Regulation and Functional Significance of 5-Hydroxymethylcytosine in Cancer
Abstract
1. Introduction
2. Interplay between 5-Hydroxymethylcytosine and Cancer
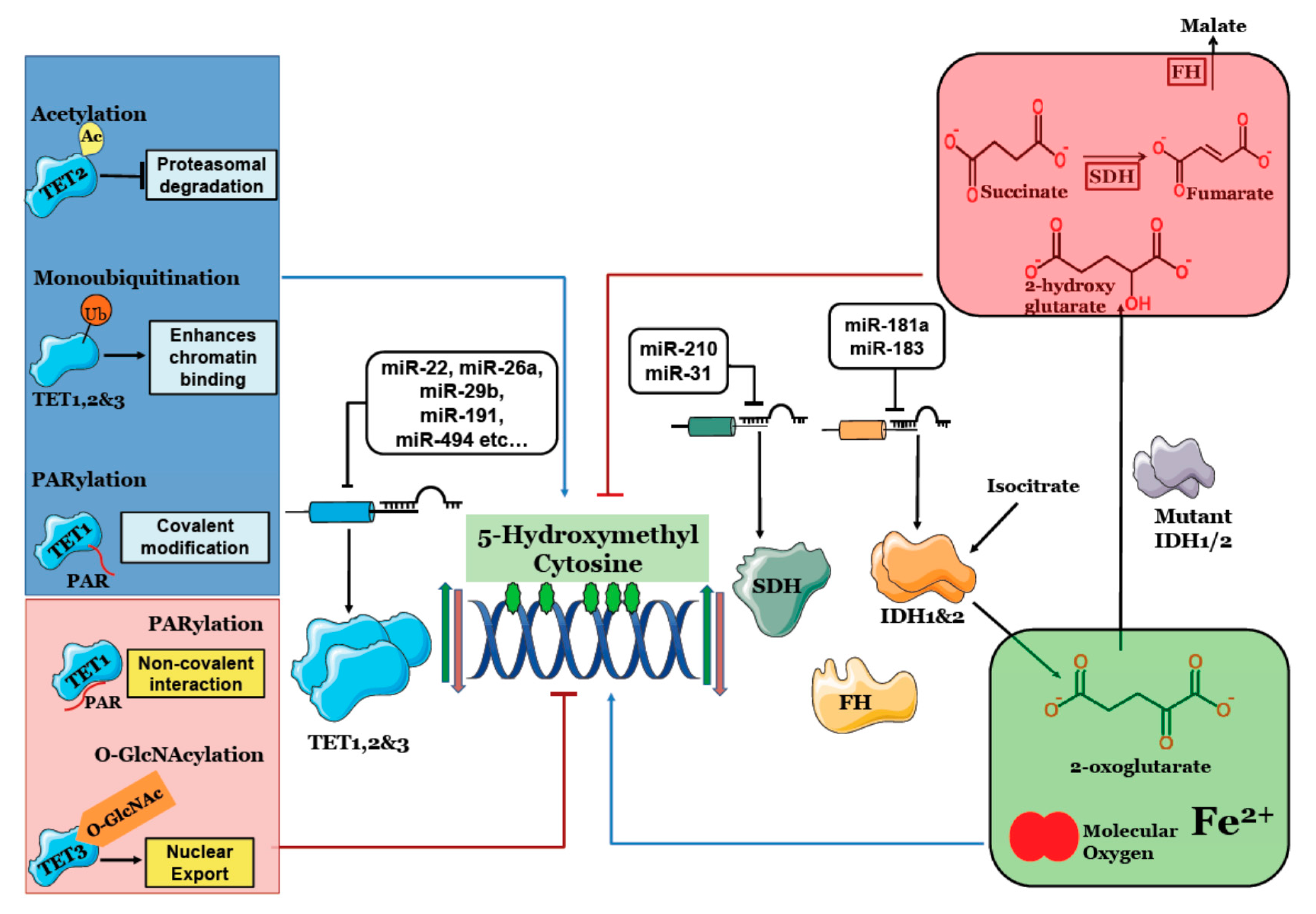
3. Effect of Cellular Factors on 5-Hydroxymethylcytosine
4. Hypoxia-Induced 5-Hydroxymethylcytosine Changes Enable Cancer Progression
5. Heavy Metals Deregulate 5-Hydroxymethylcytosine
6. Oxidative Stress on 5-Hydroxymethylcytosine
7. Carcinogens Deregulate 5-Hydroxymethylcytosine
8. Nutrients Enable 5-Hydroxymethylcytosine Regulation
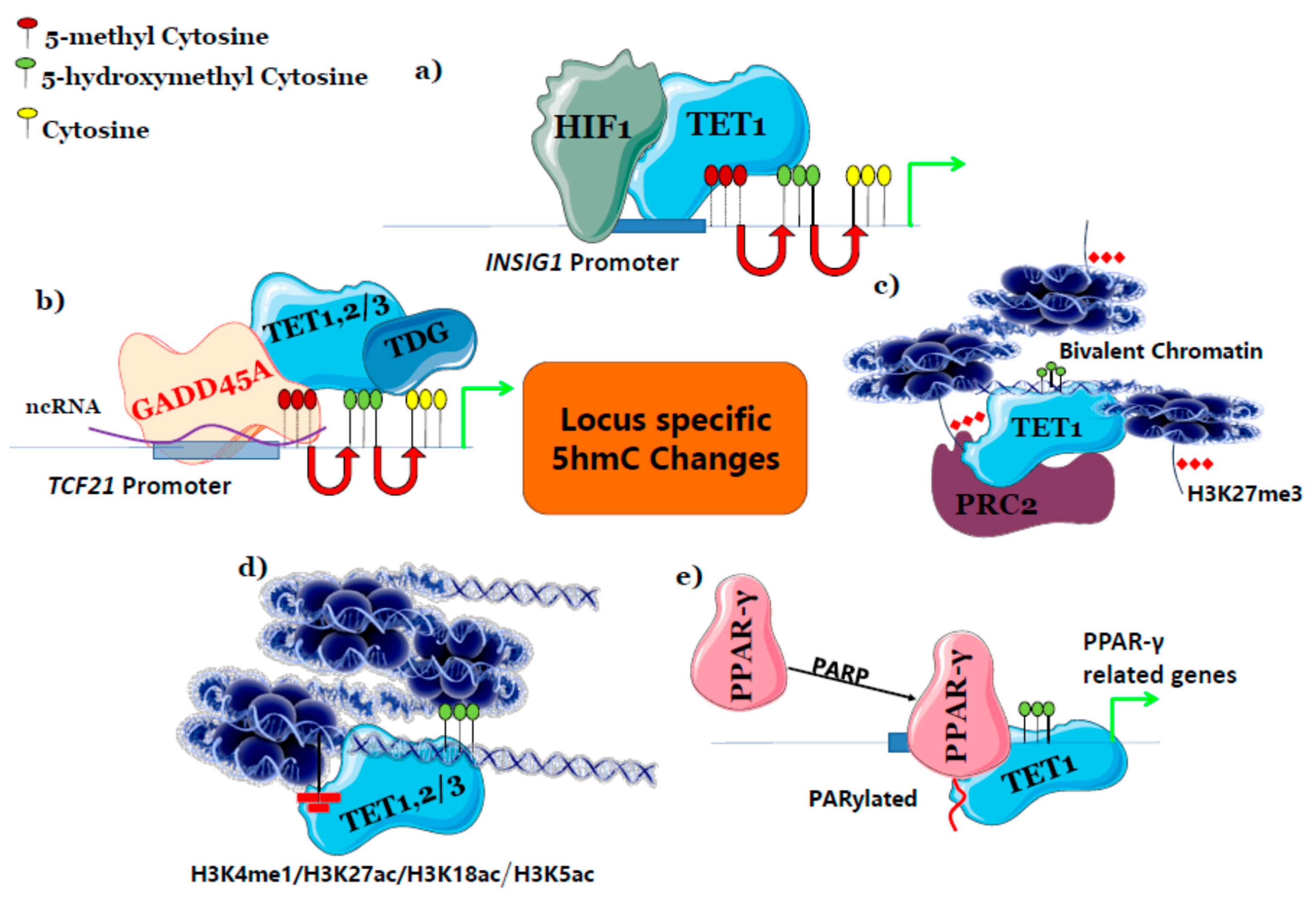
9. 5-Hydroxymethylcytosine Changes are Locus-Specific
10. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Bhutani, N.; Burns, D.M.; Blau, H.M. DNA demethylation dynamics. Cell 2011, 146, 866–872. [Google Scholar] [CrossRef] [PubMed]
- Nestor, C.E.; Ottaviano, R.; Reddington, J.; Sproul, D.; Reinhardt, D.; Dunican, D.; Katz, E.; Dixon, M.J.; Harrison, D.J.; Meehan, R.R. Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Res. 2012, 22, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Münzel, M.; Globisch, D.; Carell, T. 5-hydroxymethylcytosine, the sixth base of the genome. Angew. Chem. Int. Ed. 2011, 50, 6460–6468. [Google Scholar]
- Pastor, W.A.; Aravind, L.; Rao, A. Tetonic shift: Biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 2013, 14, 341–356. [Google Scholar] [CrossRef] [PubMed]
- Kriaucionis, S.; Tahiliani, M. Expanding the epigenetic landscape: Novel modifications of cytosine in genomic DNA. Cold Spring Harb. Perspect. Biol. 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [CrossRef] [PubMed]
- Mohr, F.; Döhner, K.; Buske, C.; Rawat, V. Tet genes: New players in DNA demethylation and important determinants for stemness. Exp. Hematol. 2011, 39, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Kinney, S.; Chin, H.; Vaisvila, R.; Bitinaite, J.; Zheng, Y.; Estève, P.-O.; Feng, S.; Stroud, H.; Jacobsen, S.E.; Pradhan, S. Tissue-specific distribution and dynamic changes of 5-hydroxymethylcytosine in mammalian genomes. J. Biol. Chem. 2011, 286, 24685–24693. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, M. Distribution of 5-hydroxymethylcytosine in different human tissues. J. Nucleic Acids 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Iurlaro, M.; Ficz, G.; Oxley, D.; Raiber, E.-A.; Bachman, M.; Booth, M.J.; Andrews, S.; Balasubramanian, S.; Reik, W. A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biol. 2013, 14, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Spruijt, C.G.; Gnerlich, F.; Smits, A.H.; Pfaffeneder, T.; Jansen, P.; Bauer, C.; Münzel, M.; Wagner, M.; Müller, M.; Khan, F.; et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 2013, 152, 1146–1159. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, H.; Liu, Y.; Upadhyay, A.K.; Chang, Y.; Howerton, S.B.; Vertino, P.M.; Zhang, X.; Cheng, X. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012, 40, 4841–4849. [Google Scholar] [CrossRef] [PubMed]
- Ji, D.; Lin, K.; Song, J.; Wang, Y. Effects of TET-induced oxidation products of 5-methylcytosine on Dnmt1- and DNMT3a-mediated cytosine methylation. Mol. BioSyst. 2014, 10, 1749–1752. [Google Scholar] [CrossRef] [PubMed]
- Bachman, M.; Uribe-Lewis, S.; Yang, X.; Williams, M.; Murrell, A.; Balasubramanian, S. 5-hydroxymethylcytosine is a predominantly stable DNA modification. Nat. Chem. 2014, 6, 1049–1055. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, K.D.; Helin, K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016, 30, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, G.P.; Xiong, W.; Hahn, M.A.; Jin, S.-G. The role of 5-hydroxymethylcytosine in human cancer. Cell Tissue Res. 2014, 356, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Losman, J.A.; Kaelin, W.G. What a difference a hydroxyl makes: Mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 2013, 27, 836–852. [Google Scholar] [CrossRef] [PubMed]
- Bardella, C.; Pollard, P.J.; Tomlinson, I. SDH mutations in cancer. Biochim. Biophys. Acta Bioenerg. 2011, 1807, 1432–1443. [Google Scholar] [CrossRef] [PubMed]
- King, A.; Selak, M.A.; Gottlieb, E. Succinate dehydrogenase and fumarate hydratase: Linking mitochondrial dysfunction and cancer. Oncogene 2006, 25, 4675–4682. [Google Scholar] [CrossRef] [PubMed]
- Kroeze, L.I.; van der Reijden, B.A.; Jansen, J.H. 5-hydroxymethylcytosine: An epigenetic mark frequently deregulated in cancer. Biochim. Biophys. Acta Rev. Cancer 2015, 1855, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Lan, L. 5-hydroxymethylcytosine: A new insight into epigenetics in cancer. Cancer Biol. Ther. 2014, 15, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Mariani, C.J.; Madzo, J.; Moen, E.L.; Yesilkanal, A.; Godley, L.A. Alterations of 5-hydroxymethylcytosine in human cancers. Cancers 2013, 5, 786–814. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Zhang, J.; Guo, Z.; Ma, Q.; Xu, Z.; Zhou, Y.; Xu, Z.; Li, Z.; Liu, Y.; Ye, X.; et al. Loss of 5-hydroxymethylcytosine is linked to gene body hypermethylation in kidney cancer. Cell Res. 2016, 26, 103–118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Liu, Y.; Zhang, Z.; Li, J.; Wan, Y.; Zhang, L.; Wang, Y.; Li, X.; Xu, Y.; Fu, X.; et al. 5-hydroxymethylcytosine loss is associated with poor prognosis for patients with WHO grade II diffuse astrocytomas. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Yu, Y.; Luo, M.; Zhang, Z.; Shi, S.; Feng, X.; Chen, Z.; He, J. Loss of 5-hydroxymethylcytosine is an independent unfavorable prognostic factor for esophageal squamous cell carcinoma. PLoS ONE 2016, 11, e0153100. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Wu, K.; Ji, M.; Jin, W.; He, N.; Shi, B.; Hou, P. Decreased 5-hydroxymethylcytosine (5-hmC) is an independent poor prognostic factor in gastric cancer patients. J. Biomed. Nanotechnol. 2013, 9, 1607–1616. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Liu, L.; Chen, X.; Shen, J.; Shan, J.; Xu, Y.; Yang, Z.; Wu, L.; Xia, F.; Bie, P.; et al. Decrease of 5-hydroxymethylcytosine is associated with progression of hepatocellular carcinoma through downregulation of TET1. PLoS ONE 2013, 8, e62828. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.-W.; Li, G.-C.; Chen, C.-H.; Yeh, M.-H.; Huang, J.-S.; Tseng, H.-H.; Fu, T.-Y.; Liou, H.-H.; Pan, H.-W.; Huang, S.-F.; et al. Reduction of global 5-hydroxymethylcytosine is a poor prognostic factor in breast cancer patients, especially for an ER/PR-negative subtype. Breast Cancer Res. Treat. 2015, 153, 219–234. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, K.; Shao, Y.; Sui, F.; Yang, Q.; Shi, B.; Hou, P.; Ji, M. Decreased 5-hydroxymethylcytosine (5-hmC) predicts poor prognosis in early-stage laryngeal squamous cell carcinoma. Am. J. Cancer Res. 2016, 6, 1089. [Google Scholar] [PubMed]
- Delhommeau, F.; Dupont, S.; Valle, V.; James, C.; Trannoy, S.; Massé, A.; Kosmider, O.; Couedic, J.-P.; Robert, F.; Alberdi, A.; et al. Mutation in TET2 in myeloid cancers. N. Engl. J. Med. 2009, 360, 2289–2301. [Google Scholar] [CrossRef] [PubMed]
- Reitman, Z.J.; Yan, H. Isocitrate dehydrogenase 1 and 2 mutations in cancer: Alterations at a crossroads of cellular metabolism. J. Natl. Cancer Inst. 2010, 102, 932–941. [Google Scholar] [CrossRef] [PubMed]
- Sciacovelli, M.; Frezza, C. Oncometabolites: Unconventional triggers of oncogenic signalling cascades. Free Radic. Biol. Med. 2016, 100, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Nowicki, S.; Gottlieb, E. Oncometabolites: Tailoring our genes. FEBS J. 2015, 282, 2796–2805. [Google Scholar] [CrossRef] [PubMed]
- Ghiam, A.F.; Cairns, R.A.; Thoms, J.; Pra, D.A.; Oncogene, A.-O. IDH mutation status in prostate cancer. Oncogene 2012, 31, 3826. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Dong, Q.; Zhang, C.; Kuan, P.F.; Liu, Y.; Oncogene, J.-W.R. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene 2013, 32, 3091–3100. [Google Scholar] [CrossRef] [PubMed]
- Killian, J.K.; Kim, S.Y.; Miettinen, M.; Smith, C.; Merino, M.; Tsokos, M.; Quezado, M.; Smith, W.I.; Jahromi, M.S.; Xekouki, P.; et al. Succinate dehydrogenase mutation underlies global epigenomic divergence in gastrointestinal stromal tumor. Cancer Discov. 2013, 3, 648–657. [Google Scholar] [CrossRef] [PubMed]
- Ricketts, C.J.; Shuch, B.; Vocke, C.D.; Metwalli, A.R.; Bratslavsky, G.; Middelton, L.; Yang, Y.; Wei, M.H.; Pautler, S.E.; Peterson, J.; et al. Succinate dehydrogenase kidney cancer: An aggressive example of the warburg effect in cancer. J. Urol. 2012, 188, 2063–2071. [Google Scholar] [CrossRef] [PubMed]
- Lehtonen, H.J.; Kiuru, M.; Ylisaukko-oja, S.K.; Salovaara, R.; Herva, R.; Koivisto, P.A.; Vierimaa, O.; Aittomäki, K.; Pukkala, E.; Launonen, V.; et al. Increased risk of cancer in patients with fumarate hydratase germline mutation. J. Med. Genet. 2006, 43, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Song, C.-X.; Huang, H.; Frankenberger, C.A.; Sankarasharma, D.; Gomes, S.; Chen, P.; Chen, J.; Chada, K.K.; He, C.; et al. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc. Natl. Acad. Sci. USA 2013, 110, 9920–9925. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.-Z.; Chen, S.-F.; Nieh, S.; Benner, C.; Ger, L.-P.; Jan, C.-I.; Ma, L.; Chen, C.-H.; Hishida, T.; Chang, H.-T.; et al. Hypoxia drives breast tumor malignancy through a TET-TNFα-p38-MAPK signaling axis. Cancer Res. 2015, 75, 3912–3924. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Poliseno, L.; Song, M.; Ala, U.; Webster, K.; Ng, C.; Beringer, G.; Brikbak, N.J.; Yuan, X.; Cantley, L.C.; et al. microRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell 2013, 154, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Matthias, W.; Liou, W.; Pulverer, W.; Singer, C.F.; Rappaport-Fuerhauser, C.; Kandioler, D.; Egger, G.; Weinhäusel, A. Cytosine 5-hydroxymethylation of the LZTS1 gene is reduced in breast cancer. Transl. Oncol. 2013, 6, 715–721. [Google Scholar] [CrossRef]
- Kamiya, T.; Nakahara, R.; Mori, N.; Hara, H.; Adachi, T. Ten-eleven translocation 1 functions as a mediator of SOD3 expression in human lung cancer A549 cells. Free Radic. Res. 2017, 51, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Chapman, C.G.; Mariani, C.J.; Wu, F.; Meckel, K.; Butun, F.; Chuang, A.; Madzo, J.; Bissonnette, M.B.; Kwon, J.H.; Godley, L.A. Tet-catalyzed 5-hydroxymethylcytosine regulates gene expression in differentiating colonocytes and colon cancer. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Neri, F.; Dettori, D.; Incarnato, D.; Krepelova, A.; Rapelli, S.; Maldotti, M.; Parlato, C.; Paliogiannis, P.; Oliviero, S. TET1 is a tumour suppressor that inhibits colon cancer growth by derepressing inhibitors of the wnt pathway. Oncogene 2014, 34, 4168–4176. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Jiang, X.; Li, Z.; Li, Y.; Song, C.-X.; He, C.; Sun, M.; Chen, P.; Gurbuxani, S.; Wang, J.; et al. TET1 plays an essential oncogenic role in MLL-rearranged leukemia. Proc. Natl. Acad. Sci. USA 2013, 110, 11994–11999. [Google Scholar] [CrossRef] [PubMed]
- Kroeze, L.I.; Aslanyan, M.G.; van Rooij, A.; Koorenhof-Scheele, T.N.; Massop, M.; Carell, T.; Boezeman, J.B.; Marie, J.-P.; Halkes, C.J.M.; de Witte, T.; et al. Characterization of acute myeloid leukemia based on levels of global hydroxymethylation. Blood 2014, 124, 1110–1118. [Google Scholar] [CrossRef] [PubMed]
- Takai, H.; Masuda, K.; Sato, T.; Sakaguchi, Y.; Suzuki, T.; Suzuki, T.; Koyama-Nasu, R.; Nasu-Nishimura, Y.; Katou, Y.; Ogawa, H.; et al. 5-Hydroxymethylcytosine plays a critical role in glioblastomagenesis by recruiting the CHTOP-methylosome complex. Cell Rep. 2014, 9, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hu, H.; Wang, Q.; Li, Z.; Zhu, Y.; Zhang, W.; Wang, Y.; Jiang, H.; Cheng, J. The level and clinical significance of 5-hydroxymethylcytosine in oral squamous cell carcinoma: An immunohistochemical study in 95 patients. Pathol. Res. Pract. 2017, 213, 969–974. [Google Scholar] [CrossRef] [PubMed]
- Strand, S.H.; Hoyer, S.; Lynnerup, A.-S.; Haldrup, C.; Storebjerg, T.; Borre, M.; Orntoft, T.F.; Sorensen, K.D. High levels of 5-hydroxymethylcytosine (5hmC) is an adverse predictor of biochemical recurrence after prostatectomy in ERG-negative prostate cancer. Clin. Epigenet. 2015, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.A.; Piao, M.J.; Kim, K.C.; Kang, H.K.; Chang, W.Y.; Park, I.C.; Keum, Y.S.; Surh, Y.J.; Hyun, J.W. Epigenetic modification of Nrf2 in 5-fluorouracil-resistant colon cancer cells: Involvement of TET-dependent DNA demethylation. Cell Death Dis. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Meng, W.; Lautenschlaeger, T.; Frankhouser, D.; Ye, Z.; Huebner, A.; Jin, V.; Yan, P.; Chakravarti, A. Abstract 2963: 5-hydroxymethylcytosine alterations at H3K9me3 marked genomic regions serve as potential biomarker for renal cell carcinoma patients. Cancer Res. 2015, 75. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Pradhan, K.; Campbell, N.; Mazdo, J.; Vasantkumar, A.; Maqbool, S.; Bhagat, T.D.; Gupta, S.; Suzuki, M.; Yu, Y.; et al. Altered hydroxymethylation is seen at regulatory regions in pancreatic cancer and regulates oncogenic pathways. Genome Res. 2017, 27, 1830–1842. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, X.; Lu, X.; You, L.; Song, Y.; Luo, Z.; Zhang, J.; Nie, J.; Zheng, W.; Xu, D.; et al. DNA 5-hydroxymethylcytosines from cell-free circulating DNA as diagnostic biomarkers for human cancers. bioRxiv 2017. [Google Scholar] [CrossRef]
- Haffner, M.; Pellakuru, L.; Ghosh, S.; Lotan, T.; Nelson, W.G.; Marzo, A.; Yegnasubramanian, S. Tight correlation of 5-hydroxymethylcytosine and polycomb marks in health and disease. Cell Cycle 2013, 12, 1835–1841. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Issa, J.-P.J.; Baylin, S. Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 2016, 17, 630–641. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, B.; McGovern, A.; Cui, Y.; Choudhury, S.; Cho, I.-H.; Cooper, B.; Chevassut, T.; Lossie, A.C.; Irudayaraj, J. The hypomethylating agent decitabine causes a paradoxical increase in 5-hydroxymethylcytosine in human leukemia cells. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Ohtani, H.; Zhou, W.; Ørskov, A.D.; Charlet, J.; Zhang, Y.W.; Shen, H.; Baylin, S.B.; Liang, G.; Grønbæk, K.; et al. Vitamin C increases viral mimicry induced by 5-aza-2′-deoxycytidine. Proc. Natl. Acad. Sci. USA 2016, 113, 10238–10244. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Tao, Y.; Gao, X.; Zhang, L.; Li, X.; Zou, W.; Ruan, K.; Wang, F.; Xu, G.-L.; Hu, R. A CRISPR-based approach for targeted DNA demethylation. Cell Discov. 2016, 2. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, S.R.; Cui, Y.; Lubecka, K.; Stefanska, B.; Irudayaraj, J. CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter. Oncotarget 2016, 7, 46545–46556. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.W.; Wang, Z.; Xie, W.; Cai, Y.; Xia, L.; Easwaran, H.; Luo, J.; Yen, R.-W.; Li, Y.; Baylin, S.B. Acetylation enhances TET2 function in protecting against abnormal DNA methylation during oxidative stress. Mol. Cell 2017, 65, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Lv, L.; Nakagawa, M.; Yu, Y.; Yu, C.; D’Alessio, A.C.; Nakayama, K.; Fan, H.-Y.; Chen, X.; Xiong, Y. CRL4(VprBP) E3 ligase promotes monoubiquitylation and chromatin binding of TET dioxygenases. Mol. Cell 2015, 57, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Ciccarone, F.; Valentini, E.; Zampieri, M.; Caiafa, P. 5mC-hydroxylase activity is influenced by the PARylation of TET1 enzyme. Oncotarget 2015, 6, 24333–24347. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Liu, X.; Gao, W.; Li, P.; Hou, J.; Li, J.; Wong, J. Differential regulation of the ten-eleven translocation (TET) family of dioxygenases by O-linked β-N-acetylglucosamine transferase (OGT). J. Biol. Chem. 2014, 289, 5986–5996. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Lin, H.; Xu, H.; Zhang, L.; Cheng, L.; Wen, B.; Shou, J.; Guan, K.; Xiong, Y.; Ye, D. TET-catalyzed 5-methylcytosine hydroxylation is dynamically regulated by metabolites. Cell Res. 2014, 24, 1017–1020. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.-Y.; Kwak, S.; Lee, S.; Kim, H.; Lee, S.; Kim, J.-H.; Kim, Y.; Jeon, Y.; Chung, D.; Jin, X.; et al. Psat1-dependent fluctuations in α-ketoglutarate affect the timing of ESC differentiation. Cell Metab. 2016, 24, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.-H.; Liang, S.; Guo, J.; Choi, J.-W.; Kim, N.-H.; Lu, W.-F.; Cui, X.-S. Analysis of ferrous on ten-eleven translocation activity and epigenetic modifications of early mouse embryos by fluorescence microscopy. Microsc. Microanal. 2016, 22, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Camarena, V.; Sant, D.W.; Huff, T.; Mustafi, S.; Muir, R.K.; Renslo, A.R.; Monje, P.; Wang, G. cAMP signaling regulates DNA demethylation by augmenting the intracellular labile ferrous iron pool. bioRxiv 2017. [Google Scholar] [CrossRef]
- Bauer, C.; Göbel, K.; Nagaraj, N.; Colantuoni, C.; Wang, M.; Müller, U.; Kremmer, E.; Rottach, A.; Leonhardt, H. Phosphorylation of TET proteins is regulated via O-glcnacylation by the O-linked N-acetylglucosamine transferase (OGT). J. Biol. Chem. 2015, 290, 4801–4812. [Google Scholar] [CrossRef] [PubMed]
- Tu, J.; Ng, S.H.; Luk, A.C.; Liao, J.; Jiang, X.; Feng, B.; Lun Mak, K.K.; Rennert, O.M.; Chan, W.Y.; Lee, T.L. MicroRNA-29b/Tet1 regulatory axis epigenetically modulates mesendoderm differentiation in mouse embryonic stem cells. Nucleic Acids Res. 2015, 43, 7805–7822. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Jin, L.; Wang, X.; Luo, A.; Hu, J.; Zheng, X.; Tsark, W.M.; Riggs, A.D.; Ku, H.T.; Huang, W. MicroRNA-26a targets ten eleven translocation enzymes and is regulated during pancreatic cell differentiation. Proc. Natl. Acad. Sci. USA 2013, 110, 17892–17897. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Guo, S.; Chen, S.; Mastriano, S.J.; Liu, C.; D’Alessio, A.C.; Hysolli, E.; Guo, Y.; Yao, H.; Megyola, C.M.; et al. An extensive network of TET2-targeting microRNAs regulates malignant hematopoiesis. Cell Rep. 2013, 5, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Lu, Z.; Gao, Y.; Ye, L.; Song, T.; Zhang, X. MiR-520b suppresses proliferation of hepatoma cells through targeting ten-eleven translocation 1 (TET1) mRNA. Biochem. Biophys. Res. Commun. 2015, 460, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Chuang, K.-H.H.; Whitney-Miller, C.L.; Chu, C.-Y.Y.; Zhou, Z.; Dokus, M.K.; Schmit, S.; Barry, C.T. MicroRNA-494 is a master epigenetic regulator of multiple invasion-suppressor microRNAs by targeting ten eleven translocation 1 in invasive human hepatocellular carcinoma tumors. Hepatology 2015, 62, 466–480. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhou, Z.Q.; Yang, Z.R.; Tong, D.N.; Guan, J.; Shi, B.J.; Nie, J.; Ding, X.T.; Li, B.; Zhou, G.W.; et al. MicroRNA-191 acts as a tumor promoter by modulating the TET1-p53 pathway in intrahepatic cholangiocarcinoma. Hepatology 2017, 66, 136–151. [Google Scholar] [CrossRef] [PubMed]
- Parikh, A.; Lee, C.; Joseph, P.; Marchini, S.; Baccarini, A.; Kolev, V.; Romualdi, C.; Fruscio, R.; Shah, H.; Wang, F.; et al. MicroRNA-181a has a critical role in ovarian cancer progression through the regulation of the epithelial-mesenchymal transition. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Puissegur, M.P.; Mazure, N.M.; Bertero, T.; Pradelli, L.; Grosso, S.; Robbe-Sermesant, K.; Maurin, T.; Lebrigand, K.; Cardinaud, B.; Hofman, V.; et al. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 2011, 18, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Sasayama, T.; Tanaka, K.; Nakamizo, S.; Nishihara, M.; Mizukawa, K.; Kohta, M.; Koyama, J.; Miyake, S.; Taniguchi, M.; et al. MicroRNA-183 upregulates HIF-1α By targeting isocitrate dehydrogenase 2 (IDH2) in glioma cells. J. Neurooncol. 2013, 111, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Edmonds, M.D.; Boyd, K.L.; Moyo, T.; Mitra, R.; Duszynski, R.; Arrate, M.P.; Chen, X.; Zhao, Z.; Blackwell, T.S.; Andl, T.; et al. MicroRNA-31 initiates lung tumorigenesis and promotes mutant KRAS-driven lung cancer. J. Clin. Investig. 2016, 126, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Ito, K.; Ala, U.; Kats, L.; Webster, K.; Sun, S.; Jongen-Lavrencic, M.; Manova-Todorova, K.; Teruya-Feldstein, J.; Avigan, D.E.; et al. The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell 2013, 13, 87–101. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Yin, D.; Zhang, Y.; Yu, L.; Li, X.-D.; Zhou, Z.-J.; Zhou, S.-L.; Gao, D.-M.; Hu, J.; Jin, C.; et al. MicroRNA-29a induces loss of 5-hydroxymethylcytosine and promotes metastasis of hepatocellular carcinoma through a TET-SOCS1-MMP9 signaling axis. Cell Death Dis. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Liu, H.; Liu, Y.; Zhang, J.; Li, H.; Liu, W.; Cao, G.; Xv, P.; Zhang, J.; Lv, C.; et al. Mir-30a as potential therapeutics by targeting TET1 through regulation of drp-1 promoter hydroxymethylation in idiopathic pulmonary fibrosis. Int. J. Mol. Sci. 2017, 18, 633. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.-P.; Chen, H.-F.; Chen, S.-Y.; Cheng, W.-C.; Wang, H.-W.; Shen, Z.-J.; Song, C.; Teng, S.-C.; He, C.; Wu, K.-J. TET1 regulates hypoxia-induced epithelial-mesenchymal transition by acting as a co-activator. Genome Biol. 2014, 15, 513. [Google Scholar] [CrossRef] [PubMed]
- Mariani, C.J.; Vasanthakumar, A.; Madzo, J.; Yesilkanal, A.; Bhagat, T.; Yu, Y.; Bhattacharyya, S.; Wenger, R.H.; Cohn, S.L.; Nanduri, J.; et al. TET1-mediated hydroxymethylation facilitates hypoxic gene induction in neuroblastoma. Cell Rep. 2014, 7, 1343–1352. [Google Scholar] [CrossRef] [PubMed]
- Thienpont, B.; Steinbacher, J.; Zhao, H.; D’Anna, F.; Kuchnio, A.; Ploumakis, A.; Ghesquière, B.; Dyck, L.; Boeckx, B.; Schoonjans, L.; et al. Tumour hypoxia causes DNA hypermethylation by reducing TET activity. Nature 2016, 537, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Wise, D.R.; Ward, P.S.; Shay, J.E.S.; Cross, J.R.; Gruber, J.J.; Sachdeva, U.M.; Platt, J.M.; DeMatteo, R.G.; Simon, C.M.; Thompson, C.B. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc. Natl. Acad. Sci. USA 2011, 108, 19611–19616. [Google Scholar] [CrossRef] [PubMed]
- Kickingereder, P.; Sahm, F.; Radbruch, A.; Wick, W.; Heiland, S.; von Deimling, A.; Bendszus, M.; Wiestler, B. IDH mutation status is associated with a distinct hypoxia/angiogenesis transcriptome signature which is non-invasively predictable with rCBV imaging in human glioma. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Her, Y.F.; Nelson-Holte, M.; Maher, L. Oxygen concentration controls epigenetic effects in models of familial paraganglioma. PLoS ONE 2015, 10, e0127471. [Google Scholar] [CrossRef] [PubMed]
- Laukka, T.; Mariani, C.J.; Ihantola, T.; Cao, J.Z.; Hokkanen, J.; Kaelin, W.G.; Godley, L.A.; Koivunen, P. Fumarate and succinate regulate expression of hypoxia-inducible genes via TET enzymes. J. Biol. Chem. 2015, 291, 4256–4265. [Google Scholar] [CrossRef] [PubMed]
- Tamás, M.J.; Sharma, S.K.; Ibstedt, S.; Jacobson, T.; Christen, P. Heavy metals and metalloids as a cause for protein misfolding and aggregation. Biomolecules 2014, 4, 252–267. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, A. Zinc finger proteins as potential targets for toxic metal ions: Differential effects on structure and function. Antioxid. Redox Signal. 2001, 3, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Martinez, V.D.; Vucic, E.A.; Becker-Santos, D.D.; Gil, L.; Lam, W.L. Arsenic exposure and the induction of human cancers. J. Toxicol. 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Reichard, J.F.; Puga, A. Effects of arsenic exposure on DNA methylation and epigenetic gene regulation. Epigenomics 2010, 2, 87–104. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Jiang, J.; Li, L.; Amato, N.J.; Wang, Z.; Wang, Y. Arsenite targets the zinc finger domains of TET proteins and inhibits TET-mediated oxidation of 5-methylcytosine. Environ. Sci. Technol. 2015, 49, 11923–11931. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.W.; Klein, C.B.; Kargacin, B.; Salnikow, K.; Kitahara, J.; Dowjat, K.; Zhitkovich, A.; Christie, N.T.; Costa, M. Carcinogenic nickel silences gene expression by chromatin condensation and DNA methylation: A new model for epigenetic carcinogens. Mol. Cell. Biol. 1995, 15, 2547–2557. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.; Mo, J.; Dai, J.; Wang, H. Nickel(II) inhibits TET-mediated 5-methylcytosine oxidation by high affinity displacement of the cofactor iron(II). ACS Chem. Biol. 2017, 12, 1494–1498. [Google Scholar] [CrossRef] [PubMed]
- Arita, A.; Costa, M. Epigenetics in metal carcinogenesis: Nickel, arsenic, chromium and cadmium. Metall. Integr. Biomet. Sci. 2009, 1, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Waris, G.; Ahsan, H. Reactive oxygen species: Role in the development of cancer and various chronic conditions. J. Carcinog. 2006, 5, 14. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Ni, X. Ros-mediated DNA methylation pattern alterations in carcinogenesis. Curr. Drug Targets 2015, 16, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.P.; Hunter, J.M.; Lempiäinen, H.; Müller, A.; Terranova, R.; Moggs, J.G.; Meehan, R.R. Dynamic changes in 5-hydroxymethylation signatures underpin early and late events in drug exposed liver. Nucleic Acids Res. 2013, 41, 5639–5654. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, W.N.; Chevalier, D.M.; Phelps, J.Y.; Cantor, A.M.; Padilla-Banks, E.; Newbold, R.R.; Archer, T.K.; Kinyamu, K.H.; Williams, C.J. Persistently altered epigenetic marks in the mouse uterus after neonatal estrogen exposure. Mol. Endocrinol. 2013, 27, 1666–1677. [Google Scholar] [CrossRef] [PubMed]
- Lian, C.; Xu, S.; Guo, W.; Yan, J.; Frank, M.; Liu, R.; Liu, C.; Chen, Y.; Murphy, G.F.; Chen, T. Decrease of 5-hydroxymethylcytosine in rat liver with subchronic exposure to genotoxic carcinogens riddelliine and aristolochic acid. Mol. Carcinog. 2015, 54, 1503–1507. [Google Scholar] [CrossRef] [PubMed]
- Coulter, J.B.; O’Driscoll, C.M.; Bressler, J.P. Hydroquinone increases 5-hydroxymethylcytosine formation through ten eleven translocation 1 (TET1) 5-methylcytosine dioxygenase. J. Biol. Chem. 2013, 288, 28792–28800. [Google Scholar] [CrossRef] [PubMed]
- Fortmann, S.P.; Burda, B.U.; Senger, C.A.; Lin, J.S.; Whitlock, E.P. Vitamin and mineral supplements in the primary prevention of cardiovascular disease and cancer: An updated systematic evidence review for the U.S. Preventive services task force. Ann. Intern. Med. 2013, 159, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Verrax, J.; Beck, R.; Dejeans, N.; Glorieux, C. Redox-active quinones and ascorbate: An innovative cancer therapy that exploits the vulnerability of cancer cells to oxidative stress. Anti-Cancer Agents Med. Chem. 2011, 11, 213–221. [Google Scholar] [CrossRef]
- Blaschke, K.; Ebata, K.T.; Karimi, M.M.; Zepeda-Martínez, J.A.; Goyal, P.; Mahapatra, S.; Tam, A.; Laird, D.J.; Hirst, M.; Rao, A.; et al. Vitamin C induces TET-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 2013, 500, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Minor, E.A.; Court, B.L.; Young, J.I.; Wang, G. Ascorbate induces ten-eleven translocation (TET) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J. Biol. Chem. 2013, 288, 13669–13674. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Yang, Y.; Wang, X.; Chong, Z.; Yin, R.; Song, S.-H.; Zhao, C.; Li, C.; Huang, H.; Sun, B.-F.; et al. Redox-active quinones induces genome-wide DNA methylation changes by an iron-mediated and TET-dependent mechanism. Nucleic Acids Res. 2014, 42, 1593–1605. [Google Scholar] [CrossRef] [PubMed]
- Prasad, A.S.; Beck, F.W.J.; Snell, D.C.; Kucuk, O. Zinc in cancer prevention. Nutr. Cancer 2009, 61, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Zhao, Y.-P.; Jiang, Y.-G.; Wang, R.-W.; Ma, Z. Restoring the metabolic disturbance of zinc: May not only contribute to the prevention of esophageal squamous cell cancer. Med. Hypotheses 2008, 71, 957–959. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.M.; Kristal, B.S.; Effron, M.S.; Shestopalov, A.I.; Ullucci, P.A.; Sheu, K.F.; Blass, J.P.; Cooper, A.J.L. Zn2+ inhibits α-ketoglutarate-stimulated mitochondrial respiration and the isolated α-ketoglutarate dehydrogenase complex. J. Biol. Chem. 2000, 275, 13441–13447. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.J.; Kim, M.R.; Chen, Y.S.; Yang, J.Y.; Oncogene, C.-C.J. Retinoic acid directs breast cancer cell state changes through regulation of TET2-pkcζ pathway. Oncogene 2017, 36, 3193–3206. [Google Scholar] [CrossRef] [PubMed]
- Hassan, H.M.; Kolendowski, B.; Isovic, M.; Reports, B.-K. Regulation of active DNA demethylation through RAR-mediated recruitment of a TET/TDG complex. Cell Rep. 2017, 19, 1685–1697. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.; Guyot, R.; Samarut, J.; Flamant, F.; Wong, J.; Gauthier, K. Methylcytosine dioxygenase TET3 interacts with thyroid hormone nuclear receptors and stabilizes their association to chromatin. Proc. Natl. Acad. Sci. USA 2017, 114, 8229–8234. [Google Scholar] [CrossRef] [PubMed]
- Arab, K.; Park, Y.; Lindroth, A.M.; Schäfer, A.; Oakes, C.; Weichenhan, D.; Lukanova, A.; Lundin, E.; Risch, A.; Meister, M.; et al. Long noncoding RNA tarid directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol. Cell 2014, 55, 604–614. [Google Scholar] [CrossRef] [PubMed]
- Neri, F.; Incarnato, D.; Krepelova, A.; Rapelli, S.; Pagnani, A.; Zecchina, R.; Parlato, C.; Oliviero, S. Genome-wide analysis identifies a functional association of TET1 and polycomb repressive complex 2 in mouse embryonic stem cells. Genome Biol. 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Putiri, E.L.; Tiedemann, R.L.; Thompson, J.J.; Liu, C.; Ho, T.; Choi, J.-H.; Robertson, K.D. Distinct and overlapping control of 5-methylcytosine and 5-hydroxymethylcytosine by the TET proteins in human cancer cells. Genome Biol. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Szulwach, K.E.; Li, X.; Li, Y.; Song, C.X.; Han, J.W.; Kim, S.; Namburi, S.; Hermetz, K.; Kim, J.J.; Rudd, M.K.; et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011, 7, e1002154. [Google Scholar] [CrossRef] [PubMed]
- Fujiki, K.; Shinoda, A.; Kano, F.; Sato, R.; Shirahige, K.; Murata, M. Pparγ-induced parylation promotes local DNA demethylation by production of 5-hydroxymethylcytosine. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Kafer, G.; Li, X.; Horii, T.; Suetake, I.; Tajima, S.; Hatada, I.; Carlton, P. 5-hydroxymethylcytosine marks sites of DNA damage and promotes genome stability. Cell Rep. 2016, 14, 1283–1292. [Google Scholar] [CrossRef] [PubMed]
| Mutation | Cancer Types | 5hmC Levels | References |
|---|---|---|---|
| IDH1 | Grade II gliomas and secondary Glioblastoma, AML, Acute B lymphoblastic Leukemia, Paraganglioma, melanoma, Intrahepatic cholangiocarcinoma, cartilageous, thyroid, colon, Prostate and follicular cancer. | Lowered | [32,35,36] |
| IDH2 | Gliomas, AML, Paraganglioma, melanoma, cartilageous, Intrahepatic cholangiocarcinoma and colon cancer. | Lowered | [32,36] |
| SDHB/C/D | HPGL/PCC, pancreatic neuroendocrine tumours, renal tumorigenesis and gastric gastrointestinal stromal tumors. | Lowered | [19,37,38] |
| FH | leiomyomas of the skin and the uterus (fibroids, myomas), renal cell carcinoma. | Lowered | [20,39] |
| Cancer Type | Locus/Region | Role or Effect | References |
|---|---|---|---|
| Breast cancer | Loss of 5hmC in HOXA | High Mobility Group AT-Hook 2 (HMGA2) promotes breast cancer cell invasion in part through inhibition of TET1-mediated demethylation and HOXA gene expression. | [40] |
| Breast cancer | Loss of 5hmC in miR200 promoter region | miR-22 overexpression targets TET1, 2 & 3 mRNA and inhibit demethylation of mir-200 and enable EMT, invasiveness and metastasis. | [42] |
| Breast cancer | Loss of 5hmC in LZTS1 gene | Decreases of 5hmC levels in LZTS1 region down-regulates its expression that enhance cancer progression and metastasis. | [43] |
| Breast cancer | Gain of 5hmC in TNFA gene | Increased TNFA expression is essential for BTIC properties by TNFα-p38-MAPK signalling. | [41] |
| Colon cancer | Loss of 5hmC or hypermethylation in DDK3 genes | Down-regulation of TET1 and DNA hydroxymethylation mediated by TET1 controlling the WNT signaling is a key player in tumor growth. | [46] |
| Colon cancer | The gain of 5hmC in Nuclear factor-erythroid 2-related factor 2 (Nrf2) favors resistance towards 5FU | 5-FU resistance in CRC involves the upregulation of Nrf2 and HO-1 expression via epigenetic modifications of DNA demethylation. | [52] |
| Colon cancer | Loss of 5hmC in CA2, FMN2, PDCD4, PKIB and SLC26A2. Gain of 5hmC in BMP7, NKD2, TESC and TGFBI | Though loss of 5hmC a hallmark of cancer, locus-specific changes also play a role in colorectal cancer. | [45] |
| Lung cancer | Loss of 5hmC in SOD3 gene | SOD3 which has been downregulated in A549, THP1 and other cancer cell lines has shown to be upregulated by overexpression of TET1CD which demethylates SOD3 promoter. | [44] |
| Post-Translational Modification | Regulatory Protein | Function | Effect on 5hmC | Cell Type/Model Cell Line | Reference |
|---|---|---|---|---|---|
| Acetylation of TET2 at K110 | p300 acetylates TET2 under oxidative stress, while HDAC1/2 deacetylates TET2 | Increases TET2 half-life and activity especially at hypermethylated sites under oxidative stress. | Increased | Ovarian and Colorectal cancer cell line (A2780 and HCT116) | [66] |
| Monoubiquitination | CRL4VprBP ubiquitylates TET1 (K1212),TET2 (K1299) and TET3 (K983) | Promotes TET DNA binding | Increased | MEF | [67] |
| PARylation (Covalent) | PARP-1 covalently links PAR to TET1 | Increased TET1 activity | Increased | HEK293 | [68] |
| PARylation (Non-Covalent) | Non-covalent binding of poly ADP-ribose to TET1 catalytic domain. | Decreases DNA binding | Decreased | HEK293 | [68] |
| O-GlcNAcylation | OGT O-GlcNAcylates TET3 | Enables nuclear export | Decreased | Not cell type specific | [69] |
| microRNA | Target | Role | Level of 5hmC | Reference |
|---|---|---|---|---|
| miR-181a | IDH1 | Increased miR-181a lowers IDH1 thereby lowers lipid synthesis and increases lipid oxidation enzymes | ND * | [77] |
| miR-183 | IDH2 | Lowers IDH2 and increases HIF1A and VEGF. upregulated in malignant gliomas | ND * | [79] |
| miR-210 | SDHD | Overexpression of miR-210 induced cell death and activates HIF1α in late stages of lung cancer | ND * | [78] |
| miR-31 | SDHA | Modulates mitochondrial metabolism that enables iPSC reprogramming. | ND * | [80] |
| miR-22 | TET2 | Increased in MDS and leukemia and increased expression linked to poor survival. | reduced | [81] |
| miR-125b, miR-29b, miR-29c, miR-101, miR-7, etc (more than 30) | TET2 | Increased in myeloid malignancies. | reduced | [73] |
| miR-26a | TET1, 2 & 3 | Promotes pancreatic cell differentiation. | reduced | [72] |
| miR-29b | TET1 | Increases sharply during embryonic body formation and favors mesendoderm lineage differentiation. | reduced | [71] |
| miR-22 | TET2 | TET2 reduction favors EMT and metastasis by 5hmC reduction in miR 200b promoter region in breast cancer | reduced | [42] |
| miR-29a | TET1 | Increased miR-29a reduces TET1 and enhances metastasis in HCC. | reduced | [82] |
| miR-191 | TET1 | Increased in Intrahepatic cholangiocarcinoma; enables proliferation, migration and invasion | reduced | [76] |
| miR-30a | TET1 | Increased in pulmonary fibrosis, as a potential therapeutic target. | reduced | [83] |
| miR-520b | TET1 | Decreased in HCC and enables proliferation | ND * | [74] |
| miR-494 | TET1 | Increased in HCC and enables invasion | reduced | [75] |
| Conditions | 5hmC Locus | Functions | References |
|---|---|---|---|
| Hypoxia | Promotor, TSS and genic regions of hypoxia-inducible genes like Vascular Endothelial Growth Factor A (VEGFA), BCL2 Interacting Protein 3 (BINP3), Enolase 1 (ENO1). | Enables EMT, angiogenesis and metastasis | [84] |
| Phenobarbital | Proximal Promotor regions of Cyp and Gst genes at initial exposure. During prolonged exposure Wisp, Cxcr7 and loss in Ndrf2 gene | A short-term exposure facilitates gene expression of xenobiotics metabolism and prolonged exposure induces HCC. | [101] |
| Drug-induced ROS | Promotors of Nrf2 genes | Cell survival and drug resistance | [52] |
| Elevated plasma glucose | Promotor regions of Hexokinase in liver | Glucose metabolism | [62] |
| DNA damage by aphidicolin and micro-irradiation | Marks sites of DNA damage | Facilitates repair mechanism and genomic integrity | [121] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arvinden, V.R.; Deva Magendhra Rao, A.K.; Rajkumar, T.; Mani, S. Regulation and Functional Significance of 5-Hydroxymethylcytosine in Cancer. Epigenomes 2017, 1, 19. https://doi.org/10.3390/epigenomes1030019
Arvinden VR, Deva Magendhra Rao AK, Rajkumar T, Mani S. Regulation and Functional Significance of 5-Hydroxymethylcytosine in Cancer. Epigenomes. 2017; 1(3):19. https://doi.org/10.3390/epigenomes1030019
Chicago/Turabian StyleArvinden, Vittal Rangan, Arunagiri Kuha Deva Magendhra Rao, Thangarajan Rajkumar, and Samson Mani. 2017. "Regulation and Functional Significance of 5-Hydroxymethylcytosine in Cancer" Epigenomes 1, no. 3: 19. https://doi.org/10.3390/epigenomes1030019
APA StyleArvinden, V. R., Deva Magendhra Rao, A. K., Rajkumar, T., & Mani, S. (2017). Regulation and Functional Significance of 5-Hydroxymethylcytosine in Cancer. Epigenomes, 1(3), 19. https://doi.org/10.3390/epigenomes1030019





