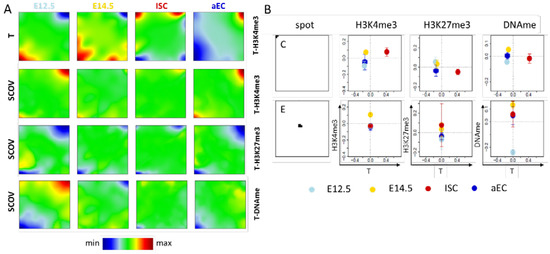
Figure A1.
Additional details of the T-H3K4me3 SOM. (A) Shown here are the transcription portrait of the T-H3K4me3 SOM and the SCOV portraits of the T-H3K4me3 SOM, the T-H3K27me3 SOM and the T-DNAme SOM. H3K4me3 and H3K27me3 show the expected positive and negative correlation with transcription, respectively. DNA methylation shows a negative correlation except for progenitors at E.12, where positive correlation is also observed; and (B) In ISCs spot C genes are transcribed higher than in all other cell types. However, the epigenetic profiles are very similar. Spot E genes show a hypo-methylation in progenitors of E12.5 and a hyper-methylation in those of E14.5, indicating rapid DNA methylation changes during these two days.
Figure A1.
Additional details of the T-H3K4me3 SOM. (A) Shown here are the transcription portrait of the T-H3K4me3 SOM and the SCOV portraits of the T-H3K4me3 SOM, the T-H3K27me3 SOM and the T-DNAme SOM. H3K4me3 and H3K27me3 show the expected positive and negative correlation with transcription, respectively. DNA methylation shows a negative correlation except for progenitors at E.12, where positive correlation is also observed; and (B) In ISCs spot C genes are transcribed higher than in all other cell types. However, the epigenetic profiles are very similar. Spot E genes show a hypo-methylation in progenitors of E12.5 and a hyper-methylation in those of E14.5, indicating rapid DNA methylation changes during these two days.
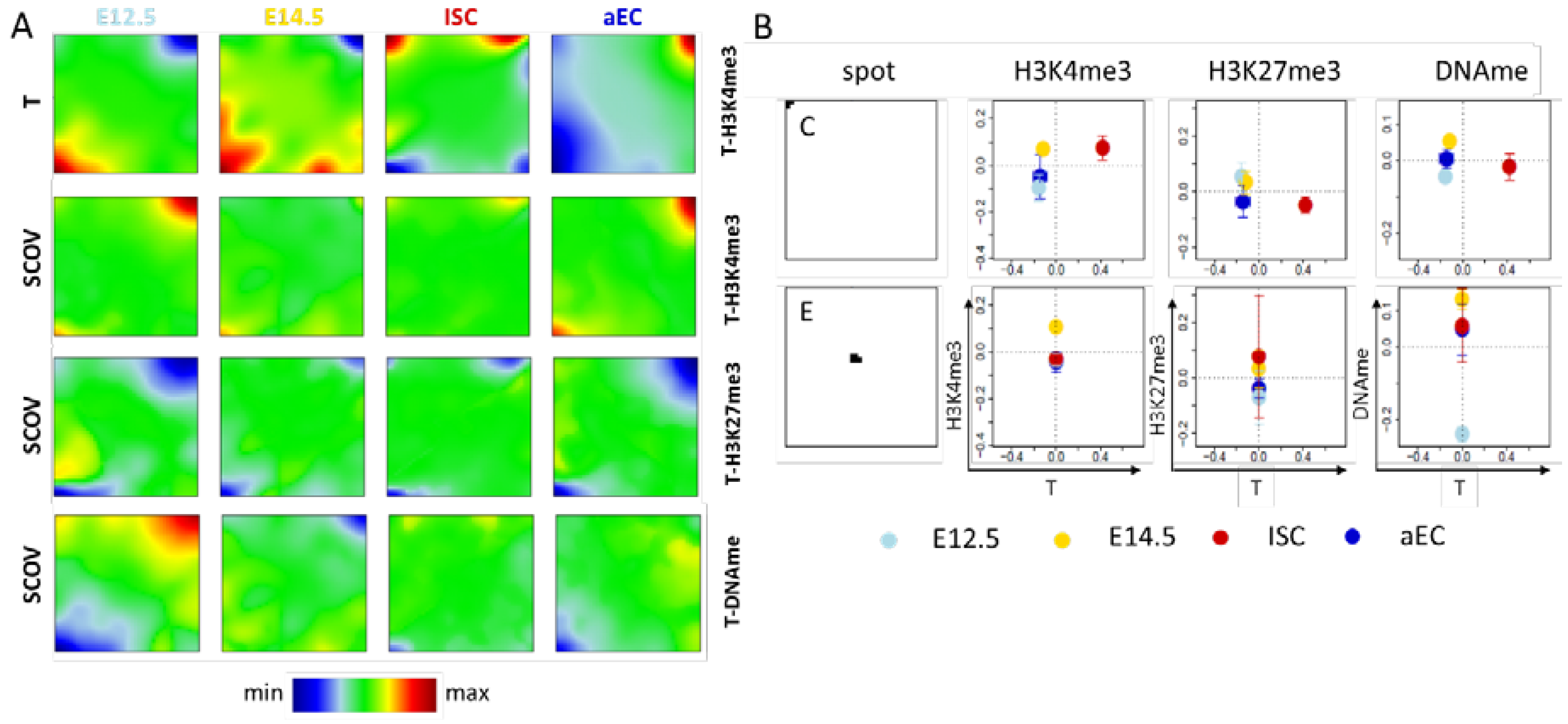
Figure A2.
Additional details of the T-H3K4me3 SOM. (A) Distribution of genes of selected GO sets throughout the SOM. Genes of the ‘antigen processing and presentation’ and ‘brush border’ set show a clear enrichment in spot B, while the large set of ‘multicellular organism development’ shows a major enrichment in the center and a secondary enrichment in spot A; and (B) Spot averages of the absolute values of selected spots for the four different cell types.
Figure A2.
Additional details of the T-H3K4me3 SOM. (A) Distribution of genes of selected GO sets throughout the SOM. Genes of the ‘antigen processing and presentation’ and ‘brush border’ set show a clear enrichment in spot B, while the large set of ‘multicellular organism development’ shows a major enrichment in the center and a secondary enrichment in spot A; and (B) Spot averages of the absolute values of selected spots for the four different cell types.
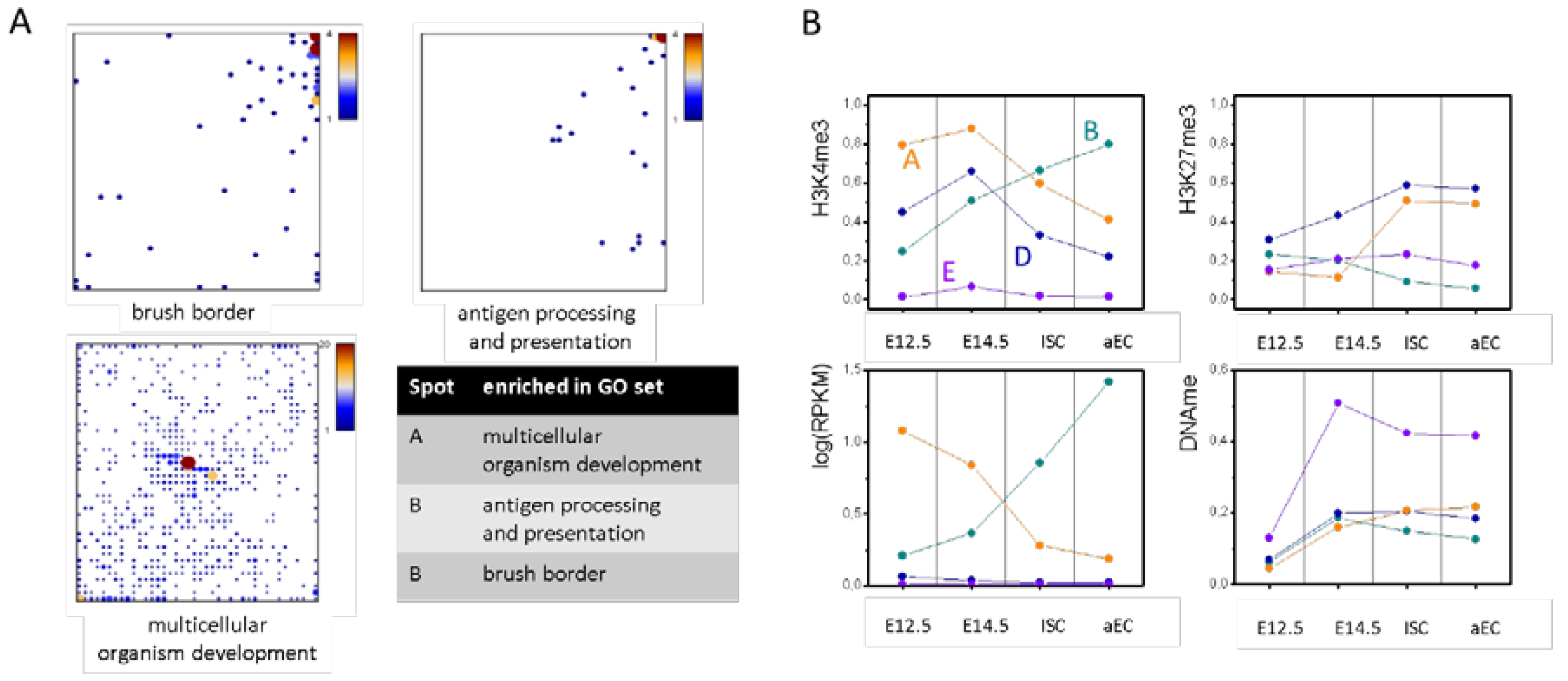
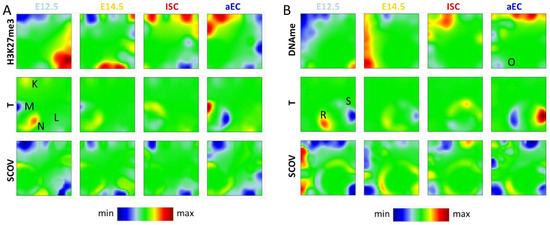
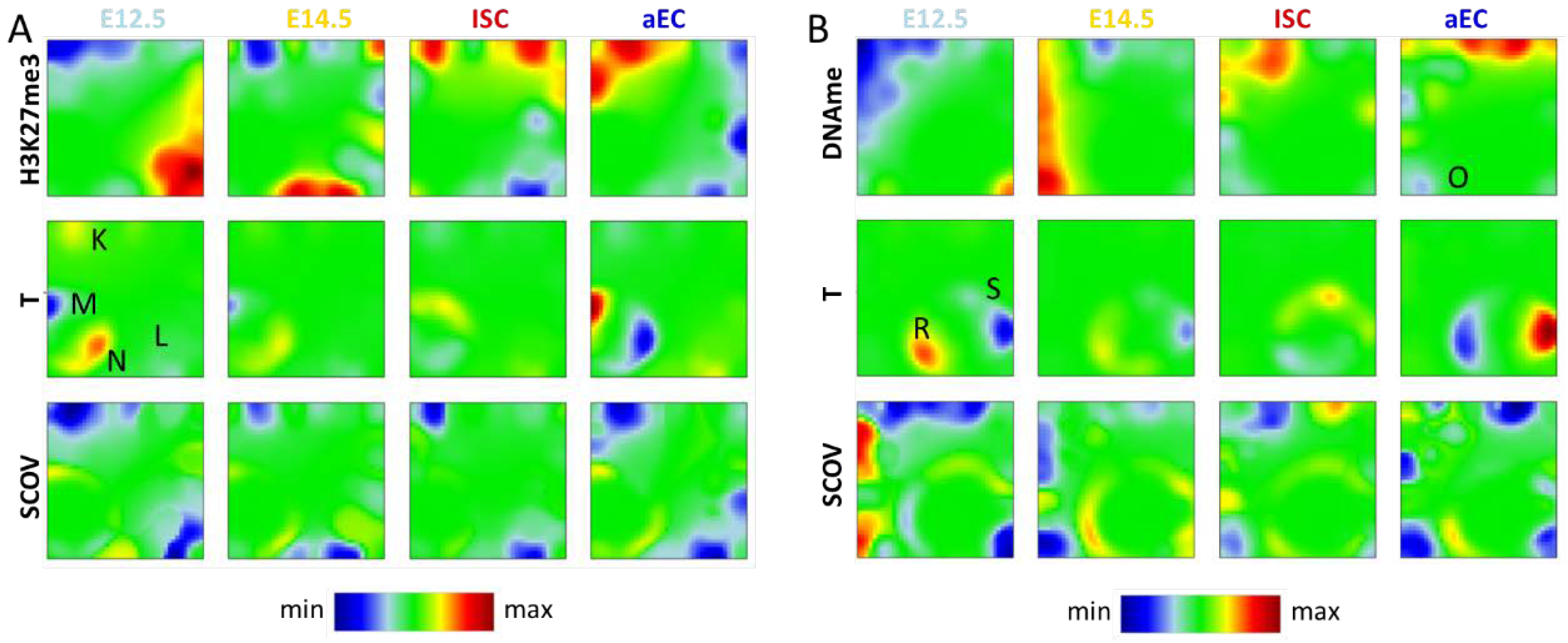
Figure A3.
Additional types of SOM analysis. (A) H3K27me3-T SOM. Spot K and L genes refer to regulation type I showing clear correlations between H3K27me3 and transcription. Spot M and N comprise genes of regulation type III changing transcription but not H3K27me3; and (B) DNAme-T SOM. Spot O genes undergo DNA de-methylation. On average, they appear as transcriptionally stable. Accordingly, they refer to regulation type II. Spot R and S genes change transcription without changing DNA methylation, representative for regulation type III.
Figure A3.
Additional types of SOM analysis. (A) H3K27me3-T SOM. Spot K and L genes refer to regulation type I showing clear correlations between H3K27me3 and transcription. Spot M and N comprise genes of regulation type III changing transcription but not H3K27me3; and (B) DNAme-T SOM. Spot O genes undergo DNA de-methylation. On average, they appear as transcriptionally stable. Accordingly, they refer to regulation type II. Spot R and S genes change transcription without changing DNA methylation, representative for regulation type III.
Appendix B. Algorithm to Cluster Transcriptional Profiles
The transcriptional profile of a gene is described as a combination of 12 dimensions (4 cell types × 3 replicates):
In order to cluster these profiles, we use the k-median clustering method which is a variant of the k-means clustering [37]. It uses the median of each dimension instead of the mean to center each cluster according to its associated genes. Genes are assigned to the nearest cluster center. Their distance to the respective cluster centers is calculated by the least square method. The cluster center represents the individual dimensions median of the genes associated to this cluster.
In an iterative process, each gene is assigned to its nearest cluster. Afterwards the cluster centers Ci are recalculated depending on the genes assigned to this cluster during this iteration. Subsequently, the genes are assigned again to their nearest cluster. In case, the cluster assignment is stable between two iterations, the algorithm terminates.
We modified the k-median algorithm, as follows: In each iteration, we assign a gene to a cluster only, if its distance to this (nearest) cluster is lower than dmin. Before the first iteration, we define a single center C1 as the median of all dimensions of any gene in this data set. Afterwards, we allow the composition of new clusters, while using a minimum distance restriction: In case, the gene’s distance to all available clusters is greater than d > 1.5 × dmin (dinit criterion) a new cluster center is introduced, using the gene’s dimensions as initial center Ci. In case, 1.5dmin > d > dmin the gene is considered as an outlier in this step. Therefore, we extend the gene/cluster assignment step as follows: (1) assign all genes to the available clusters, if possible, otherwise mark it as outlier (2) check for any outlier, if they fulfill the dinit criterion, in case they do, introduce a new center (3) assign the remaining outliers to the new centers, if possible (4) recalculate the cluster centers and go to (1). Our algorithm finally terminates, if the composition of Ci remains stable or after imax = 250 iterations. At the termination usually some outlier genes still remain as they do not fit to any existing cluster. Moreover, all cluster Ci that consist of less than 3 genes (nC) are dissolved and their genes are assigned to the remaining clusters, if possible. Otherwise, these genes are added to the group of outliers. The parameter settings are given in Table A1.

Table A1.
Cluster Parameters.
Table A1.
Cluster Parameters.
| Parameter | Description | Value |
|---|---|---|
| dmin | Minimum distance (least square method) to assign a gene to a cluster | 1.8 (default) 3 (HI-LO, LO-LO) |
| dinit | Minimum distance to all available clusters to create a new cluster | 1.5 × dmin |
| imax | Max number of iterations | 250 |
| nC | Minimum number of genes in a cluster | 3 |
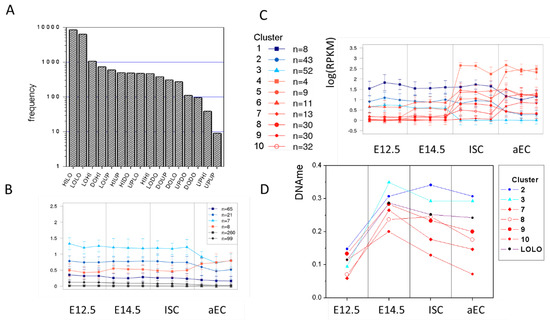
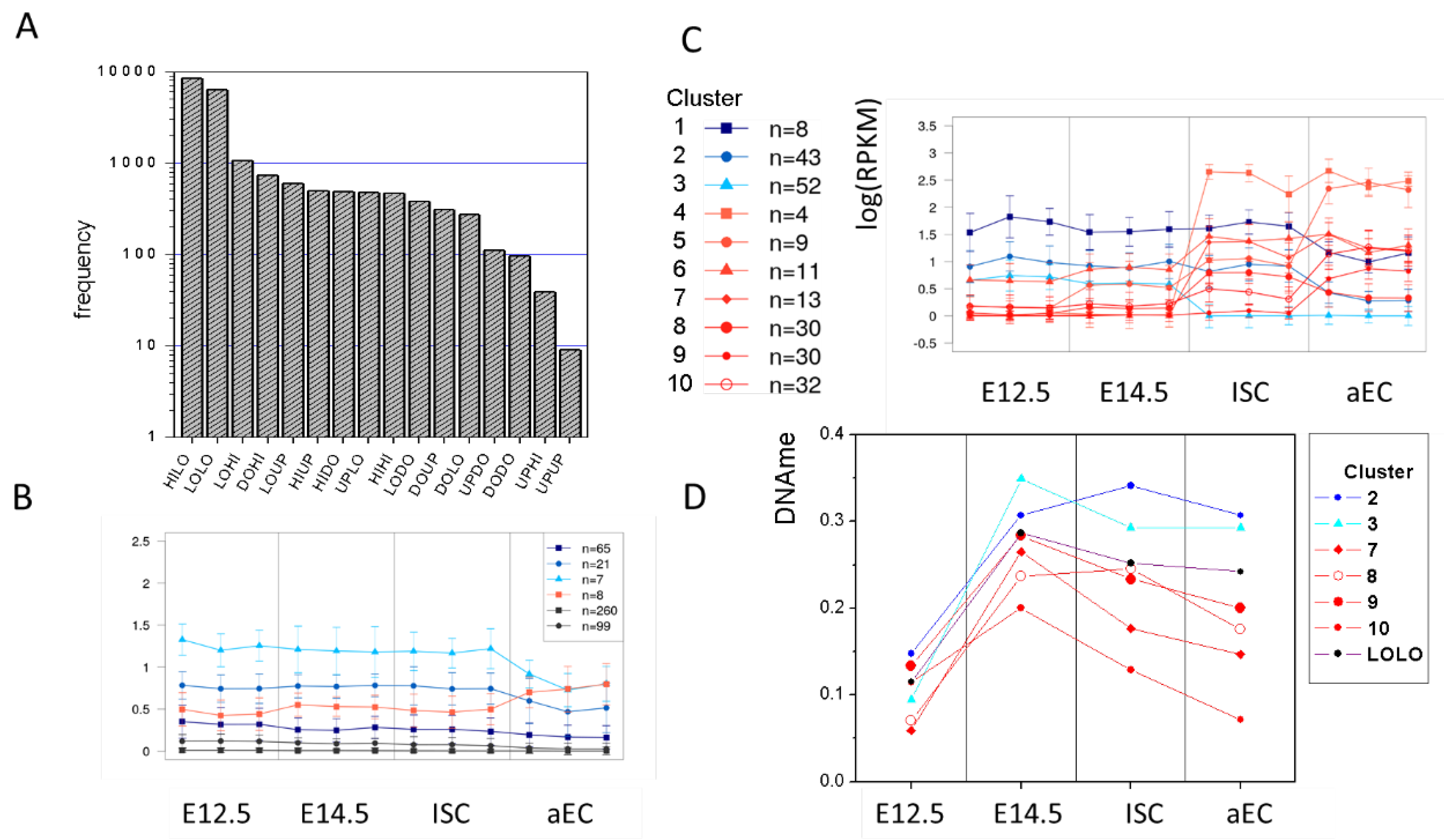
Figure A4.
Properties of genes with stable histone modification profiles. (A) Distribution of the transition states. Most frequent are HILO (stable K4) and LOLO (stable unmodified) states. HIHI states are as frequent as most of the transition states. (B) Clusters of HIHI genes with similar transcriptional profiles. The number of genes per cluster is indicated. Most HIHI genes are transcribed at low levels. However, a few genes are transcribed at higher levels and become down-regulated during ISC specification and differentiation. (C) Clusters of LOLO genes with similar transcriptional profiles. Many of them become transcriptionally activated in adult cells. (D) Repressed (cluster 2,3) and activated (cluster 7–10) LOLO genes differ in their DNA methylation profiles. The result for the entire LOLO set is integrated (black symbols).
Figure A4.
Properties of genes with stable histone modification profiles. (A) Distribution of the transition states. Most frequent are HILO (stable K4) and LOLO (stable unmodified) states. HIHI states are as frequent as most of the transition states. (B) Clusters of HIHI genes with similar transcriptional profiles. The number of genes per cluster is indicated. Most HIHI genes are transcribed at low levels. However, a few genes are transcribed at higher levels and become down-regulated during ISC specification and differentiation. (C) Clusters of LOLO genes with similar transcriptional profiles. Many of them become transcriptionally activated in adult cells. (D) Repressed (cluster 2,3) and activated (cluster 7–10) LOLO genes differ in their DNA methylation profiles. The result for the entire LOLO set is integrated (black symbols).
References
- Mikkelsen, T.S.; Ku, M.; Jaffe, D.B.; Issac, B.; Lieberman, E.; Giannoukos, G.; Alvarez, P.; Brockman, W.; Kim, T.K.; Koche, R.P.; et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 2007, 448, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Cui, K.; Zang, C.; Roh, T.Y.; Schones, D.E.; Childs, R.W.; Peng, W.; Zhao, K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell 2009, 4, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Atlasi, Y.; Stunnenberg, H.G. The interplay of epigenetic marks during stem cell differentiation and development. Nat. Rev. Genet. 2017, 18, 643–658. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, M.; Mulder, K.W.; Denissov, S.; Pijnappel, W.W.; van Schaik, F.M.; Varier, R.A.; Baltissen, M.P.; Stunnenberg, H.G.; Mann, M.; Timmers, H.T. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 2007, 131, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Pasini, D.; Malatesta, M.; Jung, H.R.; Walfridsson, J.; Willer, A.; Olsson, L.; Skotte, J.; Wutz, A.; Porse, B.; Jensen, O.N.; et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res. 2010, 38, 4958–4969. [Google Scholar] [CrossRef] [PubMed]
- Buratowski, S.; Kim, T. The role of cotranscriptional histone methylations. Cold Spring Harb. Symp. Quant. Biol. 2010, 75, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Jermann, P.; Hoerner, L.; Burger, L.; Schübeler, D. Short sequences can efficiently recruit histone H3 lysine 27 trimethylation in the absence of enhancer activity and DNA methylation. Proc. Natl. Acad. Sci. USA 2014, 111, E3415–E3421. [Google Scholar] [CrossRef] [PubMed]
- Mutskov, V.; Felsenfeld, G. Silencing of transgene transcription precedes methylation of promoter DNA and histone H3 lysine 9. EMBO J. 2004, 23, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Bintu, L.; Yong, J.; Antebi, Y.E.; McCue, K.; Kazuki, Y.; Uno, N.; Oshimura, M.; Elowitz, M.B. Dynamics of epigenetic regulation at the single-cell level. Science 2016, 351, 720–724. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.P.; Skene, P.J.; Selfridge, J.; Clouaire, T.; Guy, J.; Webb, S.; Kerr, A.R.; Deaton, A.; Andrews, R.; James, K.D.; et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 2010, 464, 1082–1086. [Google Scholar] [CrossRef] [PubMed]
- Ooi, S.K.; Qiu, C.; Bernstein, E.; Li, K.; Jia, D.; Yang, Z.; Erdjument-Bromage, H.; Tempst, P.; Lin, S.P.; Allis, C.D.; et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 2007, 448, 714–717. [Google Scholar] [CrossRef] [PubMed]
- Rush, M.; Appanah, R.; Lee, S.; Lam, L.L.; Goyal, P.; Lorincz, M.C. Targeting of EZH2 to a defined genomic site is sufficient for recruitment of Dnmt3a but not de novo DNA methylation. Epigenetics 2009, 4, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Binder, H.; Steiner, L.; Przybilla, J.; Rohlf, T.; Prohaska, S.; Galle, J. Transcriptional regulation by histone modifications, towards a theory of chromatin re-organization during stem cell differentiation. Phys. Biol. 2013, 10, 026006. [Google Scholar] [CrossRef] [PubMed]
- Thalheim, T.; Herberg, M.; Loeffler, M.; Galle, J. The Regulatory Capacity of Bivalent Genes—A Theoretical Approach. Int. J. Mol. Sci. 2017, 18, 1069. [Google Scholar] [CrossRef] [PubMed]
- Hamidouche, Z.; Rother, K.; Przybilla, J.; Krinner, A.; Clay, D.; Hopp, L.; Fabian, C.; Stolzing, A.; Binder, H.; Charbord, P.; et al. Bistable Epigenetic States Explain Age-Dependent Decline in Mesenchymal Stem Cell Heterogeneity. Stem Cells 2017, 35, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Przybilla, J.; Rohlf, T.; Loeffler, M.; Galle, J. Understanding epigenetic changes in aging stem cells—A computational model approach. Aging Cell 2014, 13, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Przybilla, J.; Hopp, L.; Lübbert, M.; Loeffler, M.; Galle, J. Targeting DNA hypermethylation, Computational modeling of DNA demethylation treatment of acute myeloid leukemia. Epigenetics 2017, 12, 886–896. [Google Scholar] [CrossRef] [PubMed]
- Thalheim, T.; Herberg, M.; Galle, J. Linking DNA Damage and Age-Related Promoter DNA Hyper-Methylation in the Intestine. Genes (Basel) 2018, 9, 17. [Google Scholar] [CrossRef] [PubMed]
- Steiner, L.; Hopp, L.; Wirth, H.; Galle, J.; Binder, H.; Prohaska, S.J.; Rohlf, T. A global genome segmentation method for exploration of epigenetic patterns. PLoS ONE 2012, 7, e46811. [Google Scholar] [CrossRef] [PubMed]
- Hopp, L.; Löffler-Wirth, H.; Galle, J.; Binder, H. Combined SOM-portrayal of transcription and DNA methylation landscapes disentangles modes of epigenetic regulation in glioblastoma. Epigenomics 2018, 10, 745–764. [Google Scholar] [CrossRef] [PubMed]
- Kazakevych, J.; Sayols, S.; Messner, B.; Krienke, C.; Soshnikova, N. Dynamic changes in chromatin states during specification and differentiation of adult intestinal stem cells. Nucleic Acids Res. 2017, 45, 5770–5784. [Google Scholar] [CrossRef] [PubMed]
- Araki, Y.; Wang, Z.; Zang, C.; Wood, W.H., 3rd; Schones, D.; Cui, K.; Roh, T.Y.; Lhotsky, B.; Wersto, R.P.; Peng, W.; et al. Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity 2009, 30, 912–925. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P.D. PANTHER version 11, expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017, 45, D183–D189. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Cooper, S.; Brockdorff, N. The interplay of histone modifications—Writers that read. EMBO Rep. 2015, 16, 1467–1481. [Google Scholar] [CrossRef] [PubMed]
- Binder, H.; Wirth, H.; Galle, J. Gene expression density profiles characterize modes of genomic regulation: Theory and experiment. J. Biotechnol. 2010, 149, 98–114. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Maurano, M.T.; Wang, H.; John, S.; Shafer, A.; Canfield, T.; Lee, K.; Stamatoyannopoulos, J.A. Role of DNA Methylation in Modulating Transcription Factor Occupancy. Cell Rep. 2015, 12, 1184–1195. [Google Scholar] [CrossRef] [PubMed]
- Elango, N.; Yi, S.V. DNA methylation and structural and functional bimodality of vertebrate promoters. Mol. Biol. Evol. 2008, 25, 1602–1608. [Google Scholar] [CrossRef] [PubMed]
- Gates, LA.; Foulds, C.E.; O’Malley, B.W. Histone Marks in the ‘Driver’s Seat’, Functional Roles in Steering the Transcription Cycle. Trends Biochem. Sci. 2017, 42, 977–989. [Google Scholar] [CrossRef] [PubMed]
- Tsankov, A.M.; Gu, H.; Akopian, V.; Ziller, M.J.; Donaghey, J.; Amit, I.; Gnirke, A.; Meissner, A. Transcription factor binding dynamics during human ES cell differentiation. Nature 2015, 518, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Tan, Y.; Huang, T.; Ding, G.; Tu, Z.; Liu, L.; Li, Y.; Dai, H.; Xie, L. TF-centered downstream gene set enrichment analysis, Inference of causal regulators by integrating TF-DNA interactions and protein post-translational modifications information. BMC Bioinform. 2010, 11 (Suppl. 11), S5. [Google Scholar] [CrossRef] [PubMed]
- Budden, D.M.; Hurley, D.G.; Cursons, J.; Markham, J.F.; Davis, M.J.; Crampin, E.J. Predicting expression, the complementary power of histone modification and transcription factor binding data. Epigenetics Chromatin 2014, 7, 36. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Bucher, P. Predicting transcription factor site occupancy using DNA sequence intrinsic and cell-type specific chromatin features. BMC Bioinform. 2016, 17 (Suppl. 1), S4. [Google Scholar] [CrossRef] [PubMed]
- Teschendorff, A.E.; Relton, C.L. Statistical and integrative system-level analysis of DNA methylation data. Nat. Rev. Genet. 2018, 19, 129–147. [Google Scholar] [CrossRef] [PubMed]
- Ecker, S.; Pancaldi, V.; Valencia, A.; Beck, S.; Paul, D.S. Epigenetic and Transcriptional Variability Shape Phenotypic Plasticity. Bioessays 2018, 40, 1700148. [Google Scholar] [CrossRef] [PubMed]
- Loeffler-Wirth, H.; Kalcher, M.; Binder, H. oposSOM: R-package for high-dimensional portraying of genome-wide expression landscapes on Bioconductor. Bioinformatics 2015, 31, 3225–3227. [Google Scholar] [CrossRef] [PubMed]
- MacQueen, J.B. Some Methods for classification and Analysis of Multivariate Observations. In Proceedings of the 5th Berkeley Symposium on Mathematical Statistics and Probability; University of California Press: Berkeley, CA, USA, 1967; pp. 281–297. [Google Scholar]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).