ARS2, a Cofactor of CBC, Promotes Meiotic Silencing by Unpaired DNA
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Methods and Genotypes
2.2. Assays for Growth, Sexual Development, and MSUD Suppression
2.3. Strain Construction and Confirmation
2.4. Bimolecular Fluorescence Complementation (BiFC)
2.5. Photography and Microscopy
3. Results
3.1. ARS2 Plays a Role in MSUD
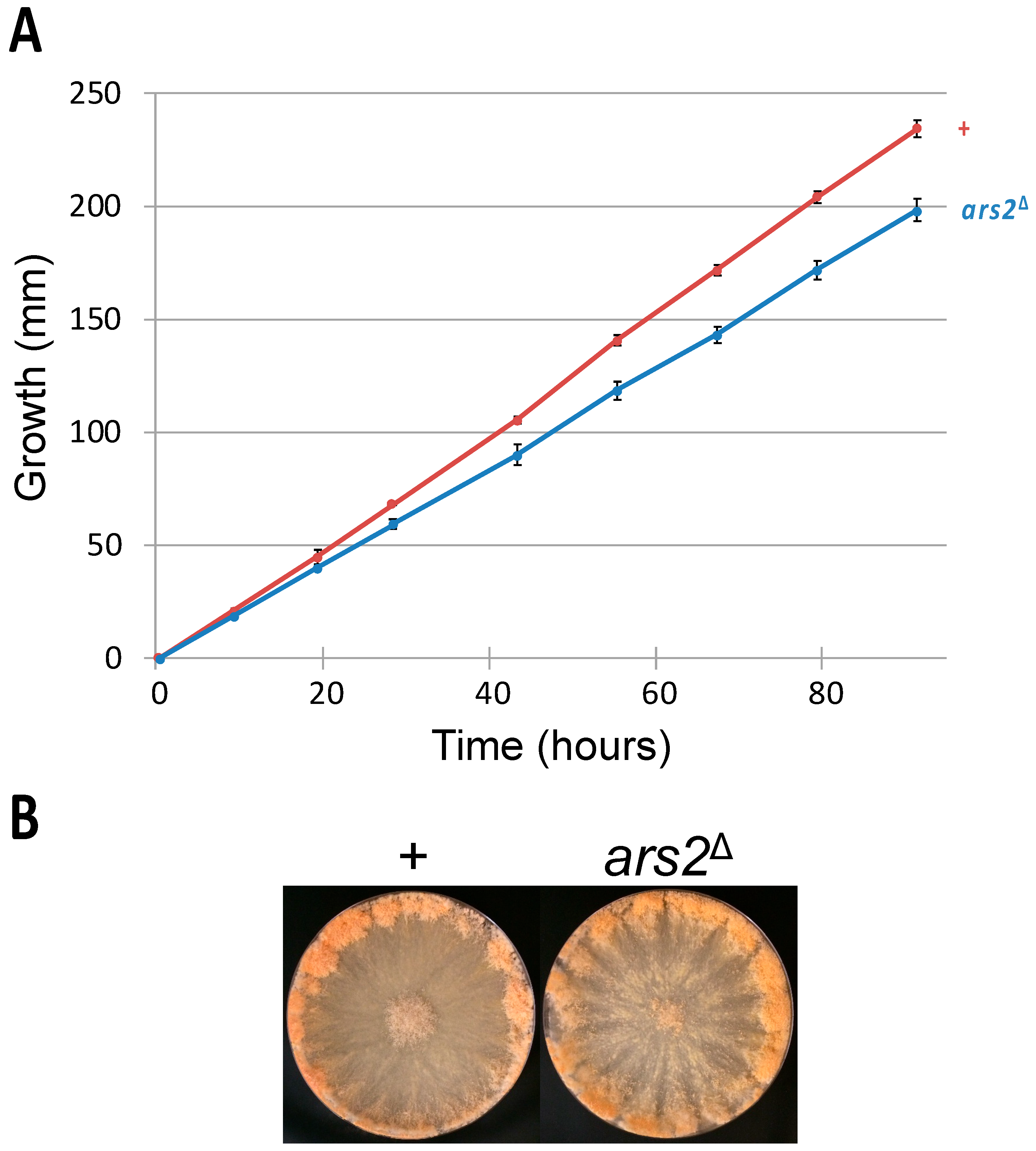
3.2. ARS2 Is Involved in Vegetative Growth
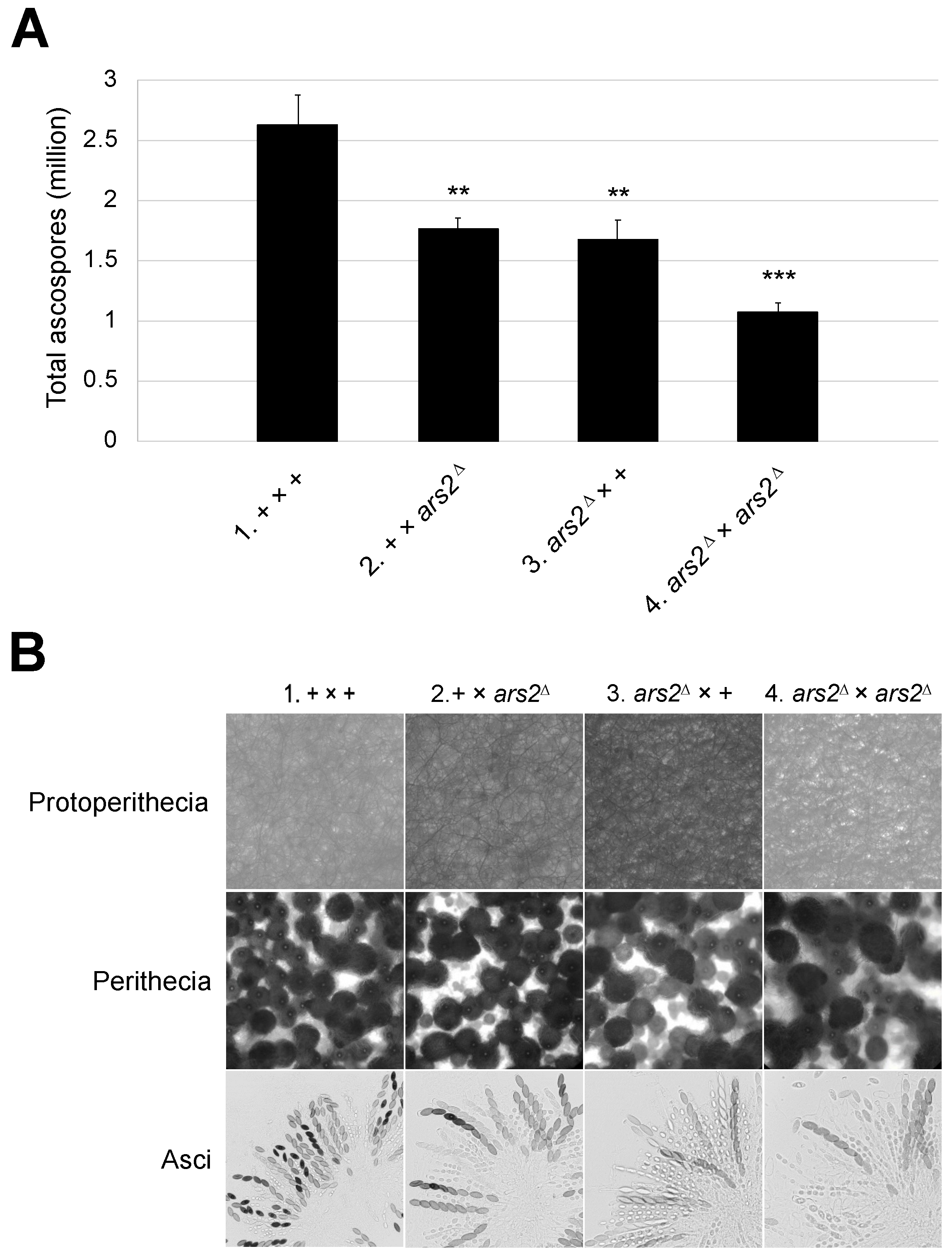
3.3. Mutation in ars2 Affects Sexual Development
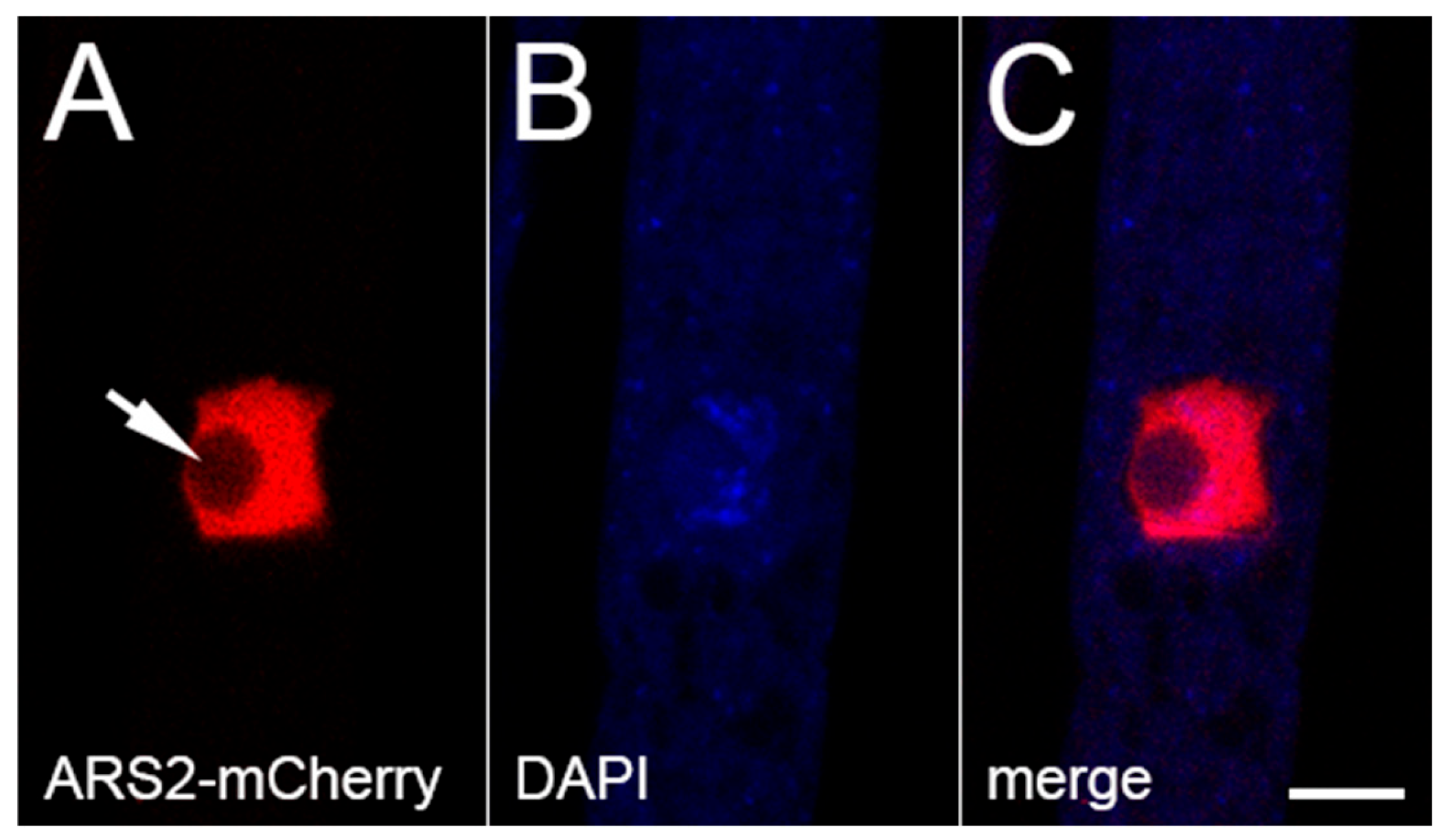
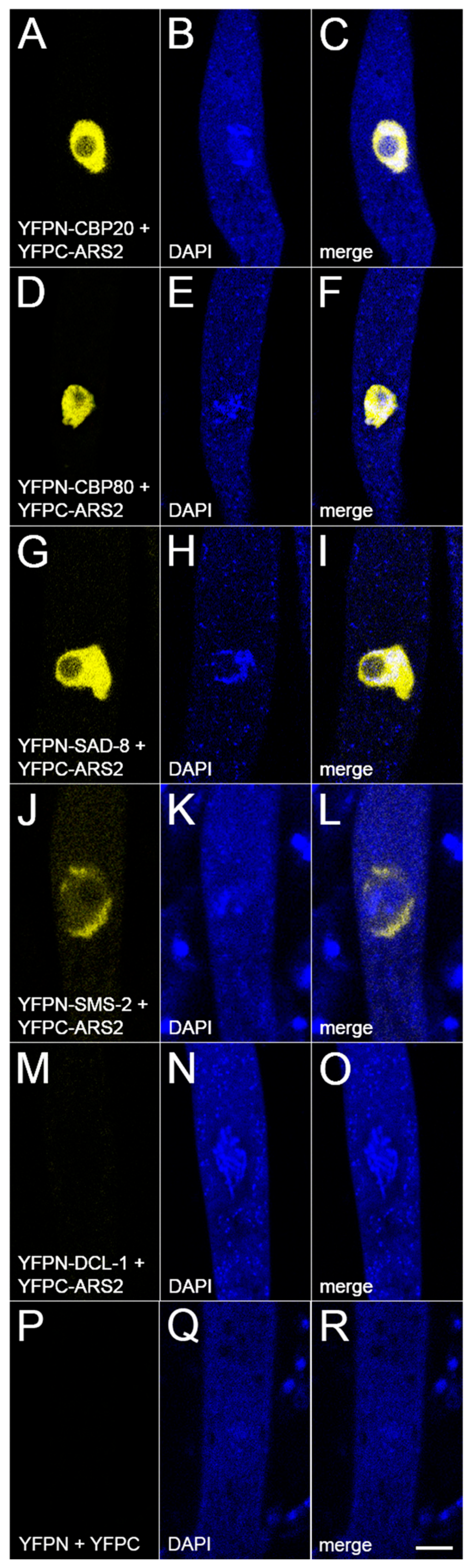
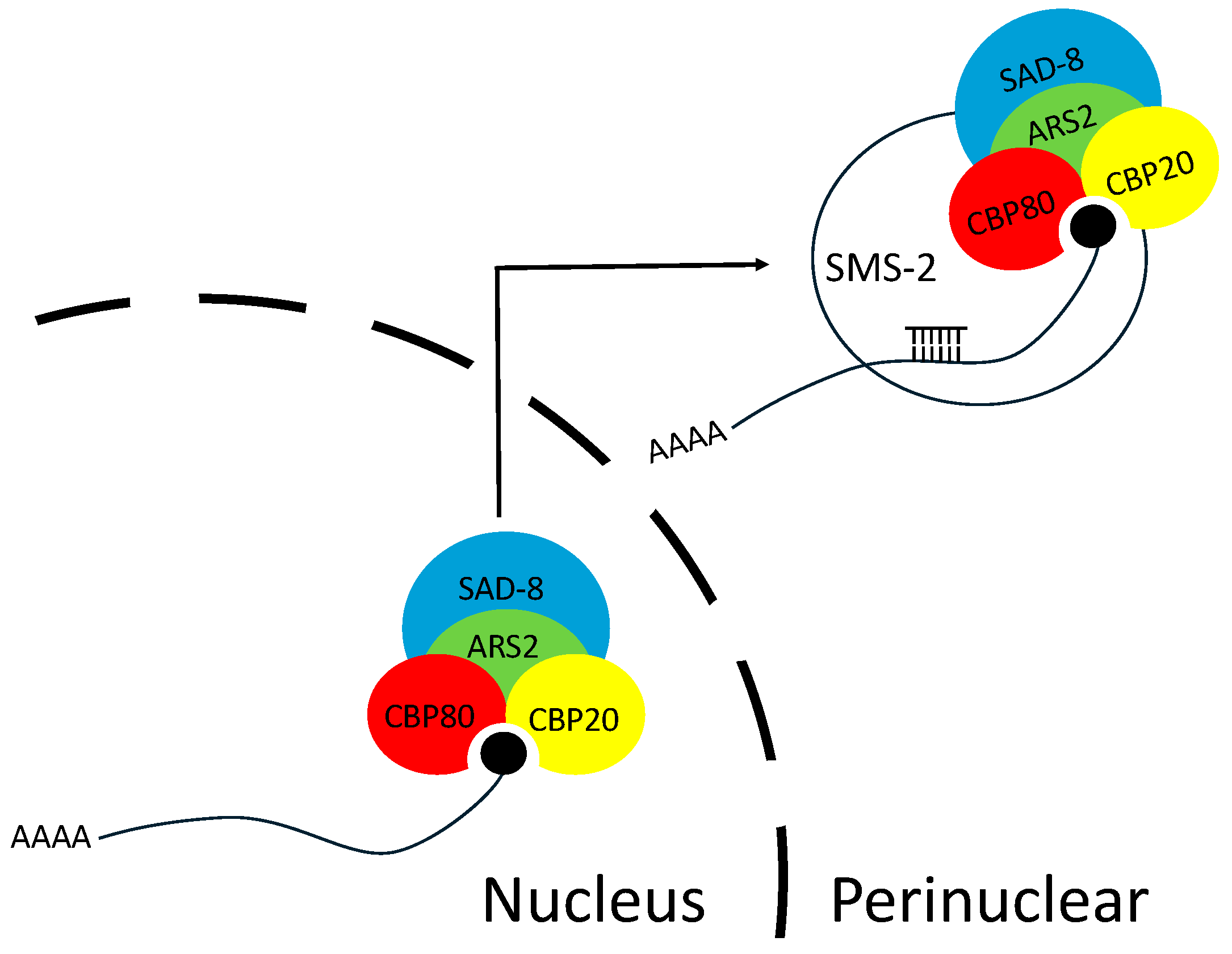
3.4. ARS2 Is Associated with CBC
3.5. ARS2 Interacts with NCBP3 and the SMS-2 Argonaute
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Springer, M.L. Genetic control of fungal differentiation: The three sporulation pathways of Neurospora crassa. Bioessays 1993, 15, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Gladyshev, E. Repeat-induced point mutation and other genome defense mechanisms in fungi. Microbiol. Spectr. 2017, 5, FUNK-0042-2017. [Google Scholar] [CrossRef] [PubMed]
- Aramayo, R.; Metzenberg, R.L. Meiotic transvection in fungi. Cell 1996, 86, 103–113. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shiu, P.K.T.; Raju, N.B.; Zickler, D.; Metzenberg, R.L. Meiotic silencing by unpaired DNA. Cell 2001, 107, 905–916. [Google Scholar] [CrossRef]
- Hammond, T.M. Sixteen years of meiotic silencing by unpaired DNA. Adv. Genet. 2017, 97, 1–42. [Google Scholar]
- Wang, Y.; Smith, K.M.; Taylor, J.W.; Freitag, M.; Stajich, J.E. Endogenous small RNA mediates meiotic silencing of a novel DNA transposon. G3 (Bethesda) 2015, 5, 1949–1960. [Google Scholar] [CrossRef]
- Samarajeewa, D.A.; Sauls, P.A.; Sharp, K.J.; Smith, Z.J.; Xiao, H.; Groskreutz, K.M.; Malone, T.L.; Boone, E.C.; Edwards, K.A.; Shiu, P.K.T.; et al. Efficient detection of unpaired DNA requires a member of the Rad54-like family of homologous recombination proteins. Genetics 2014, 198, 895–904. [Google Scholar] [CrossRef]
- Rhoades, N.; Nguyen, T.S.; Witz, G.; Cecere, G.; Hammond, T.; Mazur, A.K.; Gladyshev, E. Recombination-independent recognition of DNA homology for meiotic silencing in Neurospora crassa. Proc. Natl. Acad. Sci. USA 2021, 118, e2108664118. [Google Scholar] [CrossRef]
- Shiu, P.K.T.; Metzenberg, R.L. Meiotic silencing by unpaired DNA: Properties, regulation and suppression. Genetics 2002, 161, 1483–1495. [Google Scholar] [CrossRef]
- Hammond, T.M.; Xiao, H.; Boone, E.C.; Perdue, T.D.; Pukkila, P.J.; Shiu, P.K.T. SAD-3, a putative helicase required for meiotic silencing by unpaired DNA, interacts with other components of the silencing machinery. G3 (Bethesda) 2011, 1, 369–376. [Google Scholar] [CrossRef]
- Alexander, W.G.; Raju, N.B.; Xiao, H.; Hammond, T.M.; Perdue, T.D.; Metzenberg, R.L.; Pukkila, P.J.; Shiu, P.K.T. DCL-1 colocalizes with other components of the MSUD machinery and is required for silencing. Fungal Genet. Biol. 2008, 45, 719–727. [Google Scholar] [CrossRef]
- Hammond, T.M.; Spollen, W.G.; Decker, L.M.; Blake, S.M.; Springer, G.K.; Shiu, P.K.T. Identification of small RNAs associated with meiotic silencing by unpaired DNA. Genetics 2013, 194, 279–284. [Google Scholar] [CrossRef]
- Xiao, H.; Alexander, W.G.; Hammond, T.M.; Boone, E.C.; Perdue, T.D.; Pukkila, P.J.; Shiu, P.K.T. QIP, a protein that converts duplex siRNA into single strands, is required for meiotic silencing by unpaired DNA. Genetics 2010, 186, 119–126. [Google Scholar] [CrossRef]
- Lee, D.W.; Pratt, R.J.; McLaughlin, M.; Aramayo, R. An argonaute-like protein is required for meiotic silencing. Genetics 2003, 164, 821–828. [Google Scholar] [CrossRef]
- Shiu, P.K.T.; Zickler, D.; Raju, N.B.; Ruprich-Robert, G.; Metzenberg, R.L. SAD-2 is required for meiotic silencing by unpaired DNA and perinuclear localization of SAD-1 RNA-directed RNA polymerase. Proc. Natl. Acad. Sci. USA 2006, 103, 2243–2248. [Google Scholar] [CrossRef]
- Decker, L.M.; Boone, E.C.; Xiao, H.; Shanker, B.S.; Boone, S.F.; Kingston, S.L.; Lee, S.A.; Hammond, T.M.; Shiu, P.K.T. Complex formation of RNA silencing proteins in the perinuclear region of Neurospora crassa. Genetics 2015, 199, 1017–1021. [Google Scholar] [CrossRef]
- Sy, V.T.; Boone, E.C.; Xiao, H.; Vierling, M.M.; Schmitz, S.F.; Ung, Q.; Trawick, S.S.; Hammond, T.M.; Shiu, P.K.T. A DEAD-box RNA helicase mediates meiotic silencing by unpaired DNA. G3 (Bethesda) 2023, 13, jkad083. [Google Scholar] [CrossRef]
- Decker, L.M.; Xiao, H.; Boone, E.C.; Vierling, M.M.; Shanker, B.S.; Kingston, S.L.; Boone, S.F.; Haynes, J.B.; Shiu, P.K.T. The nuclear cap-binding complex mediates meiotic silencing by unpaired DNA. G3 (Bethesda) 2017, 7, 1149–1155. [Google Scholar] [CrossRef]
- Gonatopoulos-Pournatzis, T.; Cowling, V.H. Cap-binding complex (CBC). Biochem. J. 2014, 457, 231–242. [Google Scholar] [CrossRef]
- Kataoka, N. The Nuclear Cap-Binding Complex, a multitasking binding partner of RNA polymerase II transcripts. J. Biochem. 2024, 175, 9–15. [Google Scholar] [CrossRef]
- Boone, E.C.; Xiao, H.; Vierling, M.M.; Decker, L.M.; Sy, V.T.; Kennedy, R.F.; Bonham, M.A.; Schmitz, S.F.; John, A.M.; Hammond, T.M.; et al. An NCBP3-domain protein mediates meiotic silencing by unpaired DNA. G3 (Bethesda) 2020, 10, 1919–1927. [Google Scholar] [CrossRef]
- Rambout, X.; Maquat, L.E. NCBP3: A multifaceted adaptive regulator of gene expression. Trends Biochem. Sci. 2021, 46, 87–96. [Google Scholar] [CrossRef]
- Dubiez, E.; Pellegrini, E.; Finderup Brask, M.; Garland, W.; Foucher, A.E.; Huard, K.; Heick Jensen, T.; Cusack, S.; Kadlec, J. Structural basis for competitive binding of productive and degradative co-transcriptional effectors to the nuclear cap-binding complex. Cell Rep. 2024, 43, 113639. [Google Scholar] [CrossRef]
- Dunlap, J.C.; Borkovich, K.A.; Henn, M.R.; Turner, G.E.; Sachs, M.S.; Glass, N.L.; McCluskey, K.; Plamann, M.; Galagan, J.E.; Birren, B.W.; et al. Enabling a community to dissect an organism: Overview of the Neurospora functional genomics project. Adv. Genet. 2007, 57, 49–96. [Google Scholar] [PubMed]
- McCluskey, K.; Wiest, A.; Plamann, M. The Fungal Genetics Stock Center: A repository for 50 years of fungal genetics research. J. Biosci. 2010, 35, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Basenko, E.Y.; Shanmugasundram, A.; Böhme, U.; Starns, D.; Wilkinson, P.A.; Davison, H.R.; Crouch, K.; Maslen, G.; Harb, O.S.; Amos, B.; et al. What is new in FungiDB: A web-based bioinformatics platform for omics-scale data analysis for fungal and oomycete species. Genetics 2024, 227, iyae035. [Google Scholar] [CrossRef] [PubMed]
- Vogel, H.J. A convenient growth medium for Neurospora (Medium N). Microbial. Genet. Bull. 1956, 13, 42–43. [Google Scholar]
- Westergaard, M.; Mitchell, H.K.; Neurospora, V. A synthetic medium favoring sexual reproduction. Am. J. Bot. 1947, 34, 573–577. [Google Scholar] [CrossRef]
- Turner, G.E. Phenotypic analysis of Neurospora crassa gene deletion strains. Methods Mol. Biol. 2011, 722, 191–198. [Google Scholar]
- Xiao, H.; Hammond, T.M.; Shiu, P.K.T. Suppressors of meiotic silencing by unpaired DNA. Noncoding RNA 2019, 5, 14. [Google Scholar] [CrossRef]
- Margolin, B.S.; Freitag, M.; Selker, E.U. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet. Newsl. 1997, 44, 34–36. [Google Scholar] [CrossRef]
- Henderson, S.T.; Eariss, G.A.; Catcheside, D.E.A. Reliable PCR amplification from Neurospora crassa genomic DNA obtained from conidia. Fungal Genet. Newsl. 2005, 52, 24. [Google Scholar] [CrossRef]
- Hu, C.D.; Chinenov, Y.; Kerppola, T.K. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 2002, 9, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Bardiya, N.; Alexander, W.G.; Perdue, T.D.; Barry, E.G.; Metzenberg, R.L.; Pukkila, P.J.; Shiu, P.K.T. Characterization of interactions between and among components of the meiotic silencing by unpaired DNA machinery in Neurospora crassa using bimolecular fluorescence complementation. Genetics 2008, 178, 593–596. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hammond, T.M.; Xiao, H.; Rehard, D.G.; Boone, E.C.; Perdue, T.D.; Pukkila, P.J.; Shiu, P.K.T. Fluorescent and bimolecular-fluorescent protein tagging of genes at their native loci in Neurospora crassa using specialized double-joint PCR plasmids. Fungal Genet. Biol. 2011, 48, 866–873. [Google Scholar] [CrossRef] [PubMed]
- Lykke-Andersen, S.; Rouvière, J.O.; Jensen, T.H. ARS2/SRRT: At the nexus of RNA polymerase II transcription, transcript maturation and quality control. Biochem. Soc. Trans. 2021, 49, 1325–1336. [Google Scholar] [CrossRef]
- Hammond, T.M.; Xiao, H.; Boone, E.C.; Decker, L.M.; Lee, S.A.; Perdue, T.D.; Pukkila, P.J.; Shiu, P.K.T. Novel proteins required for meiotic silencing by unpaired DNA and siRNA generation in Neurospora crassa. Genetics 2013, 194, 91–100. [Google Scholar] [CrossRef]
- Kim, D.U.; Hayles, J.; Kim, D.; Wood, V.; Park, H.O.; Won, M.; Yoo, H.S.; Duhig, T.; Nam, M.; Palmer, G.; et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 2010, 28, 617–623. [Google Scholar] [CrossRef]
- Lobbes, D.; Rallapalli, G.; Schmidt, D.D.; Martin, C.; Clarke, J. SERRATE: A new player on the plant microRNA scene. EMBO Rep. 2006, 7, 1052–1058. [Google Scholar] [CrossRef]
- Oh, S.W.; Kingsley, T.; Shin, H.H.; Zheng, Z.; Chen, H.W.; Chen, X.; Wang, H.; Ruan, P.; Moody, M.; Hou, S.X. A P-element insertion screen identified mutations in 455 novel essential genes in Drosophila. Genetics 2003, 163, 195–201. [Google Scholar] [CrossRef]
- Golling, G.; Amsterdam, A.; Sun, Z.; Antonelli, M.; Maldonado, E.; Chen, W.; Burgess, S.; Haldi, M.; Artzt, K.; Farrington, S.; et al. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat. Genet. 2002, 31, 135–140. [Google Scholar] [CrossRef]
- Wilson, M.D.; Wang, D.; Wagner, R.; Breyssens, H.; Gertsenstein, M.; Lobe, C.; Lu, X.; Nagy, A.; Burke, R.D.; Koop, B.F.; et al. ARS2 is a conserved eukaryotic gene essential for early mammalian development. Mol. Cell. Biol. 2008, 28, 1503–1514. [Google Scholar] [CrossRef]
- Prigge, M.J.; Wagner, D.R. The Arabidopsis SERRATE gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 2001, 13, 1263–1279. [Google Scholar] [CrossRef]
- Samarajeewa, D.A.; Manitchotpisit, P.; Henderson, M.; Xiao, H.; Rehard, D.G.; Edwards, K.A.; Shiu, P.K.T.; Hammond, T.M. An RNA recognition motif-containing protein functions in meiotic silencing by unpaired DNA. G3 (Bethesda) 2017, 7, 2871–2882. [Google Scholar] [CrossRef]
- Bui, D.C.; Kim, J.E.; Shin, J.; Lim, J.Y.; Choi, G.J.; Lee, Y.W.; Seo, J.A.; Son, H. ARS2 plays diverse roles in DNA damage response, fungal development, and pathogenesis in the plant pathogenic fungus Fusarium graminearum. Front. Microbiol. 2019, 10, 2326. [Google Scholar] [CrossRef]
- Sabin, L.R.; Zhou, R.; Gruber, J.J.; Lukinova, N.; Bambina, S.; Berman, A.; Lau, C.K.; Thompson, C.B.; Cherry, S. Ars2 regulates both miRNA- and siRNA-dependent silencing and suppresses RNA virus infection in Drosophila. Cell 2009, 138, 340–351. [Google Scholar] [CrossRef]

| Strain | Genotype |
|---|---|
| F2-01 | fl A (FGSC 4317) |
| F2-30 | rid rΔ::hph; fl A |
| F5-36 | fl; sad-5Δ::hph a |
| F6-26 | ars-2Δ::hph rΔ::hph; fl a |
| F9-09 | ars2Δ;fl A |
| P3-08 | Oak Ridge wild type a (FGSC 2490) |
| P13-65 | rid his-3+::yfpn; mus-52∆::bar A |
| P14-04 | rid his-3+::yfpc; mus-51Δ::bar a |
| P17-70 | rΔ::hph; sad-5Δ::hph A |
| P20-49 | ars2∆::hph A (FGSC 15712) |
| P20-50 | ars2∆::hph a (FGSC 16001) |
| P27-40 | rid ars2-mCherry::hph; mus-51Δ::bar A |
| P27-41 | rid ars2-mCherry::hph; mus-51Δ::bar a |
| P27-42 | rid his-3 yfpc-ars2::hph; yfpn-cbp20::nat1 mus-52Δ::bar a |
| P27-43 | rid his-3 yfpc-ars2::hph; yfpn-cbp20::nat1; mus-51Δ::bar A |
| P27-48 | rid yfpc-ars2::hph; yfpn-sms-2::hph a |
| P27-49 | rid yfpc-ars2::hph; yfpn-sms-2::hph A |
| P30-23 | rid yfpc-ars2::hph; yfpn-cbp80::hph a |
| P30-24 | rid yfpc-ars2::hph; yfpn-cbp80::hph A |
| P30-25 | rid yfpc-ars2::hph; yfpn-sad-8::nat1 A |
| P30-26 | rid yfpc-ars2::hph; yfpn-sad-8::nat1 a |
| P31-04 | rid yfpc-ars2::hph; mus-52Δ::bar; yfpn-dcl-1::nat1 a |
| P31-05 | rid his-3 yfpc-ars2::hph; mus-52Δ::bar; yfpn-dcl-1::nat1 A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Vierling, M.M.; Sy, V.T.; Decker, L.M.; Xiao, H.; Hemaya, J.N.; Shiu, P.K.T. ARS2, a Cofactor of CBC, Promotes Meiotic Silencing by Unpaired DNA. Epigenomes 2026, 10, 6. https://doi.org/10.3390/epigenomes10010006
Vierling MM, Sy VT, Decker LM, Xiao H, Hemaya JN, Shiu PKT. ARS2, a Cofactor of CBC, Promotes Meiotic Silencing by Unpaired DNA. Epigenomes. 2026; 10(1):6. https://doi.org/10.3390/epigenomes10010006
Chicago/Turabian StyleVierling, Michael M., Victor T. Sy, Logan M. Decker, Hua Xiao, Justine N. Hemaya, and Patrick K. T. Shiu. 2026. "ARS2, a Cofactor of CBC, Promotes Meiotic Silencing by Unpaired DNA" Epigenomes 10, no. 1: 6. https://doi.org/10.3390/epigenomes10010006
APA StyleVierling, M. M., Sy, V. T., Decker, L. M., Xiao, H., Hemaya, J. N., & Shiu, P. K. T. (2026). ARS2, a Cofactor of CBC, Promotes Meiotic Silencing by Unpaired DNA. Epigenomes, 10(1), 6. https://doi.org/10.3390/epigenomes10010006






